Genome-Directed Cell Nucleus Assembly
Abstract
:Simple Summary
Abstract
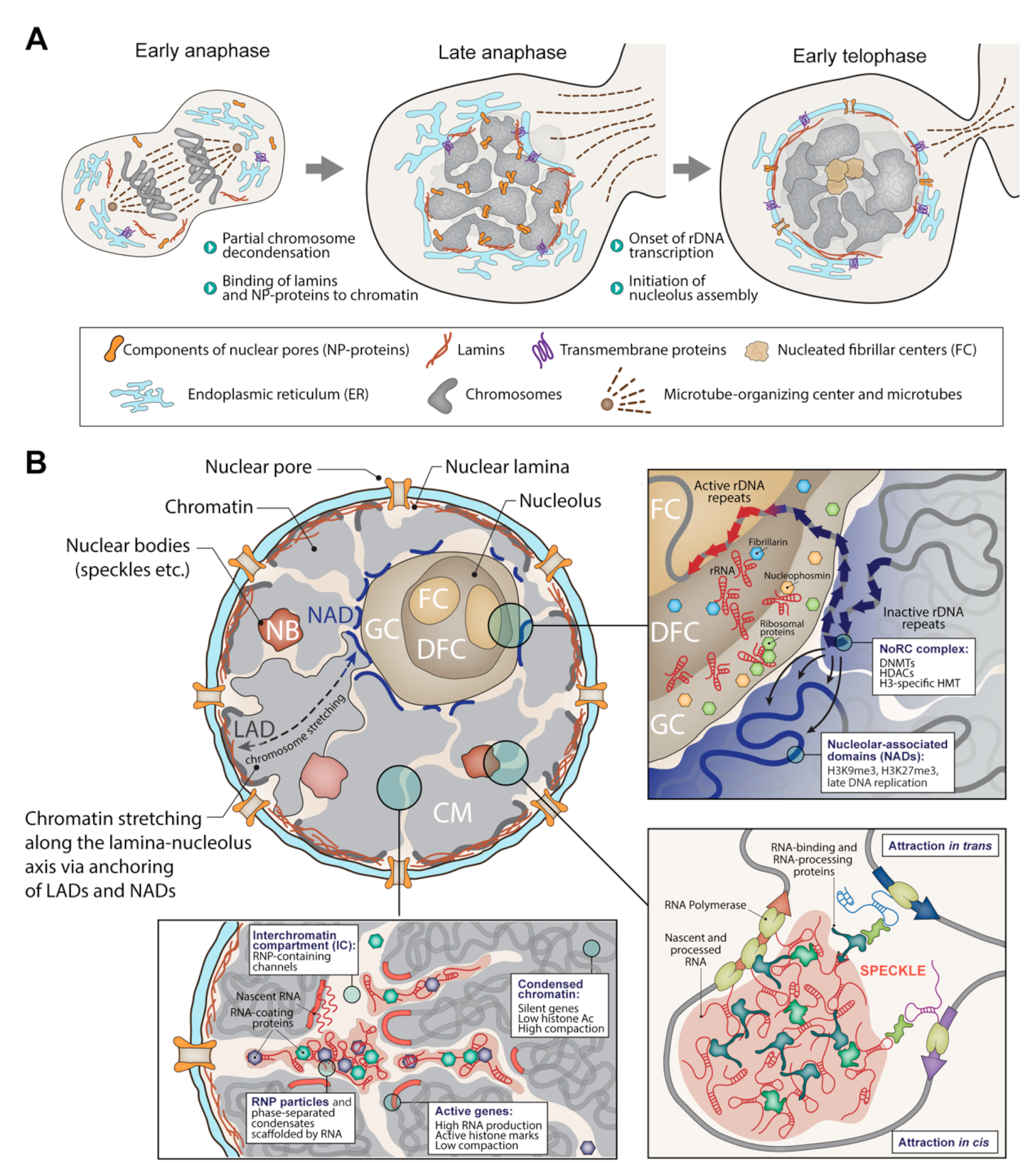
1. The Folded Genome Instructs the Assembly and Determines the Cell Nucleus’s Shape and Rigidity
2. Active and Inactive Neighborhoods in the Cell Nucleus
3. Transcription-Coupled Assembly of Functional Nuclear Compartments
3.1. The Nucleolus as a Multicomponent Phase Condensate
3.2. Liquid Condensates Containing RNA Polymerase II
3.3. Liquid-Phase Condensates Containing Enzymes Involved in Primary Transcript Processing
4. Modulation of Activity and Spatial Organization of the Genome through Mechanical Stress
5. Concluding Remarks
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Guttinger, S.; Laurell, E.; Kutay, U. Orchestrating nuclear envelope disassembly and reassembly during mitosis. Nat. Rev. Mol. Cell Biol. 2009, 10, 178–191. [Google Scholar] [CrossRef] [PubMed]
- Van Schaik, T.; Vos, M.; Peric-Hupkes, D.; Hn Celie, P.; van Steensel, B. Cell cycle dynamics of lamina-associated DNA. EMBO Rep. 2020, 21, e50636. [Google Scholar] [CrossRef] [PubMed]
- Wandke, C.; Kutay, U. Enclosing chromatin: Reassembly of the nucleus after open mitosis. Cell 2013, 152, 1222–1225. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lammerding, J.; Fong, L.G.; Ji, J.Y.; Reue, K.; Stewart, C.L.; Young, S.G.; Lee, R.T. Lamins A and C but not lamin B1 regulate nuclear mechanics. J. Biol. Chem. 2006, 281, 25768–25780. [Google Scholar] [CrossRef] [Green Version]
- Stephens, A.D.; Banigan, E.J.; Adam, S.A.; Goldman, R.D.; Marko, J.F. Chromatin and lamin A determine two different mechanical response regimes of the cell nucleus. Mol. Biol. Cell 2017, 28, 1984–1996. [Google Scholar] [CrossRef]
- Stephens, A.D.; Banigan, E.J.; Marko, J.F. Chromatin’s physical properties shape the nucleus and its functions. Curr. Opin. Cell Biol. 2019, 58, 76–84. [Google Scholar] [CrossRef]
- Stephens, A.D.; Liu, P.Z.; Banigan, E.J.; Almassalha, L.M.; Backman, V.; Adam, S.A.; Goldman, R.D.; Marko, J.F. Chromatin histone modifications and rigidity affect nuclear morphology independent of lamins. Mol. Biol. Cell 2018, 29, 220–233. [Google Scholar] [CrossRef]
- Nava, M.M.; Miroshnikova, Y.A.; Biggs, L.C.; Whitefield, D.B.; Metge, F.; Boucas, J.; Vihinen, H.; Jokitalo, E.; Li, X.; Garcia Arcos, J.M.; et al. Heterochromatin-Driven Nuclear Softening Protects the Genome against Mechanical Stress-Induced Damage. Cell 2020, 181, 800–817.e822. [Google Scholar] [CrossRef]
- Worman, H.J.; Bonne, G. “Laminopathies”: A wide spectrum of human diseases. Exp. Cell Res. 2007, 313, 2121–2133. [Google Scholar] [CrossRef] [Green Version]
- Worman, H.J.; Ostlund, C.; Wang, Y. Diseases of the nuclear envelope. Cold Spring Harb. Perspect. Biol. 2010, 2, a000760. [Google Scholar] [CrossRef]
- Ho, R.; Hegele, R.A. Complex effects of laminopathy mutations on nuclear structure and function. Clin Genet. 2019, 95, 199–209. [Google Scholar] [CrossRef]
- Booth, E.A.; Spagnol, S.T.; Alcoser, T.A.; Dahl, K.N. Nuclear stiffening and chromatin softening with progerin expression leads to an attenuated nuclear response to force. Soft. Matter. 2015, 11, 6412–6418. [Google Scholar] [CrossRef] [PubMed]
- McCord, R.P.; Nazario-Toole, A.; Zhang, H.; Chines, P.S.; Zhan, Y.; Erdos, M.R.; Collins, F.S.; Dekker, J.; Cao, K. Correlated alterations in genome organization, histone methylation, and DNA-lamin A/C interactions in Hutchinson-Gilford progeria syndrome. Genome Res. 2013, 23, 260–269. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shumaker, D.K.; Dechat, T.; Kohlmaier, A.; Adam, S.A.; Bozovsky, M.R.; Erdos, M.R.; Eriksson, M.; Goldman, A.E.; Khuon, S.; Collins, F.S.; et al. Mutant nuclear lamin A leads to progressive alterations of epigenetic control in premature aging. Proc. Natl. Acad. Sci. USA 2006, 103, 8703–8708. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cremer, T.; Cremer, M.; Hubner, B.; Silahtaroglu, A.; Hendzel, M.; Lanctot, C.; Strickfaden, H.; Cremer, C. The Interchromatin Compartment Participates in the Structural and Functional Organization of the Cell Nucleus. Bioessays 2020, 42, e1900132. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cremer, T.; Kreth, G.; Koester, H.; Fink, R.H.; Heintzmann, R.; Cremer, M.; Solovei, I.; Zink, D.; Cremer, C. Chromosome territories, interchromatin domain compartment, and nuclear matrix: An integrated view of the functional nuclear architecture. Crit. Rev. Eukaryot. Gene Expr. 2000, 10, 179–212. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Amzerin, M.; Layachi, M.; Bazine, A.; Aassab, R.; Arifi, S.; Benbrahim, Z.; Khmamouche, M.R.; Kairouani, M.; Raiss, H.; Majid, N.; et al. Cancer in Moroccan elderly: The first multicenter transverse study exploring the sociodemographic characteristics, clinical profile and quality of life of elderly Moroccan cancer patients. BMC Cancer 2020, 20, 983. [Google Scholar] [CrossRef]
- Strom, A.R.; Brangwynne, C.P. The liquid nucleome—Phase transitions in the nucleus at a glance. J. Cell Sci. 2019, 132, jcs235093. [Google Scholar] [CrossRef] [Green Version]
- Cremer, T.; Cremer, M.; Cremer, C. The 4D Nucleome: Genome Compartmentalization in an Evolutionary Context. Biochemistry 2018, 83, 313–325. [Google Scholar] [CrossRef]
- Razin, S.V.; Gavrilov, A.A. Non-coding RNAs in chromatin folding and nuclear organization. Cell Mol. Life Sci. 2021, 78, 5489–5504. [Google Scholar] [CrossRef]
- Creamer, K.M.; Kolpa, H.J.; Lawrence, J.B. Nascent RNA scaffolds contribute to chromosome territory architecture and counter chromatin compaction. Mol. Cell 2021, 81, 3509–3525.e3505. [Google Scholar] [CrossRef] [PubMed]
- Padeken, J.; Heun, P. Nucleolus and nuclear periphery: Velcro for heterochromatin. Curr. Opin. Cell Biol. 2014, 28, 54–60. [Google Scholar] [CrossRef] [PubMed]
- Bizhanova, A.; Kaufman, P.D. Close to the edge: Heterochromatin at the nucleolar and nuclear peripheries. Biochim. Biophys. Acta Gene Regul. Mech. 2021, 1864, 194666. [Google Scholar] [CrossRef] [PubMed]
- Zhao, R.; Bodnar, M.S.; Spector, D.L. Nuclear neighborhoods and gene expression. Curr. Opin. Genet. Dev. 2009, 19, 172–179. [Google Scholar] [CrossRef] [Green Version]
- Chen, Y.; Zhang, Y.; Wang, Y.; Zhang, L.; Brinkman, E.K.; Adam, S.A.; Goldman, R.; van Steensel, B.; Ma, J.; Belmont, A.S. Mapping 3D genome organization relative to nuclear compartments using TSA-Seq as a cytological ruler. J. Cell Biol. 2018, 217, 4025–4048. [Google Scholar] [CrossRef] [Green Version]
- Quinodoz, S.A.; Jachowicz, J.W.; Bhat, P.; Ollikainen, N.; Banerjee, A.K.; Goronzy, I.N.; Blanco, M.R.; Chovanec, P.; Chow, A.; Markaki, Y.; et al. RNA promotes the formation of spatial compartments in the nucleus. Cell 2021, 184, 5775–5790.e5730. [Google Scholar] [CrossRef]
- Quinodoz, S.A.; Ollikainen, N.; Tabak, B.; Palla, A.; Schmidt, J.M.; Detmar, E.; Lai, M.M.; Shishkin, A.A.; Bhat, P.; Takei, Y.; et al. Higher-Order Inter-chromosomal Hubs Shape 3D Genome Organization in the Nucleus. Cell 2018, 174, 744–757.e724. [Google Scholar] [CrossRef] [Green Version]
- Chen, Y.; Belmont, A.S. Genome organization around nuclear speckles. Curr. Opin. Genet. Dev. 2019, 55, 91–99. [Google Scholar] [CrossRef]
- Lu, J.Y.; Chang, L.; Li, T.; Wang, T.; Yin, Y.; Zhan, G.; Han, X.; Zhang, K.; Tao, Y.; Percharde, M.; et al. Homotypic clustering of L1 and B1/Alu repeats compartmentalizes the 3D genome. Cell Res. 2021, 31, 613–630. [Google Scholar] [CrossRef]
- Lu, J.Y.; Shao, W.; Chang, L.; Yin, Y.; Li, T.; Zhang, H.; Hong, Y.; Percharde, M.; Guo, L.; Wu, Z.; et al. Genomic Repeats Categorize Genes with Distinct Functions for Orchestrated Regulation. Cell Rep. 2020, 30, 3296–3311.e3295. [Google Scholar] [CrossRef] [Green Version]
- Guetg, C.; Santoro, R. Formation of nuclear heterochromatin: The nucleolar point of view. Epigenetics 2012, 7, 811–814. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Santoro, R.; Li, J.; Grummt, I. The nucleolar remodeling complex NoRC mediates heterochromatin formation and silencing of ribosomal gene transcription. Nat. Genet. 2002, 32, 393–396. [Google Scholar] [CrossRef] [PubMed]
- Santoro, R.; Grummt, I. Epigenetic mechanism of rRNA gene silencing: Temporal order of NoRC-mediated histone modification, chromatin remodeling, and DNA methylation. Mol. Cell Biol. 2005, 25, 2539–2546. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Guetg, C.; Lienemann, P.; Sirri, V.; Grummt, I.; Hernandez-Verdun, D.; Hottiger, M.O.; Fussenegger, M.; Santoro, R. The NoRC complex mediates the heterochromatin formation and stability of silent rRNA genes and centromeric repeats. EMBO J. 2010, 29, 2135–2146. [Google Scholar] [CrossRef]
- Lafontaine, D.L.J.; Riback, J.A.; Bascetin, R.; Brangwynne, C.P. The nucleolus as a multiphase liquid condensate. Nat. Rev. Mol. Cell Biol. 2021, 22, 165–182. [Google Scholar] [CrossRef] [PubMed]
- Iarovaia, O.V.; Minina, E.P.; Sheval, E.V.; Onichtchouk, D.; Dokudovskaya, S.; Razin, S.V.; Vassetzky, Y.S. Nucleolus: A Central Hub for Nuclear Functions. Trends Cell Biol. 2019, 29, 647–659. [Google Scholar] [CrossRef]
- Nemeth, A.; Conesa, A.; Santoyo-Lopez, J.; Medina, I.; Montaner, D.; Peterfia, B.; Solovei, I.; Cremer, T.; Dopazo, J.; Langst, G. Initial genomics of the human nucleolus. PLoS Genet. 2010, 6, e1000889. [Google Scholar] [CrossRef] [Green Version]
- Van Koningsbruggen, S.; Gierlinski, M.; Schofield, P.; Martin, D.; Barton, G.J.; Ariyurek, Y.; den Dunnen, J.T.; Lamond, A.I. High-resolution whole-genome sequencing reveals that specific chromatin domains from most human chromosomes associate with nucleoli. Mol. Biol. Cell 2010, 21, 3735–3748. [Google Scholar] [CrossRef] [Green Version]
- Vertii, A.; Ou, J.; Yu, J.; Yan, A.; Pages, H.; Liu, H.; Zhu, L.J.; Kaufman, P.D. Two contrasting classes of nucleolus-associated domains in mouse fibroblast heterochromatin. Genome Res. 2019, 29, 1235–1249. [Google Scholar] [CrossRef] [Green Version]
- Ragoczy, T.; Telling, A.; Scalzo, D.; Kooperberg, C.; Groudine, M. Functional redundancy in the nuclear compartmentalization of the late-replicating genome. Nucleus 2014, 5, 626–635. [Google Scholar] [CrossRef] [Green Version]
- Jackson, D.A.; Hassan, A.B.; Errington, R.J.; Cook, P.R. Visualization of focal sites of transcription within human nuclei. Embo. J. 1993, 12, 1059–1065. [Google Scholar] [CrossRef] [PubMed]
- Iborra, F.J.; Pombo, A.; Jackson, D.A.; Cook, P.R. Active RNA polymerases are localized within discrete transcription “factories” in human nuclei. J. Cell Sci. 1996, 109, 1427–1436. [Google Scholar] [CrossRef] [PubMed]
- Iborra, F.J.; Pombo, A.; McManus, J.; Jackson, D.A.; Cook, P.R. The topology of transcription by immobilized polymerases. Exp. Cell Res. 1996, 229, 167–173. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cho, W.K.; Spille, J.H.; Hecht, M.; Lee, C.; Li, C.; Grube, V.; Cisse, I.I. Mediator and RNA polymerase II clusters associate in transcription-dependent condensates. Science 2018, 361, 412–415. [Google Scholar] [CrossRef] [Green Version]
- Wei, M.; Fan, X.; Ding, M.; Li, R.; Shao, S.; Hou, Y.; Meng, S.; Tang, F.; Li, C.; Sun, Y. Nuclear actin regulates inducible transcription by enhancing RNA polymerase II clustering. Sci. Adv. 2020, 6, eaay6515. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hari-Gupta, Y.; Fili, N.; dos Santos, Á.; Cook, A.W.; Gough, R.E.; Reed, H.C.W.; Wang, L.; Aaron, J.; Venit, T.; Wait, E.; et al. Myosin VI regulates the spatial organi-sation of mammalian transcription initiation. Nat. Commun. 2022, 13, 1346. [Google Scholar] [CrossRef]
- Shrinivas, K.; Sabari, B.R.; Coffey, E.L.; Klein, I.A.; Boija, A.; Zamudio, A.V.; Schuijers, J.; Hannett, N.M.; Sharp, P.A.; Young, R.A.; et al. Enhancer Features that Drive Formation of Transcriptional Condensates. Mol. Cell 2019, 75, 549–561.e547. [Google Scholar] [CrossRef]
- Sabari, B.R.; Dall’Agnese, A.; Boija, A.; Klein, I.A.; Coffey, E.L.; Shrinivas, K.; Abraham, B.J.; Hannett, N.M.; Zamudio, A.V.; Manteiga, J.C.; et al. Coactivator condensation at super-enhancers links phase separation and gene control. Science 2018, 361. [Google Scholar] [CrossRef] [Green Version]
- Sabari, B.R.; Dall’Agnese, A.; Young, R.A. Biomolecular Condensates in the Nucleus. Trends Biochem. Sci. 2020, 45, 961–977. [Google Scholar] [CrossRef]
- Arnold, P.R.; Wells, A.D.; Li, X.C. Diversity and Emerging Roles of Enhancer RNA in Regulation of Gene Expression and Cell Fate. Front. Cell Dev. Biol. 2019, 7, 377. [Google Scholar] [CrossRef] [Green Version]
- Lee, J.H.; Wang, R.; Xiong, F.; Krakowiak, J.; Liao, Z.; Nguyen, P.T.; Moroz-Omori, E.V.; Shao, J.; Zhu, X.; Bolt, M.J.; et al. Enhancer RNA m6A methylation facilitates transcriptional condensate formation and gene activation. Mol. Cell 2021, 81, 3368–3385.e3369. [Google Scholar] [CrossRef] [PubMed]
- Pancholi, A.; Klingberg, T.; Zhang, W.; Prizak, R.; Mamontova, I.; Noa, A.; Sobucki, M.; Kobitski, A.Y.; Nienhaus, G.U.; Zaburdaev, V.; et al. RNA polymerase II clusters form in line with surface condensation on regulatory chromatin. Mol. Syst. Biol. 2021, 17, e10272. [Google Scholar] [CrossRef] [PubMed]
- Henninger, J.E.; Oksuz, O.; Shrinivas, K.; Sagi, I.; LeRoy, G.; Zheng, M.M.; Andrews, J.O.; Zamudio, A.V.; Lazaris, C.; Hannett, N.M.; et al. RNA-Mediated Feedback Control of Transcriptional Condensates. Cell 2021, 184, 207–225.e224. [Google Scholar] [CrossRef] [PubMed]
- Sharp, P.A.; Chakraborty, A.K.; Henninger, J.E.; Young, R.A. RNA in formation and regulation of transcriptional condensates. RNA 2022, 28, 52–57. [Google Scholar] [CrossRef]
- Ilik, I.A.; Aktas, T. Nuclear speckles: Dynamic hubs of gene expression regulation. FEBS J. 2021. [Google Scholar] [CrossRef] [PubMed]
- Smith, K.P.; Hall, L.L.; Lawrence, J.B. Nuclear hubs built on RNAs and clustered organization of the genome. Curr. Opin. Cell Biol. 2020, 64, 67–76. [Google Scholar] [CrossRef]
- Chalut, K.J.; Hopfler, M.; Lautenschlager, F.; Boyde, L.; Chan, C.J.; Ekpenyong, A.; Martinez-Arias, A.; Guck, J. Chromatin decondensation and nuclear softening accompany Nanog downregulation in embryonic stem cells. Biophys. J. 2012, 103, 2060–2070. [Google Scholar] [CrossRef] [Green Version]
- Le, H.Q.; Ghatak, S.; Yeung, C.Y.; Tellkamp, F.; Gunschmann, C.; Dieterich, C.; Yeroslaviz, A.; Habermann, B.; Pombo, A.; Niessen, C.M.; et al. Mechanical regulation of transcription controls Polycomb-mediated gene silencing during lineage commitment. Nat. Cell Biol. 2016, 18, 864–875. [Google Scholar] [CrossRef]
- Stephens, A.D.; Liu, P.Z.; Kandula, V.; Chen, H.; Almassalha, L.M.; Herman, C.; Backman, V.; O’Halloran, T.; Adam, S.A.; Goldman, R.D.; et al. Physicochemical mechanotransduction alters nuclear shape and mechanics via heterochromatin formation. Mol. Biol. Cell 2019, 30, 2320–2330. [Google Scholar] [CrossRef]
- Stevens, T.J.; Lando, D.; Basu, S.; Atkinson, L.P.; Cao, Y.; Lee, S.F.; Leeb, M.; Wohlfahrt, K.J.; Boucher, W.; O’Shaughnessy-Kirwan, A.; et al. 3D structures of individual mammalian genomes studied by single-cell Hi-C. Nature 2017, 544, 59–64. [Google Scholar] [CrossRef] [Green Version]
- Flyamer, I.M.; Gassler, J.; Imakaev, M.; Brandao, H.B.; Ulianov, S.V.; Abdennur, N.; Razin, S.V.; Mirny, L.A.; Tachibana-Konwalski, K. Single-nucleus Hi-C reveals unique chromatin reorganization at oocyte-to-zygote transition. Nature 2017, 544, 110–114. [Google Scholar] [CrossRef] [Green Version]
- Ulianov, S.V.; Zakharova, V.V.; Galitsyna, A.A.; Kos, P.I.; Polovnikov, K.E.; Flyamer, I.M.; Mikhaleva, E.A.; Khrameeva, E.E.; Germini, D.; Logacheva, M.D.; et al. Order and stochasticity in the folding of individual Drosophila genomes. Nat. Commun. 2021, 12, 41. [Google Scholar] [CrossRef] [PubMed]
- Kantidze, O.L.; Razin, S.V. Weak interactions in higher-order chromatin organization. Nucleic Acids Res. 2020, 48, 4614–4626. [Google Scholar] [CrossRef]
- Booth-Gauthier, E.A.; Alcoser, T.A.; Yang, G.; Dahl, K.N. Force-induced changes in subnuclear movement and rheology. Biophys. J. 2012, 103, 2423–2431. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gavrilov, A.A.; Gushchanskaya, E.S.; Strelkova, O.; Zhironkina, O.; Kireev, I.I.; Iarovaia, O.V.; Razin, S.V. Disclosure of a structural milieu for the proximity ligation reveals the elusive nature of an active chromatin hub. Nucleic Acids Res. 2013, 41, 3563–3575. [Google Scholar] [CrossRef] [PubMed]
- Ulianov, S.V.; Gavrilov, A.A.; Razin, S.V. Nuclear compartments, genome folding, and enhancer-promoter communication. Int. Rev. Cell Mol. Biol. 2015, 315, 183–244. [Google Scholar] [CrossRef]
- Fedorchak, G.R.; Kaminski, A.; Lammerding, J. Cellular mechanosensing: Getting to the nucleus of it all. Prog. Biophys. Mol. Biol. 2014, 115, 76–92. [Google Scholar] [CrossRef] [Green Version]
- Thomas, C.H.; Collier, J.H.; Sfeir, C.S.; Healy, K.E. Engineering gene expression and protein synthesis by modulation of nuclear shape. Proc. Natl. Acad. Sci. USA 2002, 99, 1972–1977. [Google Scholar] [CrossRef] [Green Version]
- Tajik, A.; Zhang, Y.; Wei, F.; Sun, J.; Jia, Q.; Zhou, W.; Singh, R.; Khanna, N.; Belmont, A.S.; Wang, N. Transcription upregulation via force-induced direct stretching of chromatin. Nat. Mater. 2016, 15, 1287–1296. [Google Scholar] [CrossRef] [Green Version]
- Vahlensieck, C.; Thiel, C.S.; Zhang, Y.; Huge, A.; Ullrich, O. Gravitational Force-Induced 3D Chromosomal Conformational Changes Are Associated with Rapid Transcriptional Response in Human T Cells. Int. J. Mol. Sci. 2021, 22, 9426. [Google Scholar] [CrossRef]
- Uhler, C.; Shivashankar, G.V. Regulation of genome organization and gene expression by nuclear mechanotransduction. Nat. Rev. Mol. Cell Biol. 2017, 18, 717–727. [Google Scholar] [CrossRef]
- Szczesny, S.E.; Mauck, R.L. The Nuclear Option: Evidence Implicating the Cell Nucleus in Mechanotransduction. J. Biomech. Eng. 2017, 139, 021006. [Google Scholar] [CrossRef] [PubMed]
- Kassianidou, E.; Kalita, J.; Lim, R.Y.H. The role of nucleocytoplasmic transport in mechanotransduction. Exp. Cell. Res. 2019, 377, 86–93. [Google Scholar] [CrossRef] [PubMed]
- Elosegui-Artola, A.; Andreu, I.; Beedle, A.E.M.; Lezamiz, A.; Uroz, M.; Kosmalska, A.J.; Oria, R.; Kechagia, J.Z.; Rico-Lastres, P.; Le Roux, A.L.; et al. Force Triggers YAP Nuclear Entry by Regulating Transport across Nuclear Pores. Cell 2017, 171, 1397–1410.e1314. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Nagarajan, M.; Uhler, C.; Shivashankar, G.V. Orientation and repositioning of chromosomes correlate with cell geometry-dependent gene expression. Mol. Biol. Cell 2017, 28, 1997–2009. [Google Scholar] [CrossRef]
- Wei, F.; Xu, X.; Zhang, C.; Liao, Y.; Ji, B.; Wang, N. Stress fiber anisotropy contributes to force-mode dependent chromatin stretching and gene upregulation in living cells. Nat. Commun. 2020, 11, 4902. [Google Scholar] [CrossRef]
- Sun, J.; Chen, J.; Amar, K.; Wu, Y.; Jiang, M.; Wang, N. LAP2beta transmits force to upregulate genes via chromatin domain stretching but not compression. Acta Biomater. 2021, in press. [Google Scholar] [CrossRef]
- Berezney, R.; Coffey, D.S. Nuclear matrix: Isolation and characterization of a framework structure from rat liver nuclei. J. Cell. Biol. 1977, 73, 616–637. [Google Scholar] [CrossRef] [Green Version]
- Berezney, R.; Mortillaro, M.J.; Ma, H.; Wei, X.; Samarabandu, J. The nuclear matrix: A structural milieu for genomic function. Int. Rev. Cytol. 1995, 162A, 1–65. [Google Scholar]
- Ma, H.; Siegel, A.J.; Berezney, R. Association of chromosome territories with the nuclear matrix. Disruption of human chromosome territories correlates with the release of a subset of nuclear matrix proteins. J. Cell Biol. 1999, 146, 531–542. [Google Scholar] [CrossRef]
- Razin, S.V.; Iarovaia, O.V.; Vassetzky, Y.S. A requiem to the nuclear matrix: From a controversial concept to 3D organization of the nucleus. Chromosoma 2014, 123, 217–224. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Razin, S.V.; Ulianov, S.V. Genome-Directed Cell Nucleus Assembly. Biology 2022, 11, 708. https://doi.org/10.3390/biology11050708
Razin SV, Ulianov SV. Genome-Directed Cell Nucleus Assembly. Biology. 2022; 11(5):708. https://doi.org/10.3390/biology11050708
Chicago/Turabian StyleRazin, Sergey V., and Sergey V. Ulianov. 2022. "Genome-Directed Cell Nucleus Assembly" Biology 11, no. 5: 708. https://doi.org/10.3390/biology11050708
APA StyleRazin, S. V., & Ulianov, S. V. (2022). Genome-Directed Cell Nucleus Assembly. Biology, 11(5), 708. https://doi.org/10.3390/biology11050708





