Transcriptome Analysis and GC-MS Profiling of Key Fatty Acid Biosynthesis Genes in Akebia trifoliata (Thunb.) Koidz Seeds
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Material
2.2. Dynamic Changes in ASO Content and FA Composition
2.3. cDNA Library Construction and Sequence Analysis and Alignment
2.4. Bioinformatic Analysis
2.5. Quantitative Analysis
2.6. Statistical Analysis
3. Results
3.1. Dynamic Changes in A. trifoliata Fruit Weight and Seed Oil Content
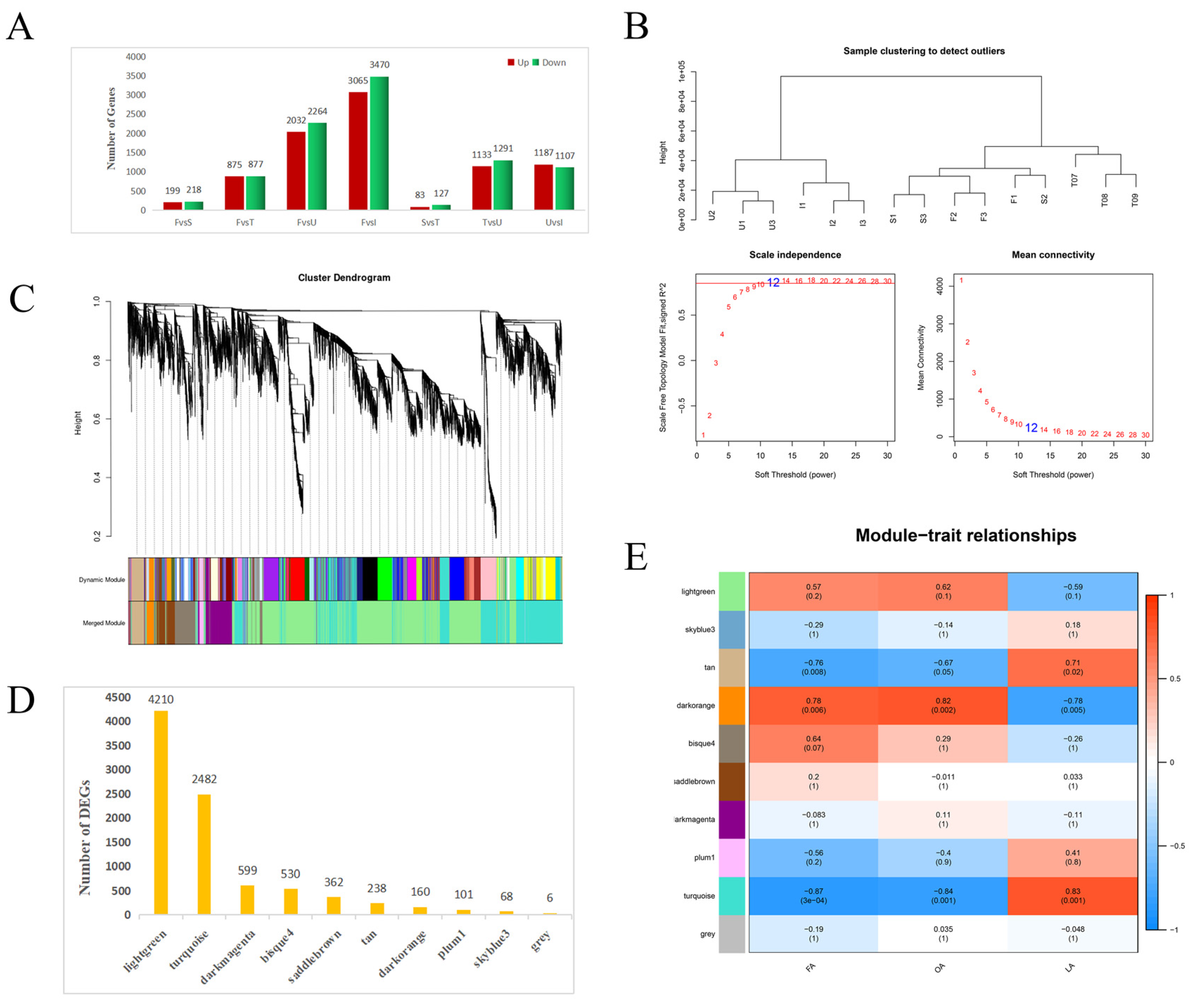
3.2. Transcriptome Sequencing
3.3. Analysis of DEGs
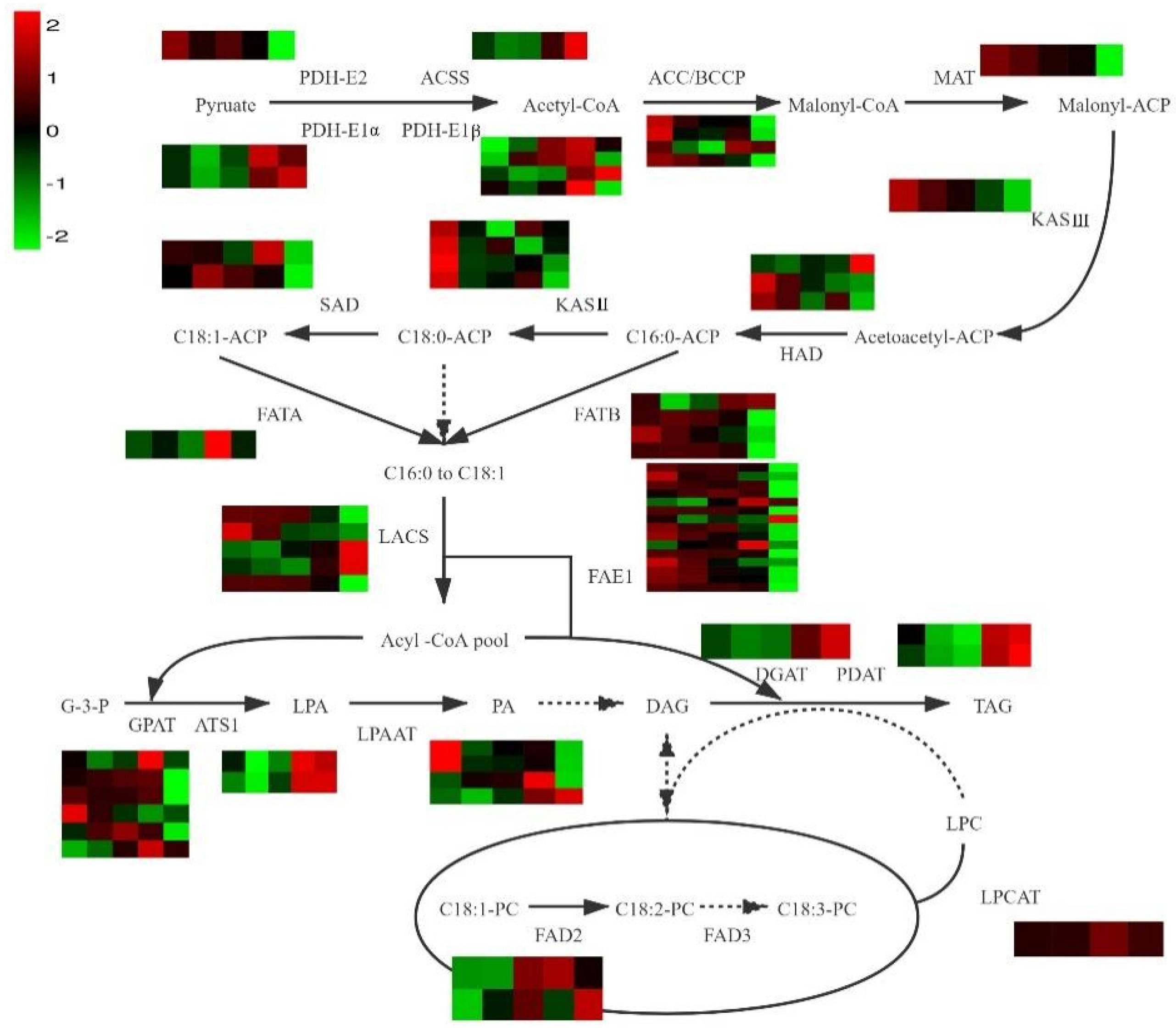
3.4. Identification of Genes Involved in FA Biosynthesis
3.5. Identification of Genes Involved in Unsaturated FA Biosynthesis
3.6. Identification of Genes Involved in TAG Biosynthesis
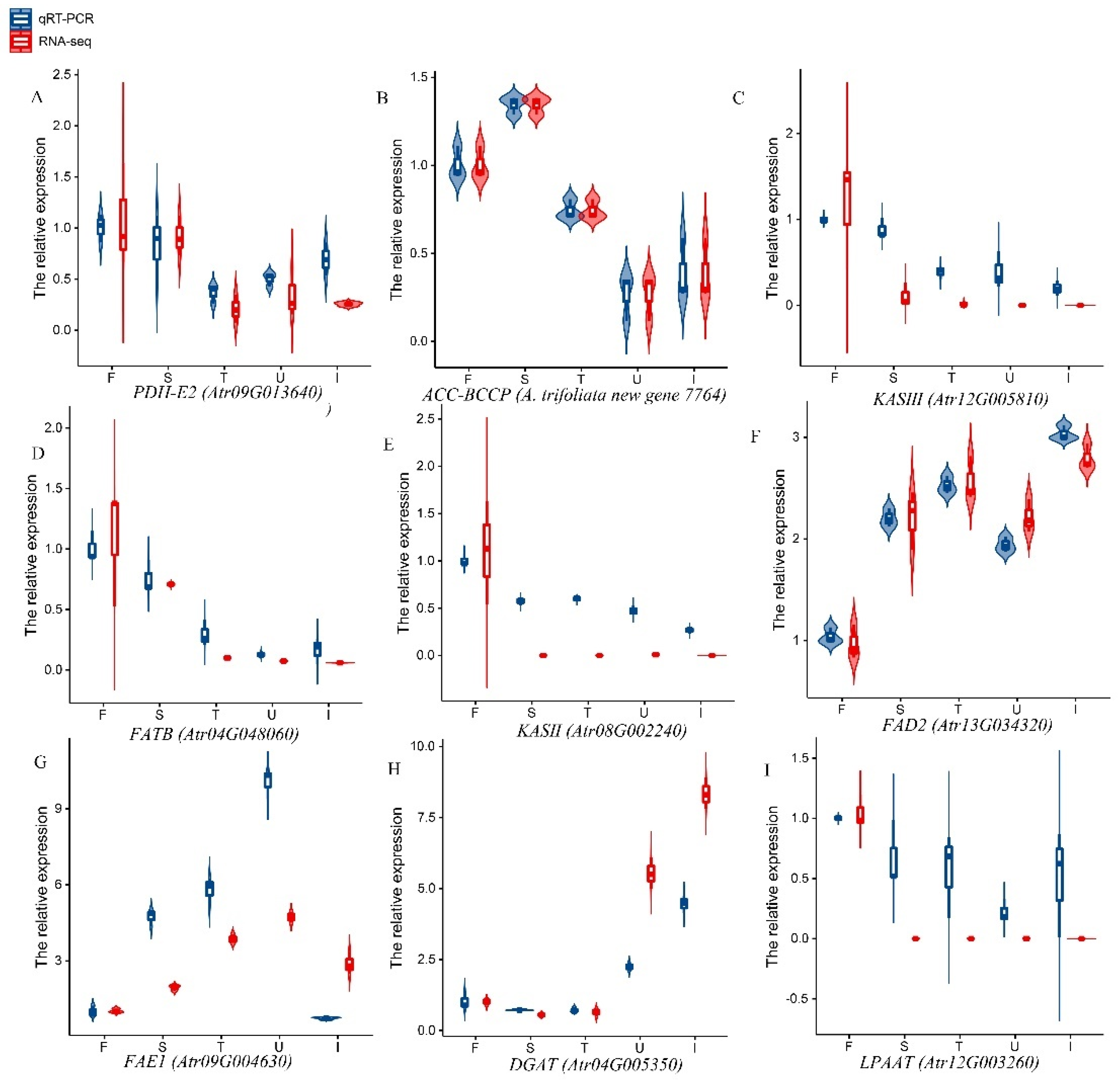
3.7. qPCR Analysis of Lipid-Related Genes
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Bos, H.L.; Meesters, K.P.H.; Conijn, S.G.; Corré, W.J.; Patel, M.K. Comparing biobased products from oil crops versus sugar crops with regard to non-renewable energy use, GHG emissions and land use. Ind. Crop. Prod. 2016, 84, 366–374. [Google Scholar] [CrossRef]
- Park, K.; Sanjaya, S.A.; Quach, T.; Cahoon, E.B. Toward sustainable production of value-added bioenergy and industrial oils in oilseed and biomass feedstocks. GCB Bioenergy 2021, 10, 1610–1623. [Google Scholar] [CrossRef]
- Chhetri, A.; Watts, K.; Islam, M. Waste Cooking Oil as an Alternate Feedstock for Biodiesel Production. Energies 2008, 1, 3–18. [Google Scholar] [CrossRef] [Green Version]
- Matsuzawa, Y.; Higashi, Y.; Takano, K.; Takahashi, M.; Yamada, Y.; Okazaki, Y.; Nakabayashi, R.; Saito, K.; Tsugawa, H. Food Lipidomics for 155 Agricultural Plant Products. J. Agric. Food Chem. 2021, 32, 8981–8990. [Google Scholar] [CrossRef]
- Xue, Z.; Li, S.; Yu, W.; Gao, X.; Zheng, X.; Yu, Y.; Kou, X. Research advancement and commercialization of microalgae edible oil: A review. J. Sci. Food Agric. 2021, 14, 5763–5774. [Google Scholar] [CrossRef]
- Olafisoye, O.B.; Oguntibeju, O.O.; Osibote, O.A. Trace elements and radionuclides in palm oil, soil, water, and leaves from oil palm plantations: A review. Crit. Rev. Food Sci. Nutr. 2017, 7, 1295–1315. [Google Scholar] [CrossRef] [PubMed]
- Metzger, J.O.; Bornscheuer, U. Lipids as renewable resources: Current state of chemical and biotechnological conversion and diversification. Appl. Microbiol. Biotechnol. 2006, 1, 13–22. [Google Scholar] [CrossRef]
- Xie, J.; Li, X.H.; Zhang, C.J.; Ouyang, H.N.; Xiao, Y.P. Distribution of Akebia trifoliata (Thunb.) Koidz wild resources. J. Shaanxi Norm. Univ. 2006, 34, 272–274. [Google Scholar]
- Council of Europe Convention on the Elaboration of a European Pharmacopoeia. Akebia stem. In European Pharmacopoeia; The Council of Europe: St. Trasbourg, France, 2021; Volume 9, pp. 5985–5986. [Google Scholar]
- Zhou, X.; Zhang, L.B.; Peng, Y.H.; Jiang, L.J.; Chen, J.Z.; Yu, P.Y.; Feng, X.C.; Li, P.W.; Xiang, M. Prospect of breeding of improved varieties and propagation technology of Akebia trifoliata. Mol. Plant Breed. 2021, 19, 1632–1639. [Google Scholar]
- Zhong, Y.; Wang, Y.; Sun, Z.; Niu, J.; Shi, Y.; Huang, K.; Chen, J.; Chen, J.; Luan, M. Genetic Diversity of a Natural Population of Akebia trifoliata (Thunb.) Koidz and Extraction of a Core Collection Using Simple Sequence Repeat Markers. Front. Genet. 2021, 12, 716498. [Google Scholar] [CrossRef]
- Zhong, Y.; Zhang, Z.; Chen, J.; Niu, J.; Shi, Y.; Wang, Y.; Chen, T.; Sun, Z.; Chen, J.; Luan, M. Physicochemical properties, content, composition and partial least squares models of A. trifoliata seeds oil. Food Chem. X 2021, 12, 100131. [Google Scholar] [CrossRef] [PubMed]
- Jiang, Y.L.; Zhou, X.F.; Zheng, Y.R.; Wang, D.F.; Deng, Y.; Zhao, Y.Y. Impact of ultrasonication/shear emulsifying/microwaveassisted enzymatic extraction on rheological, structural, and functional properties of Akebia trifoliata (Thunb.) Koidz. seed protein isolates. Food Hydrocoll. 2021, 112, 106355. [Google Scholar] [CrossRef]
- Zhao, S.Q.; Hu, J.N.; Zhu, X.M.; Bai, C.Q.; Peng, H.L.; Xiong, H.; Hu, J.W.; Zhao, Q. Characteristics and Feasibility of Trans-Free Plastic Fats through Lipozyme TL IM-Catalyzed Interesterification of Palm Stearin and Akebia trifoliata Variety Australis Seed Oil. J. Agric. Food Chem. 2014, 14, 3293–3300. [Google Scholar] [CrossRef]
- Zhou, N. Research on Extraction and Bioactivity of Akebia trifoliate Seed Oil. Master’s Thesis, Central South University of Forestry and Technology, Chasha, China, 2018. [Google Scholar]
- Huang, H.; Liang, J.; Tan, Q.; Ou, L.; Li, X.; Zhong, C.; Huang, H.; Møller, I.M.; Wu, X.; Song, S. Insights into triterpene synthesis and unsaturated fatty-acid accumulation provided by chromosomal-level genome analysis of Akebia trifoliata subsp. australis. Hortic. Res. 2021, 1, 33. [Google Scholar] [CrossRef]
- Huang, R.M.; Zhou, Y.; Zhang, J.P.; Ji, F.Y.; Jin, F.; Fan, W.; Pei, D. Transcriptome Analysis of Walnut (Juglans regia L.) Embryos Reveals Key Developmental Stages and Genes Involved in Lipid Biosynthesis and Polyunsaturated Fatty Acid Metabolism. J. Agric. Food Chem. 2021, 1, 377–396. [Google Scholar] [CrossRef] [PubMed]
- Niu, J.; Sun, Z.; Shi, Y.; Huang, K.; Zhong, Y.; Chen, J.; Chen, J.; Luan, M. Comparative Analysis of Akebia trifoliata Fruit Softening at Different Flesh Ripening Stages Using Tandem Mass Tag Technology. Front. Nutr. 2021, 8, 684271. [Google Scholar] [CrossRef]
- Sun, X.; Zhang, L.; Li, P.; Xu, B.; Ma, F.; Zhang, Q.; Zhang, W. Fatty acid profiles based adulteration detection for flaxseed oil by gas chromatography mass spectrometry. LWT-Food Sci. Technol. 2015, 1, 430–436. [Google Scholar] [CrossRef]
- Zhang, L.; Li, P.; Sun, X.; Hu, W.; Wang, X.; Zhang, Q.; Ding, X. Untargeted fatty acid profiles based on the selected ion monitoring mode. Anal. Chim. Acta 2014, 839, 44–50. [Google Scholar] [CrossRef]
- Kim, D.; Langmead, B.; Salzberg, S.L. HISAT: A fast spliced aligner with low memory requirements. Nat. Methods 2015, 4, 357–360. [Google Scholar] [CrossRef] [Green Version]
- Pertea, M.; Pertea, G.M.; Antonescu, C.M.; Chang, T.C.; Mendell, J.T.; Salzberg, S.L. StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nat. Biotechnol. 2015, 3, 290–295. [Google Scholar] [CrossRef] [Green Version]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 12, 550. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Langfelder, P.; Horvath, S. WGCNA: An R package for weighted correlation network analysis. BMC Bioinform. 2008, 9, 559. [Google Scholar] [CrossRef] [Green Version]
- Yang, H. Analysis of Gene Expression on Ethylene Biosynthesis of Akebia trifoliate (Thunb.) Koidz in Ripping Process. Master’s Thesis, Guizhou University, Guiyang, China, 2006. [Google Scholar]
- Wolf, G.D.; Sirinyan, K.; Henning, W.; Merten, R.; Gizycki, U.V.; Benda, B. Primer for the Metallization of Substrate Surfaces. U.S. Patent US5378268A, 1 March 1995. [Google Scholar]
- Troncoso-Ponce, M.A.; Kilaru, A.; Cao, X.; Durrett, T.P.; Fan, J.; Jensen, J.K.; Thrower, N.A.; Pauly, M.; Wilkerson, C.; Ohlrogge, J.B. Comparative deep transcriptional profiling of four developing oilseeds. Plant J. 2011, 6, 1014–1027. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Baud, S. Seeds as oil factories. Plant Reprod. 2018, 3, 213–235. [Google Scholar] [CrossRef] [PubMed]
- Yu, X.H.; Cai, Y.; Keereetaweep, J.; Wei, K.; Chai, J.; Deng, E.; Liu, H.; Shanklin, J. Biotin attachment domain-containing proteins mediate hydroxy fatty acid-dependent inhibition of acetyl CoA carboxylase. Plant Physiol. 2021, 3, 892–901. [Google Scholar] [CrossRef]
- Prigge, S.T.; He, X.; Gerena, L.; Waters, N.C.; Reynolds, K.A. The initiating steps of a type II fatty acid synthase in Plasmodium falciparum are catalyzed by pfACP, pfMCAT, and pfKASIII. Biochemistry 2003, 4, 1160–1169. [Google Scholar] [CrossRef]
- Liu, Q.; Wu, M.; Zhang, B.; Shrestha, P.; Petrie, J.; Green, A.G.; Singh, S.P. Genetic enhancement of palmitic acid accumulation in cotton seed oil through RNAi down-regulation of ghKAS2 encoding β-ketoacyl-ACP synthase II (KASII). Plant Biotechnol. J. 2017, 1, 132–143. [Google Scholar] [CrossRef] [Green Version]
- Haun, W.; Coffman, A.; Clasen, B.M.; Demorest, Z.L.; Lowy, A.; Ray, E.; Retterath, A.; Stoddard, T.; Juillerat, A.; Cedrone, F.; et al. Improved soybean oil quality by targeted mutagenesis of the fatty acid desaturase 2 gene family. Plant Biotechnol. J. 2014, 7, 934–940. [Google Scholar] [CrossRef]
- Dmitriev, A.A.; Kezimana, P.; Rozhmina, T.A.; Zhuchenko, A.A.; Povkhova, L.V.; Pushkova, E.N.; Novakovskiy, R.O.; Pavelek, M.; Vladimirov, G.N.; Nikolaev, E.N.; et al. Genetic diversity of SAD and FAD genes responsible for the fatty acid composition in flax cultivars and lines. BMC Plant Biol. 2020, 20, 301. [Google Scholar] [CrossRef]
- Liu, F.; Xia, Y.; Wu, L.; Fu, D.; Hayward, A.; Luo, J.; Yan, X.; Xiong, X.; Fu, P.; Wu, G.; et al. Enhanced seed oil content by overexpressing genes related to triacylglyceride synthesis. Gene 2014, 2, 163–171. [Google Scholar] [CrossRef]
- Zhu, T.; Wu, S.; Zhang, D.; Li, Z.; Xie, K.; An, X.; Ma, B.; Hou, Q.; Dong, Z.; Tian, Y.; et al. Genome-wide analysis of maize GPAT gene family and cytological characterization and breeding application of ZmMs33/ZmGPAT6 gene. Theor. Appl. Genet. 2019, 7, 2137–2154. [Google Scholar] [CrossRef]
- Harris, C.A.; Haas, J.T.; Streeper, R.S.; Stone, S.J.; Kumari, M.; Yang, K.; Han, X.; Brownell, N.; Gross, R.W.; Zechner, R.; et al. DGAT enzymes are required for triacylglycerol synthesis and lipid droplets in adipocytes. J. Lipid Res. 2011, 4, 657–667. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Furmanek, T.; Demski, K.; Banaś, W.; Haslam, R.; Napier, J.; Stymne, S.; Banaś, A. The utilization of the acyl-CoA and the involvement PDAT and DGAT in the biosynthesis of erucic acid-rich triacylglycerols in Crambe seed oil. Lipids 2014, 4, 327–333. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Giuffrè, A.M.; Tellah, S.; Capocasale, M.; Zappia, C.; Latati, M.; Badiani, M.; Ounane, S.M. Seed Oil from Ten Algerian Peanut Landraces for Edible Use and Biodiesel Production. J. Oleo Sci. 2016, 65, 9–20. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Giuffrè, A.M.; Zappia, C.; Capocasale, M. Tomato seed oil: A comparison of extraction systems and solvents on its biodiesel and edible properties. Riv. Ital. Delle Sostanze Grasse 2017, 3, 149–160. [Google Scholar]
- Giuffrè, A.M.; Capocasale, M.; Zappia, C.; Poiana, M. Influence of High Temperature and Duration of Heating on the Sunflower Seed Oil Properties for Food Use and Bio-diesel Production. J. Oleo Sci. 2017, 11, 1193–1205. [Google Scholar] [CrossRef] [Green Version]
- Niu, J.; Shi, Y.; Huang, K.; Zhong, Y.; Chen, J.; Sun, Z.; Luan, M.; Chen, J. Integrative transcriptome and proteome analyses provide new insights into different stages of Akebia trifoliata fruit cracking during ripening. Biotechnol. Biofuels 2020, 13, 149. [Google Scholar] [CrossRef]
- Chia, T.Y.; Pike, M.J.; Rawsthorne, S. Storage oil breakdown during embryo development of Brassica napus (L.). J. Exp. Bot. 2005, 415, 1285–1296. [Google Scholar] [CrossRef]
- Millar, A.A.; Kunst, L. Very-long-chain fatty acid biosynthesis is controlled through the expression and specificity of the condensing enzyme. Plant J. 1997, 1, 121–131. [Google Scholar] [CrossRef] [Green Version]
- Yang, H.; Mei, W.; Wan, H.; Xu, R.; Cheng, Y. Comprehensive analysis of KCS gene family in Citrinae reveals the involvement of CsKCS2 and CsKCS11 in fruit cuticular wax synthesis at ripening. Plant Sci. 2021, 310, 110972. [Google Scholar] [CrossRef]
- Ghafoor, K.; Ahmed, I.A.M.; Özcan, M.M.; Al-Juhaimi, F.Y.; Babiker, E.E.; Azmi, I.U. An evaluation of bioactive compounds, fatty acid composition and oil quality of chia (Salvia hispanica L.) seed roasted at different temperatures. Food Chem. 2020, 333, 127531. [Google Scholar] [CrossRef] [PubMed]
- Chernova, A.I.; Gubaev, R.F.; Singh, A.; Sherbina, K.; Goryunova, S.V.; Martynova, E.U.; Goryunov, D.V.; Boldyrev, S.V.; Vanyushkina, A.A.; Anikanov, N.A.; et al. Genotyping and lipid profiling of 601 cultivated sunflower lines reveals novel genetic determinants of oil fatty acid content. BMC Genom. 2021, 1, 505. [Google Scholar] [CrossRef] [PubMed]
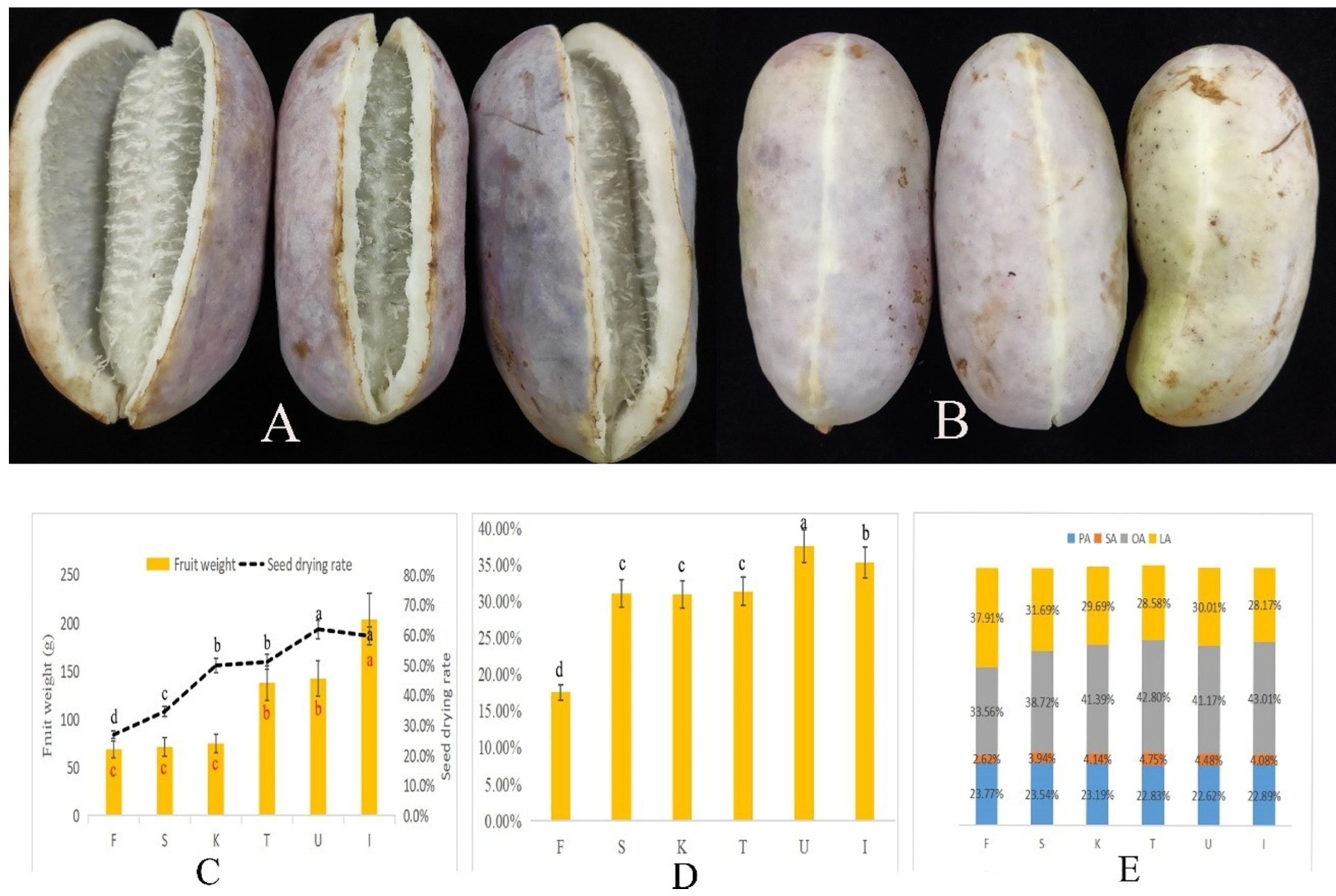

| Compositions (%) | F | S | K | T | U | I |
|---|---|---|---|---|---|---|
| C14:0 | 0.20 ab | 0.20 ab | 0.21 a | 0.18 b | 0.19 b | 0.21 a |
| C16:0 | 23.77 a | 23.54 a | 23.19 ab | 22.83 b | 22.62 b | 22.89 b |
| C16:1 | 0.48 a | 0.48 a | 0.45 b | 0.31 d | 0.40 c | 0.43 b |
| C17:0 | 0.20 a | 0.18 b | 0.13 c | 0.10 d | 0.13 c | 0.14 c |
| C17:1 | 0.12 a | 0.12 a | 0.10 b | 0.10 b | 0.10 b | 0.00 c |
| C18:0 | 2.62 d | 3.94 c | 4.14 b | 4.75 a | 4.48 ab | 4.08 bc |
| C18:1 | 33.56 d | 38.72 c | 41.39 b | 42.80 a | 41.17 b | 43.01 a |
| C18:2 | 37.91 a | 31.69 b | 29.69 c | 28.58 d | 30.01 c | 28.17 d |
| C18:3 | 0.57 a | 0.55 a | 0.30 c | 0.19 d | 0.30 c | 0.39 b |
| C20:0 | 0.33 b | 0.37 a | 0.21 c | 0.10 d | 0.32 b | 0.36 a |
| C20:1 | 0.26 b | 0.21 c | 0.19 d | 0.06 e | 0.29 b | 0.33 a |
| Enzyme | KEGG Annotation | Gene ID | Gene Expression Level | ||||
|---|---|---|---|---|---|---|---|
| F | S | T | U | I | |||
| PDH-E1α | pyruvate dehydrogenase E1 component alpha subunit [EC:1.2.4.1] | Atr14G031810 | 34.82 | 20.58 | 31.76 | 86.20 | 59.54 |
| Atr14G033430 | 12.17 | 5.83 | 8.96 | 29.54 | 49.31 | ||
| PDH-E1β | pyruvate dehydrogenase E1 component beta subunit [EC:1.2.4.1] | Atr14G023850 | 32.10 | 22.99 | 25.20 | 56.18 | 81.96 |
| Atr01G001270 | 4.77 | 9.94 | 11.60 | 13.63 | 5.10 | ||
| Atr04G024990 | 1.01 | 0.81 | 1.01 | 5.61 | 0.60 | ||
| PDH-E2 | pyruvate dehydrogenase E2 component (dihydrolipoamide acetyltransferase) [EC:2.3.1.12] | Atr09G013640 | 7.42 | 5.97 | 6.56 | 5.65 | 3.20 |
| ACC-BCCP | acetyl-CoA carboxylase biotin carboxyl carrier protein | Atr14G034290 | 41.19 | 31.02 | 27.76 | 28.30 | 16.91 |
| Atr15G033160 | 49.04 | 26.98 | 25.63 | 32.15 | 15.65 | ||
| Atr01G010810 | 10.40 | 6.40 | 4.89 | 11.74 | 10.35 | ||
| Akebia_trifoliata_newGene_7764 | 7.59 | 6.92 | 6.06 | 5.03 | 3.14 | ||
| MAT | [acyl-carrier-protein] S-malonyltransferase [EC:2.3.1.39] | Atr05G032500 | 45.07 | 41.11 | 36.37 | 34.81 | 17.76 |
| KASⅢ | 3-oxoacyl-[acyl-carrier-protein] synthase III [EC:2.3.1.180] | Atr12G005810 | 18.53 | 13.90 | 11.71 | 8.65 | 5.49 |
| HAD | 3-hydroxyacyl-[acyl-carrier-protein] dehydratase [EC:4.2.1.59] | Atr08G008530 | 34.10 | 29.97 | 37.20 | 34.65 | 89.15 |
| Atr11G026150 | 35.59 | 27.84 | 22.55 | 19.28 | 18.18 | ||
| Atr01G047740 | 27.63 | 24.27 | 16.63 | 24.09 | 14.17 | ||
| KASⅡ | 3-oxoacyl-[acyl-carrier-protein] synthase II [EC:2.3.1.179] | Atr03G013000 | 12.65 | 7.05 | 3.73 | 9.51 | 7.33 |
| Atr07G009450 | 14.85 | 7.62 | 9.82 | 5.37 | 8.21 | ||
| Atr03G057450 | 39.15 | 25.09 | 25.48 | 27.23 | 21.91 | ||
| Atr08G002240 | 31.60 | 18.59 | 20.02 | 23.37 | 13.56 | ||
| SAD | acyl-[acyl-carrier-protein] desaturase [EC:1.14.19.2 1.14.19.11 1.14.19.26] | Atr07G043510 | 529.94 | 502.14 | 374.17 | 814.09 | 246.83 |
| Atr01G045190 | 83.93 | 139.64 | 108.93 | 90.12 | 36.20 | ||
| FAD2 | acyl-lipid omega-6 desaturase (Delta-12 desaturase) [EC:1.14.19.6 1.14.19.22] | Atr13G034320 | 232.53 | 417.51 | 635.19 | 371.35 | 873.55 |
| Atr05G005700 | 3.11 | 3.04 | 8.00 | 9.05 | 5.50 | ||
| FATB | fatty acyl-ACP thioesterase B [EC:3.1.2.14 3.1.2.21] | Atr08G012910 | 107.49 | 114.40 | 109.96 | 97.92 | 21.44 |
| Atr08G012370 | 75.74 | 88.06 | 81.30 | 64.73 | 19.41 | ||
| Atr04G048060 | 3.61 | 1.66 | 0.93 | 0.52 | 0.09 | ||
| Atr02G007540 | 0.68 | 0.25 | 0.40 | 0.83 | 0.95 | ||
| FATA | fatty acyl-ACP thioesterase A [EC:3.1.2.14] | Atr07G024450 | 53.72 | 60.74 | 49.22 | 110.67 | 58.98 |
| LACS | long-chain acyl-CoA synthetase [EC:6.2.1.3] | Atr06G006720 | 36.88 | 33.60 | 48.22 | 55.27 | 100.21 |
| Atr11G013800 | 12.03 | 11.79 | 11.32 | 6.73 | 2.95 | ||
| Atr07G001260 | 38.88 | 41.14 | 37.52 | 22.96 | 2.22 | ||
| Atr11G034050 | 7.41 | 3.01 | 1.00 | 0.87 | 0.56 | ||
| Atr02G017850 | 0.98 | 0.84 | 0.74 | 1.41 | 2.65 | ||
| FAE1 | 3-ketoacyl-CoA synthase [EC:2.3.1.199] | Atr04G006760 | 96.46 | 83.59 | 80.09 | 34.99 | 0.40 |
| Atr06G016990 | 46.74 | 26.00 | 35.06 | 9.40 | 1.79 | ||
| Atr07G006560 | 40.24 | 11.52 | 3.94 | 1.80 | 0.05 | ||
| Atr01G055960 | 17.47 | 13.52 | 13.99 | 10.67 | 9.15 | ||
| Atr02G000850 | 26.95 | 14.79 | 10.00 | 15.93 | 0.06 | ||
| Atr02G061280 | 0.14 | 0.19 | 0.22 | 0.17 | 0.03 | ||
| Atr03G011460 | 1.44 | 1.09 | 2.29 | 4.91 | 3.27 | ||
| Atr04G047630 | 6.22 | 4.79 | 5.55 | 5.05 | 8.56 | ||
| Atr01G019190 | 1.28 | 0.92 | 0.94 | 0.63 | 0.04 | ||
| Atr09G004630 | 1.66 | 3.77 | 4.71 | 23.40 | 1.21 | ||
| Atr15G034500 | 37.47 | 37.03 | 36.44 | 35.01 | 23.21 | ||
| Atr05G047000 | 23.22 | 1.99 | 0.23 | 0.02 | 0.01 | ||
| Atr02G056530 | 8.14 | 5.77 | 3.63 | 2.87 | 0.60 | ||
| Atr02G056560 | 4.51 | 3.67 | 1.80 | 1.76 | 0.27 | ||
| Atr06G010840 | 14.00 | 11.02 | 11.46 | 9.99 | 1.23 | ||
| GPAT | glycerol-3-phosphate O-acyltransferase 1/2 [EC:2.3.1.15] | Atr09G000930 | 8.65 | 11.90 | 15.20 | 17.83 | 0.36 |
| Atr12G002270 | 36.13 | 28.21 | 28.59 | 35.24 | 4.56 | ||
| Atr16G001970 | 9.05 | 2.97 | 1.60 | 0.88 | 1.35 | ||
| Atr05G014350 | 42.35 | 64.68 | 83.48 | 60.68 | 19.91 | ||
| Atr05G039970 | 0.07 | 0.01 | 0.03 | 1.77 | 0.02 | ||
| Atr05G043400 | 0.01 | 0.04 | 0.46 | 6.38 | 0.52 | ||
| ATS1 | glycerol-3-phosphate O-acyltransferase [EC:2.3.1.15] | Atr03G046480 | 3.00 | 2.17 | 2.59 | 4.34 | 4.07 |
| Atr03G047060 | 5.11 | 4.56 | 5.49 | 7.37 | 7.39 | ||
| LPAAT | lysocardiolipin and lysophospholipid acyltransferase [EC:2.3.1.-2.3.1.51] | Atr05G010470 | 54.78 | 36.01 | 39.86 | 41.25 | 30.69 |
| Atr05G010480 | 41.01 | 26.86 | 27.45 | 29.32 | 21.23 | ||
| Atr12G003260 | 6.94 | 7.88 | 8.03 | 10.26 | 5.91 | ||
| Atr11G034300 | 3.22 | 2.19 | 4.17 | 9.08 | 14.07 | ||
| DGAT1 | diacylglycerol O-acyltransferase 1 [EC:2.3.1.20 2.3.1.75 2.3.1.76] | Atr04G050350 | 2.11 | 1.24 | 1.45 | 9.27 | 25.77 |
| PDAT | phospholipid:diacylglycerol acyltransferase [EC:2.3.1.158] | Atr04G004940 | 3.51 | 1.73 | 1.42 | 7.18 | 8.24 |
| Atr04G005010 | 2.77 | 1.93 | 1.70 | 5.98 | 7.71 | ||
| LPCAT | lysophospholipid acyltransferase [EC:2.3.1.51 2.3.1.23 2.3.1.-] | Atr03G002590 | 4.97 | 5.08 | 8.66 | 5.75 | 0.60 |
| Name | Subfamilies | Chromosome | Number of Amino Acids | Subcellular Location | Gene Expression Level | ||||
|---|---|---|---|---|---|---|---|---|---|
| F | S | T | U | I | |||||
| AtrFAD1 | SAD | Chr 1 | 385 | Chloroplast | 77.90 | 152.03 | 108.93 | 90.12 | 14.58 |
| AtrFAD2 | Front-end | Chr 1 | 446 | Endoplasmic reticulum | 28.73 | 13.65 | 15.16 | 25.22 | 59.98 |
| AtrFAD3 | Δ7/Δ9 | Chr 2 | 384 | Chloroplast, Endoplasmic reticulum | 4.67 | 7.86 | 16.08 | 18.82 | 18.38 |
| AtrFAD4 | Δ7/Δ9 | Chr 4 | 298 | Cell membrane, Cell wall, Chloroplast, Mitochondrion | 0.14 | 0.02 | 0.16 | 0.03 | 0.00 |
| AtrFAD5 | Δ7/Δ9 | Chr 4 | 295 | Cell membrane, Cell wall, Cell nucleus | 0.34 | 0.07 | 0.47 | 0.19 | 0.00 |
| AtrFAD6 | Δ12/ω3 | Chr 5 | 397 | Endoplasmic reticulum | 3.24 | 3.00 | 8.00 | 9.05 | 4.92 |
| AtrFAD7 | Δ12/ω3 | Chr 5 | 457 | Cell membrane, Endoplasmic reticulum | 22.62 | 20.10 | 26.67 | 18.92 | 7.95 |
| AtrFAD8 | Front-end | Chr 6 | 195 | Chloroplast, Endoplasmic reticulum | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 |
| AtrFAD9 | SAD | Chr 7 | 397 | Chloroplast | 528.17 | 461.17 | 374.17 | 814.09 | 260.38 |
| AtrFAD10 | SAD | Chr 7 | 397 | Chloroplast | 1.36 | 2.73 | 3.36 | 2.60 | 0.52 |
| AtrFAD11 | SAD | Chr 7 | 395 | Chloroplast | 0.11 | 0.34 | 0.69 | 4.65 | 1.94 |
| AtrFAD12 | SAD | Chr 7 | 336 | Chloroplast | 0.00 | 0.00 | 0.00 | 0.03 | 0.00 |
| AtrFAD13 | SAD | Chr 7 | 467 | Chloroplast | 0.01 | 0.00 | 0.35 | 0.17 | 1.67 |
| AtrFAD14 | Δ12/ω3 | Chr 8 | 347 | Chloroplast | 8.63 | 13.51 | 19.96 | 17.33 | 14.14 |
| AtrFAD15 | Δ12/ω3 | Chr 8 | 341 | Chloroplast | 7.94 | 11.55 | 17.80 | 14.74 | 13.46 |
| AtrFAD16 | SAD | Chr 9 | 397 | Chloroplast | 47.63 | 59.75 | 68.32 | 95.17 | 106.35 |
| AtrFAD17 | Δ7/Δ9 | Chr 11 | 322 | Chloroplast, Endoplasmic reticulum | 47,497.02 | 36,879.01 | 33,223.82 | 2311.01 | 87.18 |
| AtrFAD18 | Δ7/Δ9 | Chr 11 | 332 | Chloroplast, Endoplasmic reticulum | 41,957.81 | 31,117.12 | 38,430.20 | 2062.99 | 99.40 |
| AtrFAD19 | Δ12/ω3 | Chr 13 | 381 | Endoplasmic reticulum | 228.41 | 414.97 | 635.19 | 371.35 | 758.35 |
| AtrFAD20 | Δ12/ω3 | Chr 16 | 461 | Chloroplast, Endoplasmic reticulum | 1.63 | 0.99 | 0.70 | 0.42 | 1.18 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhong, Y.; Zhao, Y.; Wang, Y.; Niu, J.; Sun, Z.; Chen, J.; Luan, M. Transcriptome Analysis and GC-MS Profiling of Key Fatty Acid Biosynthesis Genes in Akebia trifoliata (Thunb.) Koidz Seeds. Biology 2022, 11, 855. https://doi.org/10.3390/biology11060855
Zhong Y, Zhao Y, Wang Y, Niu J, Sun Z, Chen J, Luan M. Transcriptome Analysis and GC-MS Profiling of Key Fatty Acid Biosynthesis Genes in Akebia trifoliata (Thunb.) Koidz Seeds. Biology. 2022; 11(6):855. https://doi.org/10.3390/biology11060855
Chicago/Turabian StyleZhong, Yicheng, Yunlei Zhao, Yue Wang, Juan Niu, Zhimin Sun, Jianhua Chen, and Mingbao Luan. 2022. "Transcriptome Analysis and GC-MS Profiling of Key Fatty Acid Biosynthesis Genes in Akebia trifoliata (Thunb.) Koidz Seeds" Biology 11, no. 6: 855. https://doi.org/10.3390/biology11060855
APA StyleZhong, Y., Zhao, Y., Wang, Y., Niu, J., Sun, Z., Chen, J., & Luan, M. (2022). Transcriptome Analysis and GC-MS Profiling of Key Fatty Acid Biosynthesis Genes in Akebia trifoliata (Thunb.) Koidz Seeds. Biology, 11(6), 855. https://doi.org/10.3390/biology11060855






