Functional Characterization and Whole-Genome Analysis of an Aflatoxin-Degrading Rhodococcus pyridinivorans Strain
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Bacterial Strains and Plasmids
2.2. The Detection of AFB1 Degradation Ability
2.3. Detection of the AFB1-Degrading Activity of Different Bacterial Components
2.4. Whole-Genome Sequencing of R. pyridinivorans 4-4
2.5. Genome Assembly and Annotation
2.6. Comparative Genomic Analysis
2.7. Liquid Chromatography Tandem Mass Spectrometry (LC-MS/MS) Analysis of Extracellular Component
2.8. Protein Spectrum Data Analysis
2.9. Data Statistics
3. Results
3.1. AFB1 Degradation Ability of R. pyridinivorans 4-4
3.2. Genomic Analysis of R. pyridinivorans 4-4
3.3. Comparative Genomic Analysis of R. pyridinivorans 4-4 and other Rhodococcus Strains
3.4. Proteomic Analysis of Extracellular Components
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Gizachew, D.; Chang, C.H.; Szonyi, B.; De La Torre, S.; Ting, W.E. Aflatoxin B1 (AFB1) production by Aspergillus flavus and Aspergillus parasiticus on ground Nyjer seeds: The effect of water activity and temperature. Int. J. Food Microbiol. 2019, 296, 8–13. [Google Scholar] [CrossRef] [PubMed]
- Wang, P.; Ma, L.; Jin, J.; Zheng, M.; Pan, L.; Zhao, Y.; Sun, X.; Liu, Y.; Xing, F. The anti-aflatoxigenic mechanism of cinnamaldehyde in Aspergillus flavus. Sci. Rep. 2019, 9, 10499. [Google Scholar] [CrossRef]
- Woloshuk, C.P.; Shim, W.B. Aflatoxins, fumonisins, and trichothecenes: A convergence of knowledge. FEMS Microbiol. Rev. 2013, 37, 94–109. [Google Scholar] [CrossRef] [Green Version]
- Theumer, M.G.; Henneb, Y.; Khoury, L.; Snini, S.P.; Tadrist, S.; Canlet, C.; Puel, O.; Oswald, I.P.; Audebert, M. Genotoxicity of aflatoxins and their precursors in human cells. Toxicol. Lett. 2018, 287, 100–107. [Google Scholar] [CrossRef] [PubMed]
- Adebo, O.A.; Njobeh, P.B.; Gbashi, S.; Nwinyi, O.C.; Mavumengwana, V. Review on microbial degradation of aflatoxins. Crit. Rev. Food Sci. Nutr. 2017, 57, 3208–3217. [Google Scholar] [CrossRef]
- Marin, S.; Ramos, A.J.; Cano-Sancho, G.; Sanchis, V. Mycotoxins: Occurrence, toxicology, and exposure assessment. Food Chem. Toxicol. 2013, 60, 218–237. [Google Scholar] [CrossRef]
- Dhakal, A.; Sbar, E. StatPearls; StatPearls Publishing LLC.: Treasure Island, FL, USA, 2022. [Google Scholar]
- Haskard, C.A.; El-Nezami, H.S.; Kankaanpää, P.E.; Salminen, S.; Ahokas, J.T. Surface binding of aflatoxin B(1) by lactic acid bacteria. Appl. Environ. Microbiol. 2001, 67, 3086–3091. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Adebo, O.A.; Njobeh, P.B.; Sidu, S.; Tlou, M.G.; Mavumengwana, V. Aflatoxin B1 degradation by liquid cultures and lysates of three bacterial strains. Int. J. Food Microbiol. 2016, 233, 11–19. [Google Scholar] [CrossRef]
- Wang, L.; Huang, W.; Sha, Y.; Yin, H.; Liang, Y.; Wang, X.; Shen, Y.; Wu, X.; Wu, D.; Wang, J. Co-Cultivation of Two Bacillus Strains for Improved Cell Growth and Enzyme Production to Enhance the Degradation of Aflatoxin B(1). Toxins 2021, 13, 435. [Google Scholar] [CrossRef]
- Alberts, J.F.; Engelbrecht, Y.; Steyn, P.S.; Holzapfel, W.H.; van Zyl, W.H. Biological degradation of aflatoxin B1 by Rhodococcus erythropolis cultures. Int. J. Food Microbiol. 2006, 109, 121–126. [Google Scholar] [CrossRef]
- Zhuang, Z.; Huang, Y.; Yang, Y.; Wang, S. Identification of AFB1-interacting proteins and interactions between RPSA and AFB1. J. Hazard. Mater. 2016, 301, 297–303. [Google Scholar] [CrossRef] [PubMed]
- Singh, J.; Mehta, A. Protein-mediated degradation of aflatoxin B(1) by Pseudomonas putida. Braz. J. Microbiol. 2019, 50, 1031–1039. [Google Scholar] [CrossRef] [PubMed]
- Taylor, M.C.; Jackson, C.J.; Tattersall, D.B.; French, N.; Peat, T.S.; Newman, J.; Briggs, L.J.; Lapalikar, G.V.; Campbell, P.M.; Scott, C.; et al. Identification and characterization of two families of F420 H2-dependent reductases from Mycobacteria that catalyse aflatoxin degradation. Mol. Microbiol. 2010, 78, 561–575. [Google Scholar] [CrossRef] [Green Version]
- Wang, J.; Ogata, M.; Hirai, H.; Kawagishi, H. Detoxification of aflatoxin B1 by manganese peroxidase from the white-rot fungus Phanerochaete sordida YK-624. FEMS Microbiol. Lett. 2011, 314, 164–169. [Google Scholar] [CrossRef] [PubMed]
- Loi, M.; Fanelli, F.; Zucca, P.; Liuzzi, V.C.; Quintieri, L.; Cimmarusti, M.T.; Monaci, L.; Haidukowski, M.; Logrieco, A.F.; Sanjust, E.; et al. Aflatoxin B₁ and M₁ Degradation by Lac2 from Pleurotus pulmonarius and Redox Mediators. Toxins 2016, 8, 245. [Google Scholar] [CrossRef] [Green Version]
- Xu, L.; Eisa Ahmed, M.F.; Sangare, L.; Zhao, Y.; Selvaraj, J.N.; Xing, F.; Wang, Y.; Yang, H.; Liu, Y. Novel Aflatoxin-Degrading Enzyme from Bacillus shackletonii L7. Toxins 2017, 9, 36. [Google Scholar] [CrossRef]
- Yehia, R.S. Aflatoxin detoxification by manganese peroxidase purified from Pleurotus ostreatus. Braz. J. Microbiol. 2014, 45, 127–133. [Google Scholar] [CrossRef] [Green Version]
- Cao, H.; Liu, D.; Mo, X.; Xie, C.; Yao, D. A fungal enzyme with the ability of aflatoxin B₁ conversion: Purification and ESI-MS/MS identification. Microbiol. Res. 2011, 166, 475–483. [Google Scholar] [CrossRef]
- Li, J.; Huang, J.; Jin, Y.; Wu, C.; Shen, D.; Zhang, S.; Zhou, R. Aflatoxin B(1) degradation by salt tolerant Tetragenococcus halophilus CGMCC 3792. Food Chem. Toxicol. 2018, 121, 430–436. [Google Scholar] [CrossRef]
- Teniola, O.D.; Addo, P.A.; Brost, I.M.; Färber, P.; Jany, K.D.; Alberts, J.F.; van Zyl, W.H.; Steyn, P.S.; Holzapfel, W.H. Degradation of aflatoxin B(1) by cell-free extracts of Rhodococcus erythropolis and Mycobacterium fluoranthenivorans sp. nov. DSM44556(T). Int. J. Food Microbiol. 2005, 105, 111–117. [Google Scholar] [CrossRef]
- Cai, M.; Qian, Y.; Chen, N.; Ling, T.; Wang, J.; Jiang, H.; Wang, X.; Qi, K.; Zhou, Y. Detoxification of aflatoxin B1 by Stenotrophomonas sp. CW117 and characterization the thermophilic degradation process. Environ. Pollut. 2020, 261, 114178. [Google Scholar] [CrossRef] [PubMed]
- Guan, S.; Ji, C.; Zhou, T.; Li, J.; Ma, Q.; Niu, T. Aflatoxin B(1) degradation by Stenotrophomonas maltophilia and other microbes selected using coumarin medium. Int. J. Mol. Sci. 2008, 9, 1489–1503. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, Y.; Zhao, C.; Zhang, D.; Zhao, M.; Zheng, D.; Lyu, Y.; Cheng, W.; Guo, P.; Cui, Z. Effective degradation of aflatoxin B(1) using a novel thermophilic microbial consortium TADC7. Bioresour. Technol. 2017, 224, 166–173. [Google Scholar] [CrossRef] [PubMed]
- Verheecke, C.; Liboz, T.; Mathieu, F. Microbial degradation of aflatoxin B1: Current status and future advances. Int. J. Food Microbiol. 2016, 237, 1–9. [Google Scholar] [CrossRef] [Green Version]
- He, S.; Feng, K.; Ding, T.; Huang, K.; Yan, H.; Liu, X.; Zhang, Z. Complete genome sequence of Bacillus licheniformis BL-010. Microb. Pathog. 2018, 118, 199–201. [Google Scholar] [CrossRef]
- Fang, Q.; Du, M.; Chen, J.; Liu, T.; Zheng, Y.; Liao, Z.; Zhong, Q.; Wang, L.; Fang, X.; Wang, J. Degradation and Detoxification of Aflatoxin B1 by Tea-Derived Aspergillus niger RAF106. Toxins 2020, 12, 777. [Google Scholar] [CrossRef]
- Ma, J.; Liu, Y.; Guo, Y.; Ma, Q.; Ji, C.; Zhao, L. Transcriptional Profiling of Aflatoxin B1-Induced Oxidative Stress and Inflammatory Response in Macrophages. Toxins 2021, 13, 401. [Google Scholar] [CrossRef]
- Li, S.; Liu, R.; Wei, G.; Guo, G.; Yu, H.; Zhang, Y.; Ishfaq, M.; Fazilani, S.A.; Zhang, X. Curcumin protects against Aflatoxin B1-induced liver injury in broilers via the modulation of long non-coding RNA expression. Ecotoxicol. Environ. Saf. 2021, 208, 111725. [Google Scholar] [CrossRef]
- Pauletto, M.; Giantin, M.; Tolosi, R.; Bassan, I.; Barbarossa, A.; Zaghini, A.; Dacasto, M. Curcumin Mitigates AFB1-Induced Hepatic Toxicity by Triggering Cattle Antioxidant and Anti-inflammatory Pathways: A Whole Transcriptomic In Vitro Study. Antioxidants 2020, 9, 1059. [Google Scholar] [CrossRef]
- Zhu, L.; Huang, C.; Yang, X.; Zhang, B.; He, X.; Xu, W.; Huang, K. Proteomics reveals the alleviation of zinc towards aflatoxin B1-induced cytotoxicity in human hepatocyes (HepG2 cells). Ecotoxicol. Environ. Saf. 2020, 198, 110596. [Google Scholar] [CrossRef]
- Liew, W.P.; Sabran, M.R.; Than, L.T.; Abd-Ghani, F. Metagenomic and proteomic approaches in elucidating aflatoxin B(1) detoxification mechanisms of probiotic Lactobacillus casei Shirota towards intestine. Food Chem. Toxicol. 2022, 160, 112808. [Google Scholar] [CrossRef]
- Li, X.; Jiang, Y.; Ma, L.; Ma, X.; Liu, Y.; Shan, J.; Ma, K.; Xing, F. Comprehensive Transcriptome and Proteome Analyses Reveal the Modulation of Aflatoxin Production by Aspergillus flavus on Different Crop Substrates. Front. Microbiol. 2020, 11, 1497. [Google Scholar] [CrossRef] [PubMed]
- Eshelli, M.; Harvey, L.; Edrada-Ebel, R.; McNeil, B. Metabolomics of the bio-degradation process of aflatoxin B1 by actinomycetes at an initial pH of 6.0. Toxins 2015, 7, 439–456. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, S.; Yang, X.; Liu, F.; Wang, X.; Zhang, X.; He, K.; Wang, H. Comprehensive Metabolomic Analysis Reveals Dynamic Metabolic Reprogramming in Hep3B Cells with Aflatoxin B1 Exposure. Toxins 2021, 13, 384. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Zhang, Y.; Zheng, N.; Guo, L.; Song, X.; Zhao, S.; Wang, J. Biological System Responses of Dairy Cows to Aflatoxin B1 Exposure Revealed with Metabolomic Changes in Multiple Biofluids. Toxins 2019, 11, 77. [Google Scholar] [CrossRef] [Green Version]
- Cserháti, M.; Kriszt, B.; Krifaton, C.; Szoboszlay, S.; Háhn, J.; Tóth, S.; Nagy, I.; Kukolya, J. Mycotoxin-degradation profile of Rhodococcus strains. Int. J. Food Microbiol. 2013, 166, 176–185. [Google Scholar] [CrossRef]
- Risa, A.; Krifaton, C.; Kukolya, J.; Kriszt, B.; Cserháti, M.; Táncsics, A. Aflatoxin B1 and Zearalenone-Detoxifying Profile of Rhodococcus Type Strains. Curr. Microbiol. 2018, 75, 907–917. [Google Scholar] [CrossRef]
- Kechin, A.; Boyarskikh, U.; Kel, A.; Filipenko, M. cutPrimers: A New Tool for Accurate Cutting of Primers from Reads of Targeted Next Generation Sequencing. J. Comput. Biol. 2017, 24, 1138–1143. [Google Scholar] [CrossRef]
- Li, H.; Durbin, R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 2009, 25, 1754–1760. [Google Scholar] [CrossRef] [Green Version]
- Hunt, M.; Silva, N.D.; Otto, T.D.; Parkhill, J.; Keane, J.A.; Harris, S.R. Circlator: Automated circularization of genome assemblies using long sequencing reads. Genome Biol. 2015, 16, 294. [Google Scholar] [CrossRef] [Green Version]
- Simão, F.A.; Waterhouse, R.M.; Ioannidis, P.; Kriventseva, E.V.; Zdobnov, E.M. BUSCO: Assessing genome assembly and annotation completeness with single-copy orthologs. Bioinformatics 2015, 31, 3210–3212. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Seemann, T. Prokka: Rapid prokaryotic genome annotation. Bioinformatics 2014, 30, 2068–2069. [Google Scholar] [CrossRef] [PubMed]
- Tatusov, R.L.; Fedorova, N.D.; Jackson, J.D.; Jacobs, A.R.; Kiryutin, B.; Koonin, E.V.; Krylov, D.M.; Mazumder, R.; Mekhedov, S.L.; Nikolskaya, A.N.; et al. The COG database: An updated version includes eukaryotes. BMC Bioinform. 2003, 4, 41. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Finn, R.D.; Bateman, A.; Clements, J.; Coggill, P.; Eberhardt, R.Y.; Eddy, S.R.; Heger, A.; Hetherington, K.; Holm, L.; Mistry, J.; et al. Pfam: The protein families database. Nucleic Acids Res. 2014, 42, D222–D230. [Google Scholar] [CrossRef] [Green Version]
- Page, A.J.; Cummins, C.A.; Hunt, M.; Wong, V.K.; Reuter, S.; Holden, M.T.; Fookes, M.; Falush, D.; Keane, J.A.; Parkhill, J. Roary: Rapid large-scale prokaryote pan genome analysis. Bioinformatics 2015, 31, 3691–3693. [Google Scholar] [CrossRef]
- Nguyen, L.T.; Schmidt, H.A.; von Haeseler, A.; Minh, B.Q. IQ-TREE: A fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol. Biol. Evol. 2015, 32, 268–274. [Google Scholar] [CrossRef]
- Wiśniewski, J.R.; Zougman, A.; Nagaraj, N.; Mann, M. Universal sample preparation method for proteome analysis. Nat. Methods 2009, 6, 359–362. [Google Scholar] [CrossRef]
- Sangare, L.; Zhao, Y.; Folly, Y.M.; Chang, J.; Li, J.; Selvaraj, J.N.; Xing, F.; Zhou, L.; Wang, Y.; Liu, Y. Aflatoxin B₁ degradation by a Pseudomonas strain. Toxins 2014, 6, 3028–3040. [Google Scholar] [CrossRef]
- Song, J.; Zhang, S.; Xie, Y.; Li, Q. Purification and characteristics of an aflatoxin B1 degradation enzyme isolated from Pseudomonas aeruginosa. FEMS Microbiol. Lett. 2019, 366, fnz034. [Google Scholar] [CrossRef]
- Jingjing, E.; Lili, M.; Zichao, C.; Rongze, M.; Qiaoling, Z.; Ruiyin, S.; Zongbai, H.; Junguo, W. Effects of buffer salts on the freeze-drying survival rate of Lactobacillus plantarum LIP-1 based on transcriptome and proteome analyses. Food Chem. 2020, 326, 126849. [Google Scholar] [CrossRef]
- Pedersen, H.B.; Josephsen, J.; Kerszman, G. Phosphate buffer and salt medium concentrations affect the inactivation of T4 phage by platinum(II) complexes. Chem. Biol. Interact. 1985, 54, 1–8. [Google Scholar] [CrossRef]
- Lapalikar, G.V.; Taylor, M.C.; Warden, A.C.; Scott, C.; Russell, R.J.; Oakeshott, J.G. F420H2-dependent degradation of aflatoxin and other furanocoumarins is widespread throughout the actinomycetales. PLoS ONE 2012, 7, e30114. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ellis, E.M.; Judah, D.J.; Neal, G.E.; Hayes, J.D. An ethoxyquin-inducible aldehyde reductase from rat liver that metabolizes aflatoxin B1 defines a subfamily of aldo-keto reductases. Proc. Natl. Acad. Sci. USA 1993, 90, 10350–10354. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Knight, L.P.; Primiano, T.; Groopman, J.D.; Kensler, T.W.; Sutter, T.R. cDNA cloning, expression and activity of a second human aflatoxin B1-metabolizing member of the aldo-keto reductase superfamily, AKR7A3. Carcinogenesis 1999, 20, 1215–1223. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cuccioloni, M.; Mozzicafreddo, M.; Barocci, S.; Ciuti, F.; Re, L.; Eleuteri, A.M.; Angeletti, M. Aflatoxin B1 misregulates the activity of serine proteases: Possible implications in the toxicity of some mycotoxin. Toxicol. Vitro 2009, 23, 393–399. [Google Scholar] [CrossRef]
- Sun, L.H.; Lei, M.Y.; Zhang, N.Y.; Zhao, L.; Krumm, C.S.; Qi, D.S. Hepatotoxic effects of mycotoxin combinations in mice. Food Chem. Toxicol. 2014, 74, 289–293. [Google Scholar] [CrossRef]
- Huang, S.; Zheng, N.; Fan, C.; Cheng, M.; Wang, S.; Jabar, A.; Wang, J.; Cheng, J. Effects of aflatoxin B1 combined with ochratoxin A and/or zearalenone on metabolism, immune function, and antioxidant status in lactating dairy goats. Asian-Australas J. Anim. Sci. 2018, 31, 505–513. [Google Scholar] [CrossRef] [Green Version]
- Feng, G.D.; He, J.; Ao, X.; Chen, D.W. Effects of maize naturally contaminated with aflatoxin B1 on growth performance, intestinal morphology, and digestive physiology in ducks. Poult. Sci. 2017, 96, 1948–1955. [Google Scholar] [CrossRef]
- Moss, E.J.; Manson, M.M.; Neal, G.E. Effect of manipulation of gamma-glutamyl transpeptidase levels on biliary excretion of aflatoxin B1 conjugates. Carcinogenesis 1984, 5, 869–874. [Google Scholar] [CrossRef]
- Youngman, L.D.; Campbell, T.C. Inhibition of aflatoxin B1-induced gamma-glutamyltranspeptidase positive (GGT+) hepatic preneoplastic foci and tumors by low protein diets: Evidence that altered GGT+ foci indicate neoplastic potential. Carcinogenesis 1992, 13, 1607–1613. [Google Scholar] [CrossRef]
- Sakamoto, M.I.; Murakami, A.E.; Fernandes, A.M.; Ospina-Rojas, I.C.; Nunes, K.C.; Hirata, A.K. Performance and serum biochemical profile of Japanese quail supplemented with silymarin and contaminated with aflatoxin B1. Poult. Sci. 2018, 97, 159–166. [Google Scholar] [CrossRef] [PubMed]
- Farombi, E.O.; Adepoju, B.F.; Ola-Davies, O.E.; Emerole, G.O. Chemoprevention of aflatoxin B1-induced genotoxicity and hepatic oxidative damage in rats by kolaviron, a natural bioflavonoid of Garcinia kola seeds. Eur. J. Cancer Prev. 2005, 14, 207–214. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Wang, W. Aflatoxin B1 impairs mitochondrial functions, activates ROS generation, induces apoptosis and involves Nrf2 signal pathway in primary broiler hepatocytes. Anim. Sci. J. 2016, 87, 1490–1500. [Google Scholar] [CrossRef] [PubMed]
- Wagner, G.; Frenzel, H.; Wefers, H.; Sies, H. Lack of effect of long-term glutathione administration on aflatoxin B1-induced hepatoma in male rats. Chem. Biol. Interact. 1985, 53, 57–68. [Google Scholar] [CrossRef]
- Chhonker, S.K.; Rawat, D.; Koiri, R.K. Protective and therapeutic effects of sildenafil and tadalafil on aflatoxin B1-induced hepatocellular carcinoma. Mol. Cell. Biochem. 2021, 476, 1195–1209. [Google Scholar] [CrossRef]
- Long, M.; Zhang, Y.; Li, P.; Yang, S.H.; Zhang, W.K.; Han, J.X.; Wang, Y.; He, J.B. Intervention of Grape Seed Proanthocyanidin Extract on the Subchronic Immune Injury in Mice Induced by Aflatoxin B1. Int. J. Mol. Sci. 2016, 17, 516. [Google Scholar] [CrossRef] [Green Version]
- Sulzberger, S.A.; Melnichenko, S.; Cardoso, F.C. Effects of clay after an aflatoxin challenge on aflatoxin clearance, milk production, and metabolism of Holstein cows. J. Dairy Sci. 2017, 100, 1856–1869. [Google Scholar] [CrossRef]
- Pate, R.T.; Paulus Compart, D.M.; Cardoso, F.C. Aluminosilicate clay improves production responses and reduces inflammation during an aflatoxin challenge in lactating Holstein cows. J. Dairy Sci. 2018, 101, 11421–11434. [Google Scholar] [CrossRef] [Green Version]
- Quezada, T.; Cuéllar, H.; Jaramillo-Juárez, F.; Valdivia, A.G.; Reyes, J.L. Effects of aflatoxin B(1) on the liver and kidney of broiler chickens during development. Comp. Biochem. Physiol. C Toxicol. Pharmacol. 2000, 125, 265–272. [Google Scholar] [CrossRef]
- Sajan, M.P.; Satav, J.G.; Bhattacharya, R.K. Activity of some respiratory enzymes and cytochrome contents in rat hepatic mitochondria following aflatoxin B1 administration. Toxicol. Lett. 1995, 80, 55–60. [Google Scholar] [CrossRef]
- Baldissera, M.D.; Souza, C.F.; Zeppenfeld, C.C.; Descovi, S.; da Silva, A.S.; Baldisserotto, B. Changes in the cerebral phosphotransfer network impair energetic homeostasis in an aflatoxin B(1)-contaminated diet. Fish Physiol. Biochem. 2018, 44, 1051–1059. [Google Scholar] [CrossRef] [PubMed]
- Ch’ih, J.J.; Ewaskiewicz, J.I.; Taggart, P.; Devlin, T.M. Nuclear translocation of aflatoxin B1—Protein complex. Biochem. Biophys. Res. Commun. 1993, 190, 186–191. [Google Scholar] [CrossRef] [PubMed]
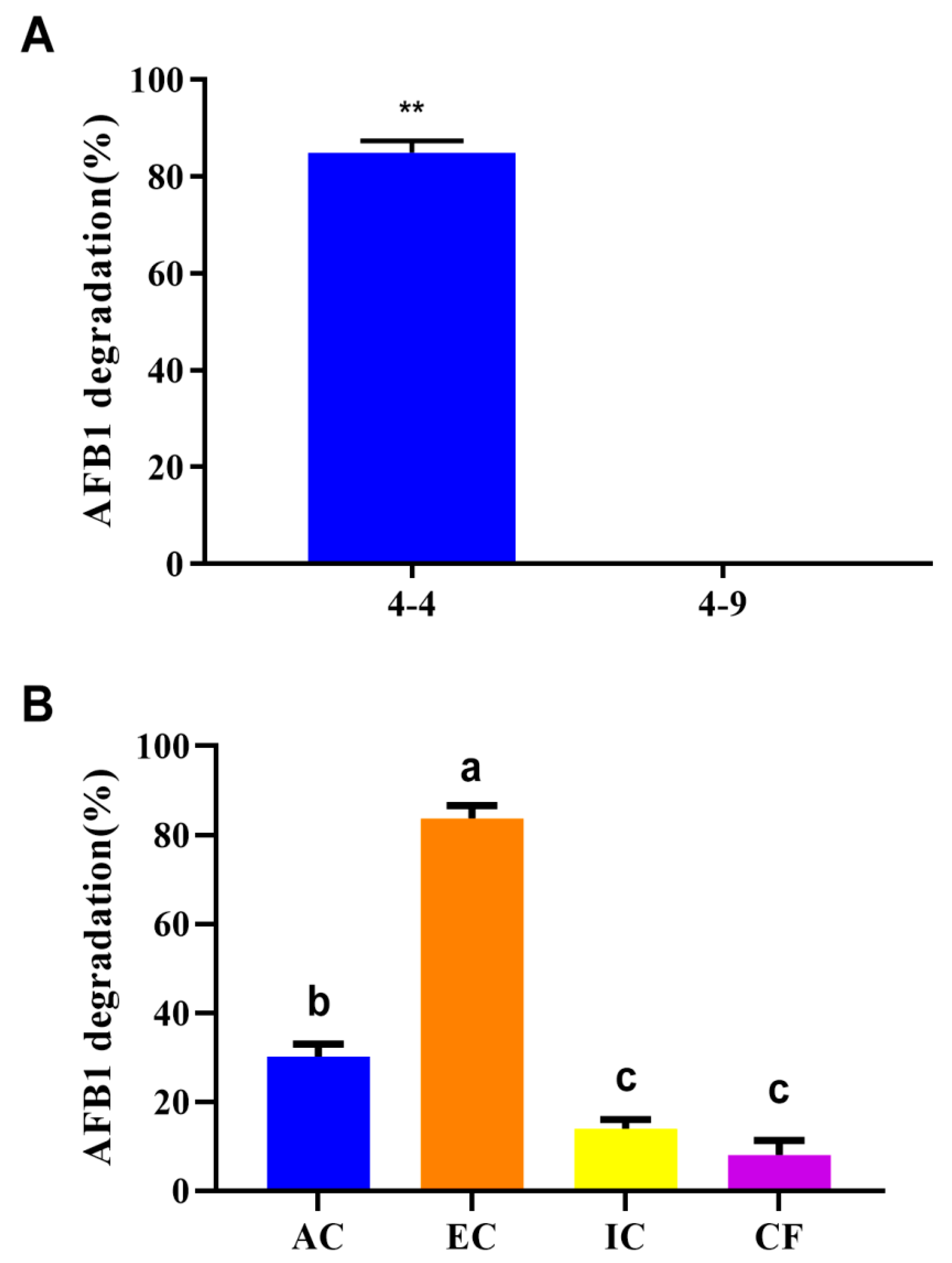

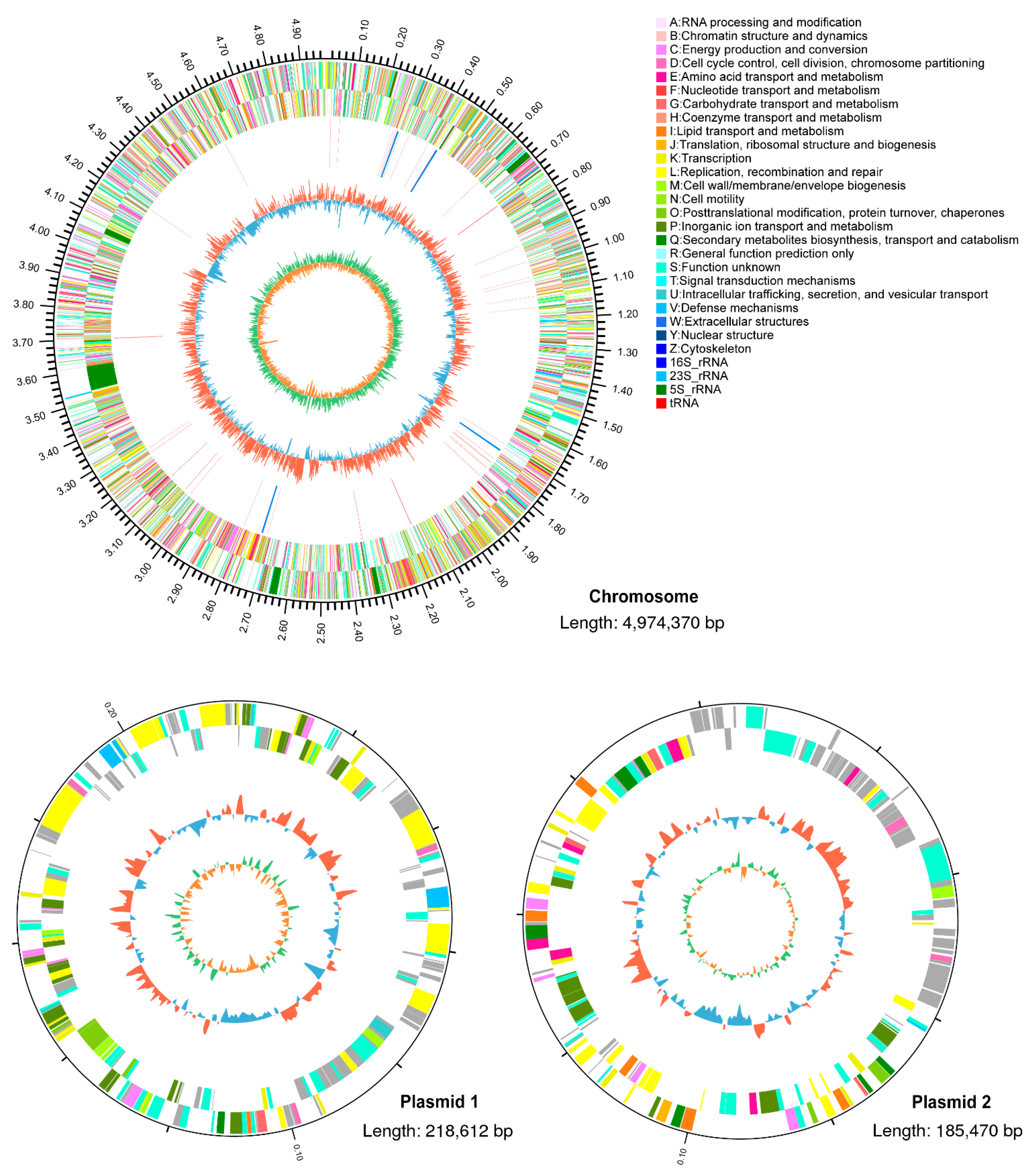
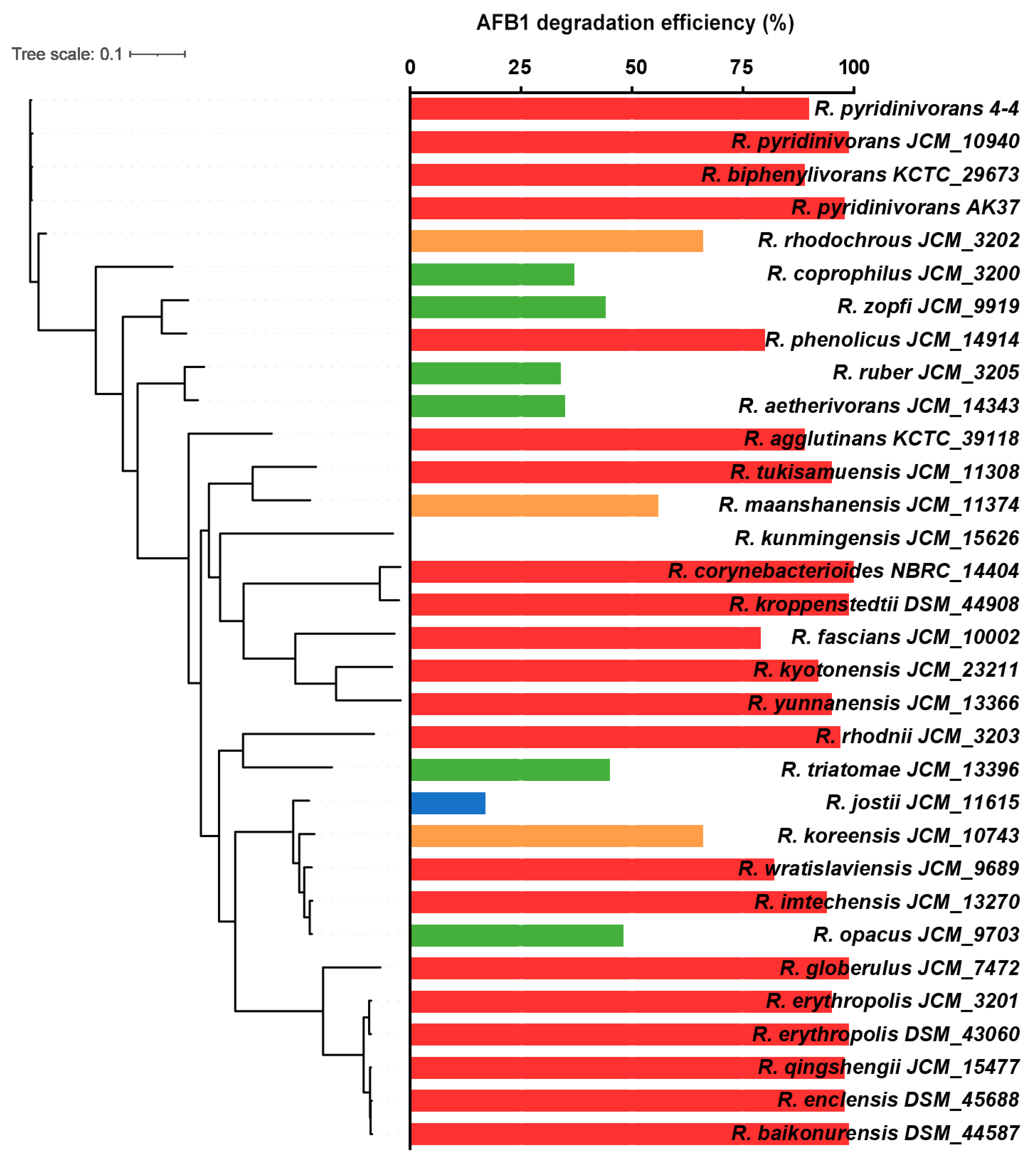
| Database | Number of Annotated Genes | Percentage |
|---|---|---|
| Nr | 4985 | 0.95 |
| Swiss-Prot | 3480 | 0.66 |
| COG | 4308 | 0.82 |
| Pfam | 3997 | 0.76 |
| GO | 3633 | 0.69 |
| KEGG | 2111 | 0.40 |
| Overall | 4986 | 0.95 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Deng, D.; Tang, J.; Liu, Z.; Tian, Z.; Song, M.; Cui, Y.; Rong, T.; Lu, H.; Yu, M.; Li, J.; et al. Functional Characterization and Whole-Genome Analysis of an Aflatoxin-Degrading Rhodococcus pyridinivorans Strain. Biology 2022, 11, 774. https://doi.org/10.3390/biology11050774
Deng D, Tang J, Liu Z, Tian Z, Song M, Cui Y, Rong T, Lu H, Yu M, Li J, et al. Functional Characterization and Whole-Genome Analysis of an Aflatoxin-Degrading Rhodococcus pyridinivorans Strain. Biology. 2022; 11(5):774. https://doi.org/10.3390/biology11050774
Chicago/Turabian StyleDeng, Dun, Jiahong Tang, Zhichang Liu, Zhimei Tian, Min Song, Yiyan Cui, Ting Rong, Huijie Lu, Miao Yu, Jinbao Li, and et al. 2022. "Functional Characterization and Whole-Genome Analysis of an Aflatoxin-Degrading Rhodococcus pyridinivorans Strain" Biology 11, no. 5: 774. https://doi.org/10.3390/biology11050774
APA StyleDeng, D., Tang, J., Liu, Z., Tian, Z., Song, M., Cui, Y., Rong, T., Lu, H., Yu, M., Li, J., Pang, R., & Ma, X. (2022). Functional Characterization and Whole-Genome Analysis of an Aflatoxin-Degrading Rhodococcus pyridinivorans Strain. Biology, 11(5), 774. https://doi.org/10.3390/biology11050774






