Sperm Phosphoproteome: Unraveling Male Infertility
Abstract
Simple Summary
Abstract
1. Introduction
2. Proteomics and Sperm Physiology
3. Phosphoproteomics Technique in Male Fertility
3.1. Phosphorylation as Post-Translational Modification of Sperm Proteins
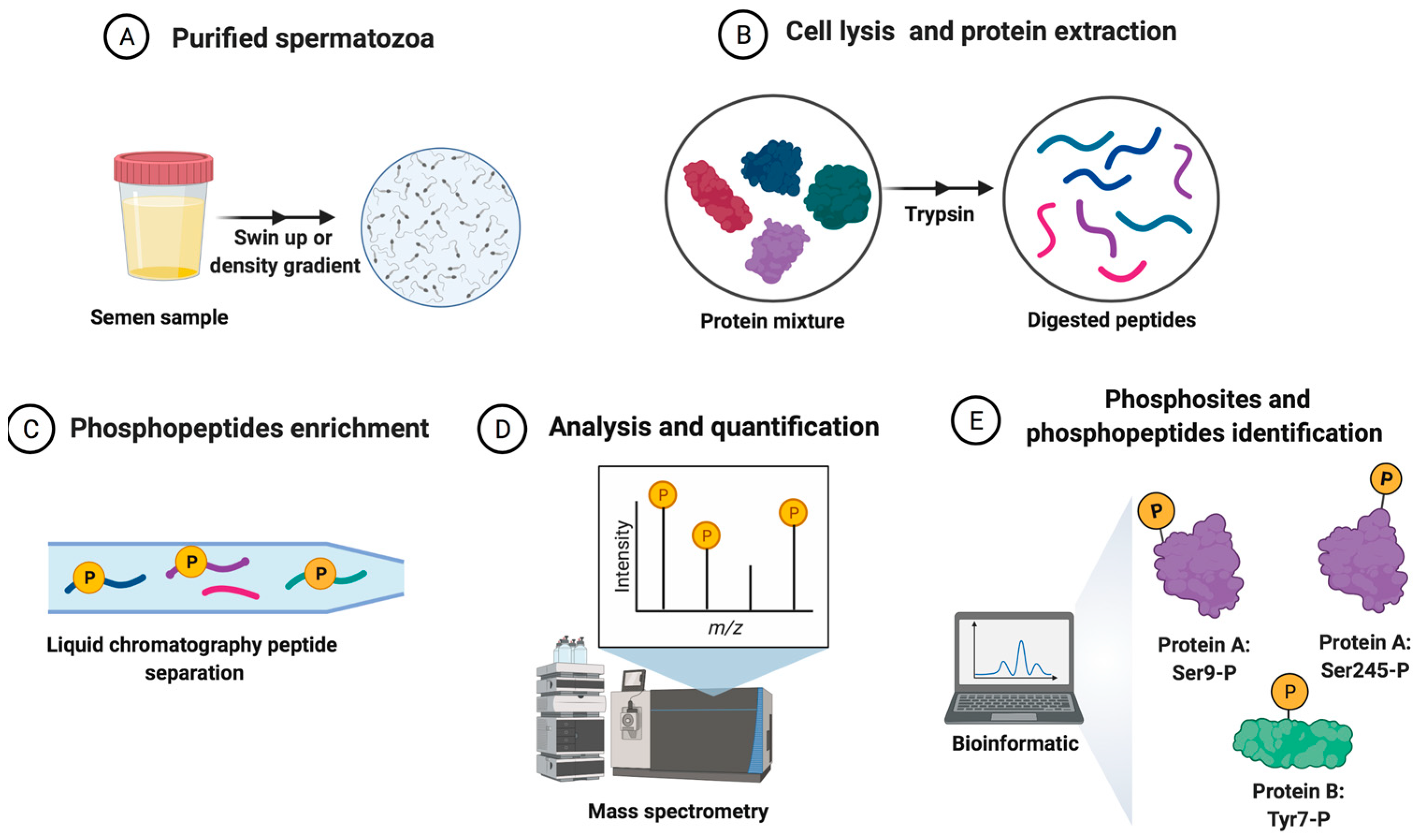
3.2. Phosphoproteomics Technique
4. Phosphoproteomics and Spermatogenesis
5. Phosphoproteomics and Sperm Motility
6. Phosphoproteomics and Sperm Capacitation
7. Outlook
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Patents
References
- Varner, D.D. Odyssey of the spermatozoon. Asian J. Androl. 2015, 17, 522–528. [Google Scholar] [CrossRef] [PubMed]
- Hetherington, L.; Schneider, E.K.; Scott, C.; DeKretser, D.; Muller, C.H.; Hondermarck, H.; Velkov, T.; Baker, M.A. Deficiency in Outer Dense Fiber 1 Is a Marker and Potential Driver of Idiopathic Male Infertility. Mol. Cell. Proteom. 2016, 15, 3685–3693. [Google Scholar] [CrossRef]
- World Human Organization. WHO Laboratory Manual for the Examination and Processing of Human Semen, 5th ed.; WHO Press: Geneva, Switzerland, 2010; p. 287. [Google Scholar]
- Esteves, S.C.; Miyaoka, R.; Agarwal, A. An update on the clinical assessment of the infertile male. Clinics 2011, 66, 691–700. [Google Scholar] [CrossRef] [PubMed]
- Eliasson, R. Semen analysis with regard to sperm number, sperm morphology and functional aspects. Asian J. Androl. 2010, 12, 26–32. [Google Scholar] [CrossRef] [PubMed]
- Sánchez, V.; Wistuba, J.; Mallidis, C. Semen analysis: Update on clinical value, current needs and future perspectives. Reproduction 2013, 146, R249–R258. [Google Scholar] [CrossRef]
- Wang, C.; Swerdloff, R.S. Limitations of semen analysis as a test of male fertility and anticipated needs from newer tests. Fertil. Steril. 2014, 102, 1502–1507. [Google Scholar] [CrossRef]
- Papillon-Smith, J.; Baker, S.E.; Agbo, C.; Dahan, M.H. Pregnancy rates with intrauterine insemination: Comparing 1999 and 2010 World Health Organization semen analysis norms. Reprod. Biomed. Online 2015, 30, 392–400. [Google Scholar] [CrossRef]
- Stern, J.E.; Luke, B.; Hornstein, M.D.; Cabral, H.; Gopal, D.; Diop, H.; Kotelchuck, M. The effect of father’s age in fertile, subfertile, and assisted reproductive technology pregnancies: A population based cohort study. J. Assist. Reprod. Genet. 2014, 31, 1437–1444. [Google Scholar] [CrossRef]
- Santiago, J.; Silva, J.V.; Howl, J.; Santos, M.A.S.; Fardilha, M. All you need to know about sperm RNAs. Hum. Reprod. Update 2021, 28, 67–91. [Google Scholar] [CrossRef]
- Samanta, L.; Swain, N.; Ayaz, A.; Venugopal, V.; Agarwal, A. Post-Translational Modifications in sperm Proteome: The Chemistry of Proteome diversifications in the Pathophysiology of male factor infertility. Biochim. Biophys. Acta 2016, 1860, 1450–1465. [Google Scholar] [CrossRef]
- Cheng, Y.M.; Hu, X.N.; Peng, Z.; Pan, T.T.; Wang, F.; Chen, H.Y.; Chen, W.Q.; Zhang, Y.; Zeng, X.H.; Luo, T. Lysine glutarylation in human sperm is associated with progressive motility. Hum. Reprod. 2019, 34, 1186–1194. [Google Scholar] [CrossRef]
- Qi, Q.; Pan, H.; Jiang, N.; Zhang, M.; Sun, S.; Wan, X.; Zhang, F.; Zhang, L.; Diao, H.; Wang, J.; et al. A novel posttranslational modification of histone, H3 S-sulfhydration, is down-regulated in asthenozoospermic sperm. J. Assist. Reprod. Genet. 2021, 38, 3175–3193. [Google Scholar] [CrossRef]
- Marchiani, S.; Tamburrino, L.; Giuliano, L.; Nosi, D.; Sarli, V.; Gandini, L.; Piomboni, P.; Belmonte, G.; Forti, G.; Baldi, E.; et al. Sumo1-ylation of human spermatozoa and its relationship with semen quality. Int. J. Androl. 2011, 34, 581–593. [Google Scholar] [CrossRef] [PubMed]
- Cheng, Y.M.; Peng, Z.; Chen, H.Y.; Pan, T.T.; Hu, X.N.; Wang, F.; Luo, T. Posttranslational lysine 2-hydroxyisobutyrylation of human sperm tail proteins affects motility. Hum. Reprod. 2020, 35, 494–503. [Google Scholar] [CrossRef]
- Sun, G.; Jiang, M.; Zhou, T.; Guo, Y.; Cui, Y.; Guo, X.; Sha, J. Insights into the lysine acetylproteome of human sperm. J. Proteom. 2014, 109, 199–211. [Google Scholar] [CrossRef] [PubMed]
- Porambo, J.R.; Salicioni, A.M.; Visconti, P.E.; Platt, M.D. Sperm phosphoproteomics: Historical perspectives and current methodologies. Expert Rev. Proteom. 2012, 9, 533–548. [Google Scholar] [CrossRef][Green Version]
- Chan, C.C.; Shui, H.A.; Wu, C.H.; Wang, C.Y.; Sun, G.H.; Chen, H.M.; Wu, G.J. Motility and protein phosphorylation in healthy and asthenozoospermic sperm. J. Proteome Res. 2009, 8, 5382–5386. [Google Scholar] [CrossRef] [PubMed]
- Barbonetti, A.; Vassallo, M.R.; Fortunato, D.; Francavilla, S.; Maccarrone, M.; Francavilla, F. Energetic metabolism and human sperm motility: Impact of CB₁ receptor activation. Endocrinology 2010, 151, 5882–5892. [Google Scholar] [CrossRef]
- Visconti, P.E.; Krapf, D.; de la Vega-Beltrán, J.L.; Acevedo, J.J.; Darszon, A. Ion channels, phosphorylation and mammalian sperm capacitation. Asian J. Androl. 2011, 13, 395405. [Google Scholar] [CrossRef]
- Castillo, J.; Knol, J.C.; Korver, C.M.; Piersma, S.R.; Pham, T.V.; de Goeij-de Haas, R.R.; van Pelt, A.M.M.; Jimenez, C.R.; Jansen, B.J.H. Human Testis Phosphoproteome Reveals Kinases as Potential Targets in Spermatogenesis and Testicular Cancer. Mol. Cell. Proteom. 2019, 18 (Suppl. S1), S132–S144. [Google Scholar] [CrossRef]
- Li, Y.; Cheng, Y.; Zhu, T.; Zhang, H.; Li, W.; Guo, Y.; Qi, Y.; Chen, X.; Zhang, J.; Sha, J.; et al. The Protein Phosphorylation Landscape of Mouse Spermatids during Spermiogenesis. Proteomics 2019, 19, e1900055. [Google Scholar] [CrossRef] [PubMed]
- Zhang, P.F.; Huang, Y.L.; Fu, Q.; He, W.T.; Xiao, K.; Zhang, M. Integrated analysis of phosphoproteome and ubiquitylome in epididymal sperm of buffalo (Bubalus bubalis). Mol. Reprod. Dev. 2021, 88, 15–33. [Google Scholar] [CrossRef] [PubMed]
- Ficarro, S.; Chertihin, O.; Westbrook, V.A.; White, F.; Jayes, F.; Kalab, P.; Marto, J.A.; Shabanowitz, J.; Herr, J.C.; Hunt, D.F.; et al. Phosphoproteome analysis of capacitated human sperm. Evidence of tyrosine phosphorylation of a kinase-anchoring protein 3 and valosin-containing protein/p97 during capacitation. J. Biol. Chem. 2003, 278, 11579–11589. [Google Scholar] [CrossRef]
- Bailey, J.L.; Tardif, S.; Dubé, C.; Beaulieu, M.; Reyes-Moreno, C.; Lefièvre, L.; Leclerc, P. Use of phosphoproteomics to study tyrosine kinase activity in capacitating boar sperm. Kinase activity and capacitation. Theriogenology 2005, 63, 599–614. [Google Scholar] [CrossRef] [PubMed]
- (Kota, V.; Dhople, V.M.; Shivaji, S. Tyrosine phosphoproteome of hamster spermatozoa: Role of glycerol-3-phosphate dehydrogenase 2 in sperm capacitation. Proteomics 2009, 9, 1809–1826. [Google Scholar] [CrossRef]
- Wang, J.; Qi, L.; Huang, S.; Zhou, T.; Guo, Y.; Wang, G.; Guo, X.; Zhou, Z.; Sha, J. Quantitative phosphoproteomics analysis reveals a key role of insulin growth factor 1 receptor (IGF1R) tyrosine kinase in human sperm capacitation. Mol. Cell. Proteom. 2015, 14, 1104–1112. [Google Scholar] [CrossRef]
- Syifa, N.; Yang, J.T.; Wu, C.S.; Lin, M.H.; Wu, W.L.; Lai, C.W.; Ku, S.H.; Liang, S.Y.; Hung, Y.C.; Chou, C.T.; et al. Phosphoproteomics and Bioinformatics Analyses Reveal Key Roles of GSK-3 and AKAP4 in Mouse Sperm Capacitation. Int. J. Mol. Sci. 2020, 21, 7283. [Google Scholar] [CrossRef]
- Parte, P.P.; Rao, P.; Redij, S.; Lobo, V.; D’Souza, S.J.; Gajbhiye, R.; Kulkarni, V. Sperm phosphoproteome profiling by ultra performance liquid chromatography followed by data independent analysis (LC-MS(E)) reveals altered proteomic signatures in asthenozoospermia. J. Proteom. 2012, 75, 5861–5871. [Google Scholar] [CrossRef]
- Urizar-Arenaza, I.; Osinalde, N.; Akimov, V.; Puglia, M.; Candenas, L.; Pinto, F.M.; Muñoa-Hoyos, I.; Gianzo, M.; Matorras, R.; Irazusta, J.; et al. Phosphoproteomic and Functional Analyses Reveal Sperm-specific Protein Changes Downstream of Kappa Opioid Receptor in Human Spermatozoa. Mol. Cell Proteom. 2019, 18 (Suppl. S1), S118–S131. [Google Scholar] [CrossRef]
- Martin-Hidalgo, D.; Serrano, R.; Zaragoza, C.; Garcia-Marin, L.J.; Bragado, M.J. Human sperm phosphoproteome reveals differential phosphoprotein signatures that regulate human sperm motility. J. Proteom. 2020, 215, 103654. [Google Scholar] [CrossRef]
- Wang, J.; Wang, M.; Hong, R.; Tang, S.; Xu, Y.; Zhao, X.; Zhou, T.; Wang, Z.; Huang, S. Quantitative phosphoproteomics reveals GSK3A substrate network is involved in the cryodamage of sperm motility. Biosci. Rep. 2021, 41, BSR20211326. [Google Scholar] [CrossRef] [PubMed]
- Intasqui, P.; Agarwal, A.; Sharma, R.; Samanta, L.; Bertolla, R.P. Towards the identification of reliable sperm biomarkers for male infertility: A sperm proteomic approach. Andrologia 2018, 50, e12919. [Google Scholar] [CrossRef] [PubMed]
- Nowicka-Bauer, K.; Malcher, A.; Włoczkowska, O.; Kamieniczna, M.; Olszewska, M.; Kurpisz, M.K. Evaluation of seminal plasma HSPA2 protein as a biomarker of human spermatogenesis status. Reprod. Biol. 2022, 22, 100597. [Google Scholar] [CrossRef] [PubMed]
- Netherton, J.; Ogle, R.A.; Hetherington, L.; Silva Balbin Villaverde, A.I.; Hondermarck, H.; Baker, M.A. Proteomic Analysis Reveals that Topoisomerase 2A is Associated with Defective Sperm Head Morphology. Mol. Cell. Proteom. 2020, 19, 444–455. [Google Scholar] [CrossRef]
- Pixton, K.L.; Deeks, E.D.; Flesch, F.M.; Moseley, F.L.; Björndahl, L.; Ashton, P.R.; Barratt, C.L.; Brewis, I.A. Sperm proteome mapping of a patient who experienced failed fertilization at IVF reveals altered expression of at least 20 proteins compared with fertile donors: Case report. Hum. Reprod. 2004, 19, 1438–1447. [Google Scholar] [CrossRef]
- Zhao, C.; Huo, R.; Wang, F.Q.; Lin, M.; Zhou, Z.M.; Sha, J.H. Identification of several proteins involved in regulation of sperm motility by proteomic analysis. Fertil. Steril. 2007, 87, 436–438. [Google Scholar] [CrossRef]
- Martínez-Heredia, J.; de Mateo, S.; Vidal-Taboada, J.M.; Ballescà, J.L.; Oliva, R. Identification of proteomic differences in asthenozoospermic sperm samples. Hum. Reprod. 2008, 23, 783–791. [Google Scholar] [CrossRef]
- Siva, A.B.; Kameshwari, D.B.; Singh, V.; Pavani, K.; Sundaram, C.S.; Rangaraj, N.; Deenadayal, M.; Shivaji, S. Proteomics-based study on asthenozoospermia: Differential expression of proteasome alpha complex. Mol. Hum. Reprod. 2010, 16, 452–462. [Google Scholar] [CrossRef]
- Thacker, S.; Yadav, S.P.; Sharma, R.K.; Kashou, A.; Willard, B.; Zhang, D.; Agarwal, A. Evaluation of sperm proteins in infertile men: A proteomic approach. Fertil. Steril. 2011, 95, 2745–2748. [Google Scholar] [CrossRef]
- Shen, S.; Wang, J.; Liang, J.; He, D. Comparative proteomic study between human normal motility sperm and idiopathic asthenozoospermia. World J. Urol. 2013, 31, 1395–1401. [Google Scholar] [CrossRef]
- Amaral, A.; Paiva, C.; Attardo Parrinello, C.; Estanyol, J.M.; Ballescà, J.L.; Ramalho- Santos, J.; Oliva, R. Identification of proteins involved in human sperm motility using highthroughput differential proteomics. J. Proteome Res. 2014, 13, 5670–5684. [Google Scholar] [CrossRef] [PubMed]
- Hashemitabar, M.; Sabbagh, S.; Orazizadeh, M.; Ghadiri, A.; Bahmanzadeh, M. A proteomic analysis on human sperm tail: Comparison between normozoospermia and asthenozoospermia. J. Assist. Reprod. Genet. 2015, 32, 853–863. [Google Scholar] [CrossRef] [PubMed]
- Saraswat, M.; Joenväärä, S.; Jain, T.; Tomar, A.K.; Sinha, A.; Singh, S.; Yadav, S.; Renkonen, R. Human Spermatozoa Quantitative Proteomic Signature Classifies Normo- and Asthenozoospermia. Mol. Cell. Proteom. 2017, 16, 57–72. [Google Scholar] [CrossRef] [PubMed]
- Moscatelli, N.; Lunetti, P.; Braccia, C.; Armirotti, A.; Pisanello, F.; De Vittorio, M.; Zara, V.; Ferramosca, A. Comparative Proteomic Analysis of Proteins Involved in Bioenergetics Pathways Associated with Human Sperm Motility. Int. J. Mol. Sci. 2019, 20, 3000. [Google Scholar] [CrossRef]
- Guo, Y.; Jiang, W.; Yu, W.; Niu, X.; Liu, F.; Zhou, T.; Zhang, H.; Li, Y.; Zhu, H.; Zhou, Z.; et al. Proteomics analysis of asthenozoospermia and identification of glucose-6-phosphate isomerase as an important enzyme for sperm motility. J. Proteom. 2019, 208, 103478. [Google Scholar] [CrossRef]
- Aitken, R.J.; Sutton, M.; Warner, P.; Richardson, D.W. Relationship between the movement characteristics of human spermatozoa and their ability to penetrate cervical mucus and zona-free hamster oocytes. J. Reprod. Fertil. 1985, 73, 441–449. [Google Scholar] [CrossRef]
- Curi, S.M.; Ariagno, J.I.; Chenlo, P.H.; Mendeluk, G.R.; Pugliese, M.N.; Sardi Segovia, L.M.; Repetto, H.E.; Blanco, A.M. Asthenozoospermia: Analysis of a large population. Arch. Androl. 2003, 49, 343–349. [Google Scholar] [CrossRef]
- Liu, X.; Teng, Z.; Wang, Z.; Zhu, P.; Song, Z.; Liu, F. Expressions of HSPA1L and HSPA9 are associated with poor sperm quality of low-motility spermatozoa in fertile men. Andrologia 2022, 54, e14321. [Google Scholar] [CrossRef]
- Meccariello, R.; Chianese, R.; Ciaramella, V.; Fasano, S.; Pierantoni, R. Molecular chaperones, cochaperones, and ubiquitination/deubiquitination system: Involvement in the production of high quality spermatozoa. Biomed. Res. Int. 2014, 2014, 561426. [Google Scholar] [CrossRef]
- Dun, M.D.; Aitken, R.J.; Nixon, B. The role of molecular chaperones in spermatogenesis and the post-testicular maturation of mammalian spermatozoa. Hum. Reprod. Update 2012, 18, 420–435. [Google Scholar] [CrossRef]
- Liu, X.X.; Shen, X.F.; Liu, F.J. Screening targeted testis-specific genes for molecular assessment of aberrant sperm quality. Mol. Med. Rep. 2016, 14, 1594–1600. [Google Scholar] [CrossRef] [PubMed]
- Nixon, B.; Bromfield, E.G.; Cui, J.; De Iuliis, G.N. Heat Shock Protein A2 (HSPA2): Regulatory Roles in Germ Cell Development and Sperm Function. Adv. Anat. Embryol. Cell Biol. 2017, 222, 67–93. [Google Scholar] [CrossRef] [PubMed]
- Radons, J. The human HSP70 family of chaperones: Where do we stand? Cell Stress Chaperones 2016, 21, 379–404. [Google Scholar] [CrossRef]
- Bracke, A.; Peeters, K.; Punjabi, U.; Hoogewijs, D.; Dewilde, S. A search for molecular mechanisms underlying male idiopathic infertility. Reprod. Biomed. Online 2018, 36, 327–339. [Google Scholar] [CrossRef]
- Légaré, C.; Droit, A.; Fournier, F.; Bourassa, S.; Force, A.; Cloutier, F.; Tremblay, R.; Sullivan, R. Investigation of male infertility using quantitative comparative proteomics. J. Proteome Res. 2014, 13, 5403–5414. [Google Scholar] [CrossRef]
- Ostrowski, L.E.; Andrews, K.; Potdar, P.; Matsuura, H.; Jetten, A.; Nettesheim, P. Cloning and characterization of KPL2, a novel gene induced during ciliogenesis of tracheal epithelial cells. Am. J. Respir. Cell Mol. Biol. 1999, 20, 675–683. [Google Scholar] [CrossRef] [PubMed]
- Tu, C.; Nie, H.; Meng, L.; Wang, W.; Li, H.; Yuan, S.; Cheng, D.; He, W.; Liu, G.; Du, J.; et al. Novel mutations in SPEF2 causing different defects between flagella and cilia bridge: The phenotypic link between MMAF and PCD. Hum. Genet. 2020, 139, 257–271. [Google Scholar] [CrossRef]
- Li, D.Y.; Yang, X.X.; Tu, C.F.; Wang, W.L.; Meng, L.L.; Lu, G.X.; Tan, Y.Q.; Zhang, Q.J.; Du, J. Sperm flagellar 2 (SPEF2) is essential for sperm flagellar assembly in humans. Asian J. Androl. 2021, 23, 1–8. [Google Scholar] [CrossRef]
- Lehti, M.S.; Sironen, A. Formation and function of sperm tail structures in association with sperm motility defects. Biol. Reprod. 2017, 97, 522–536. [Google Scholar] [CrossRef]
- Wang, S.; Wang, W.; Xu, Y.; Tang, M.; Fang, J.; Sun, H.; Sun, Y.; Gu, M.; Liu, Z.; Zhang, Z.; et al. Proteomic characteristics of human sperm cryopreservation. Proteomics 2014, 14, 298–310. [Google Scholar] [CrossRef]
- Fu, L.; An, Q.; Zhang, K.; Liu, Y.; Tong, Y.; Xu, J.; Zhou, F.; Wang, X.; Guo, Y.; Lu, W.; et al. Quantitative proteomic characterization of human sperm cryopreservation: Using data independent acquisition mass spectrometry. BMC Urol. 2019, 19, 133. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.C.; Peterson, S.E.; Loring, J.F. Protein post-translational modifications and regulation of pluripotency in human stem cells. Cell Res. 2014, 24, 143–160. [Google Scholar] [CrossRef] [PubMed]
- Khoury, G.A.; Baliban, R.C.; Floudas, C.A. Proteome-wide post-translational modification statistics: Frequency analysis and curation of the swiss-prot database. Sci. Rep. 2011, 1, 90. [Google Scholar] [CrossRef] [PubMed]
- Cohen, P. The role of protein phosphorylation in human health and disease. The Sir Hans Krebs Medal Lecture. Eur. J. Biochem. 2001, 268, 5001–5010. [Google Scholar] [CrossRef]
- Olsen, J.V.; Blagoev, B.; Gnad, F.; Macek, B.; Kumar, C.; Mortensen, P.; Mann, M. Global, in vivo, and site-specific phosphorylation dynamics in signaling networks. Cell 2006, 127, 635–648. [Google Scholar] [CrossRef]
- Adam, K.; Hunter, T. Histidine kinases and the missing phosphoproteome from prokaryotes to eukaryotes. Lab. Investig. 2018, 98, 233–247. [Google Scholar] [CrossRef]
- Beltrao, P.; Bork, P.; Krogan, N.J.; van Noort, V. Evolution and functional cross-talk of protein post-translational modifications. Mol. Syst. Biol. 2013, 9, 714. [Google Scholar] [CrossRef]
- Brohi, R.D.; Huo, L.J. Posttranslational Modifications in Spermatozoa and Effects on Male Fertility and Sperm Viability. OMICS 2017, 21, 245–256. [Google Scholar] [CrossRef]
- Low, T.Y.; Mohtar, M.A.; Lee, P.Y.; Omar, N.; Zhou, H.; Ye, M. Widening the Bottleneck of phosphoproteomics: Evolving strategies for Phosphopeptide enrichment. Mass Spectrom. Rev. 2021, 40, 309–333. [Google Scholar] [CrossRef]
- Amaral, A.; Castillo, J.; Ramalho-Santos, J.; Oliva, R. The combined human sperm proteome: Cellular pathways and implications for basic and clinical science. Hum. Reprod. Update 2014, 20, 40–62. [Google Scholar] [CrossRef]
- Larsen, M.R.; Thingholm, T.E.; Jensen, O.N.; Roepstorff, P.; Jørgensen, T.J. Highly selective enrichment of phosphorylated peptides from peptide mixtures using titanium dioxide microcolumns. Mol. Cell. Proteom. 2005, 4, 873–886. [Google Scholar] [CrossRef]
- Mitulovic, G.; Mechtler, K. HPLC techniques for proteomics analysis—A short overview of latest developments. Brief. Funct. Genom. Proteomic 2006, 5, 249–260. [Google Scholar] [CrossRef]
- Thingholm, T.E.; Larsen, M.R. The Use of Titanium Dioxide for Selective Enrichment of Phosphorylated Peptides. Methods Mol. Biol. 2016, 1355, 135–146. [Google Scholar] [CrossRef]
- Agarwal, A.; Panner Selvam, M.K.; Baskaran, S. Proteomic Analyses of Human Sperm Cells: Understanding the Role of Proteins and Molecular Pathways Affecting Male Reproductive Health. Int. J. Mol. Sci. 2020, 21, 1621. [Google Scholar] [CrossRef] [PubMed]
- Needham, E.J.; Parker, B.L.; Burykin, T.; James, D.E.; Humphrey, S.J. Illuminating the dark phosphoproteome. Sci. Signal. 2019, 12, 565. [Google Scholar] [CrossRef] [PubMed]
- Anand, S.; Samuel, M.; Ang, C.S.; Keerthikumar, S.; Mathivanan, S. Label-Based and Label-Free Strategies for Protein Quantitation. Methods Mol. Biol. 2017, 1549, 31–43. [Google Scholar] [CrossRef]
- Megger, D.A.; Pott, L.L.; Ahrens, M.; Padden, J.; Bracht, T.; Kuhlmann, K.; Eisenacher, M.; Meyer, H.E.; Sitek, B. Comparison of label-free and label-based strategies for proteome analysis of hepatoma cell lines. Biochim. Biophys. Acta 2014, 1844, 967–976. [Google Scholar] [CrossRef] [PubMed]
- Agarwal, A.; Samanta, L.; Bertolla, R.P.; Durairajanayagam, D.; Intasqui, P. Proteomics in Human Reproduction: Biomarkers for Millennials; Springer: Berlin/Heidelberg, Germany, 2016. [Google Scholar]
- Macek, B.; Mann, M.; Olsen, J.V. Global and site-specific quantitative phosphoproteomics: Principles and applications. Annu Rev. Pharmacol. Toxicol. 2009, 49, 199–221. [Google Scholar] [CrossRef]
- Aitken, R.J.; Baker, M.A. The role of proteomics in understanding sperm cell biology. Int. J. Androl. 2008, 31, 295–302. [Google Scholar] [CrossRef]
- Jordan, P.W.; Karppinen, J.; Handel, M.A. Polo-like kinase is required for synaptonemal complex disassembly and phosphorylation in mouse spermatocytes. J. Cell Sci. 2012, 125, 5061–5072. [Google Scholar] [CrossRef]
- Qi, L.; Liu, Z.; Wang, J.; Cui, Y.; Guo, Y.; Zhou, T.; Zhou, Z.; Guo, X.; Xue, Y.; Sha, J. Systematic analysis of the phosphoproteome and kinase-substrate networks in the mouse testis. Mol. Cell. Proteom. 2014, 13, 3626–3638. [Google Scholar] [CrossRef] [PubMed]
- Wei, Y.; Gao, Q.; Niu, P.; Xu, K.; Qiu, Y.; Hu, Y.; Liu, S.; Zhang, X.; Yu, M.; Liu, Z.; et al. Integrative Proteomic and Phosphoproteomic Profiling of Testis from Wip1 Phosphatase- Knockout Mice: Insights into Mechanisms of Reduced Fertility. Mol. Cell. Proteom. 2019, 18, 216–230. [Google Scholar] [CrossRef] [PubMed]
- Filipponi, D.; Muller, J.; Emelyanov, A.; Bulavin, D.V. Wip1 controls global heterochromatin silencing via ATM/BRCA1-dependent DNA methylation. Cancer Cell 2013, 24, 528–541. [Google Scholar] [CrossRef] [PubMed]
- Heller, C.G.; Clermont, Y. Spermatogenesis in man: An estimate of its duration. Science 1963, 140, 184–186. [Google Scholar] [CrossRef] [PubMed]
- Sutovsky, P.; Moreno, R.; Ramalho-Santos, J.; Dominko, T.; Thompson, W.E.; Schatten, G. A putative, ubiquitin-dependent mechanism for the recognition and elimination of defective spermatozoa in the mammalian epididymis. J. Cell Sci. 2001, 114, 1665–1675. [Google Scholar] [CrossRef]
- Wang, Y.; Wan, J.; Ling, X.; Liu, M.; Zhou, T. The human sperm proteome 2.0: An integrated resource for studying sperm functions at the level of posttranslational modification. Proteomics 2016, 16, 2597–2601. [Google Scholar] [CrossRef]
- Eskandari-Shahraki, M.; Tavalaee, M.; Deemeh, M.R.; Jelodar, G.A.; Nasr-Esfahani, M.H. Proper ubiquitination effect on the fertilisation outcome post-ICSI. Andrologia 2013, 45, 204–210. [Google Scholar] [CrossRef]
- Tash, J.S.; Means, A.R. Regulation of protein phosphorylation and motility of sperm by cyclic adenosine monophosphate and calcium. Biol. Reprod. 1982, 26, 745–763. [Google Scholar] [CrossRef]
- Luconi, M.; Baldi, E. How do sperm swim? Molecular mechanisms underlying sperm motility. Cell Mol. Biol. 2003, 49, 357–369. [Google Scholar]
- Huang, Z.; Khatra, B.; Bollen, M.; Carr, D.W.; Vijayaraghavan, S. Sperm PP1gamma2 is regulated by a homologue of the yeast protein phosphatase binding protein sds22. Biol. Reprod. 2002, 67, 1936–1942. [Google Scholar] [CrossRef]
- Chakrabarti, R.; Cheng, L.; Puri, P.; Soler, D.; Vijayaraghavan, S. Protein phosphatase PP1 gamma 2 in sperm morphogenesis and epididymal initiation of sperm motility. Asian J. Androl. 2007, 9, 445–452. [Google Scholar] [CrossRef] [PubMed]
- Visconti, P.E.; Bailey, J.L.; Moore, G.D.; Pan, D.; Olds-Clarke, P.; Kopf, G.S. Capacitation of mouse spermatozoa. I. Correlation between the capacitation state and protein tyrosine phosphorylation. Development 1995, 121, 1129–1137. [Google Scholar] [CrossRef] [PubMed]
- Yunes, R.; Doncel, G.F.; Acosta, A.A. Incidence of sperm-tail tyrosine phosphorylation and hyperactivated motility in normozoospermic and asthenozoospermic human sperm samples. Biocell 2003, 27, 29–36. [Google Scholar] [CrossRef] [PubMed]
- Huang, Z.; Danshina, P.V.; Mohr, K.; Qu, W.; Goodson, S.G.; O’Connell, T.M.; O’Brien, D.A. Sperm function, protein phosphorylation, and metabolism differ in mice lacking successive sperm-specific glycolytic enzymes. Biol. Reprod. 2017, 97, 586–597. [Google Scholar] [CrossRef][Green Version]
- Morgan, D.J.; Weisenhaus, M.; Shum, S.; Su, T.; Zheng, R.; Zhang, C.; Shokat, K.M.; Hille, B.; Babcock, D.F.; McKnight, G.S. Tissue-specific PKA inhibition using a chemical genetic approach and its application to studies on sperm capacitation. Proc. Natl. Acad. Sci. USA 2008, 105, 20740–20745. [Google Scholar] [CrossRef]
- Liu, X.; Wang, X.; Liu, F. Decreased expression of heat shock protein A4L in spermatozoa is positively related to poor human sperm quality. Mol. Reprod. Dev. 2019, 86, 379–386. [Google Scholar] [CrossRef]
- Nijs, M.; Ombelet, W. Cryopreservation of human sperm. Hum. Fertil. 2001, 4, 158–163. [Google Scholar] [CrossRef]
- Bogle, O.A.; Kumar, K.; Attardo-Parrinello, C.; Lewis, S.E.; Estanyol, J.M.; Ballescà, J.L.; Oliva, R. Identification of protein changes in human spermatozoa throughout the cryopreservation process. Andrology 2017, 5, 10–22. [Google Scholar] [CrossRef]
- Freitas, M.J.; Silva, J.V.; Brothag, C.; Regadas-Correia, B.; Fardilha, M.; Vijayaraghavan, S. Isoform-specific GSK3A activity is negatively correlated with human sperm motility. Mol. Hum. Reprod. 2019, 25, 171–183. [Google Scholar] [CrossRef]
- Vijayaraghavan, S.; Mohan, J.; Gray, H.; Khatra, B.; Carr, D.W. A role for phosphorylation of glycogen synthase kinase-3alpha in bovine sperm motility regulation. Biol Reprod. 2000, 62, 1647–1654. [Google Scholar] [CrossRef]
- Aparicio, I.M.; Bragado, M.J.; Gil, M.C.; Garcia-Herreros, M.; Gonzalez-Fernandez, L.; Tapia, J.A.; Garcia-Marin, L.J. Porcine sperm motility is regulated by serine phosphorylation of the glycogen synthase kinase-3alpha. Reproduction 2007, 134, 435–444. [Google Scholar] [CrossRef] [PubMed]
- Bhattacharjee, R.; Goswami, S.; Dudiki, T.; Popkie, A.P.; Phiel, C.J.; Kline, D.; Vijayaraghavan, S. Targeted disruption of glycogen synthase kinase 3A (GSK3A) in mice affects sperm motility resulting in male infertility. Biol. Reprod. 2015, 92, 65. [Google Scholar] [CrossRef]
- Zhu, Z.; Li, R.; Wang, L.; Zheng, Y.; Hoque, S.A.M.; Lv, Y.; Zeng, W. Glycogen Synthase Kinase-3 Regulates Sperm Motility and Acrosome Reaction via Affecting Energy Metabolism in Goats. Front. Physiol. 2019, 10, 968. [Google Scholar] [CrossRef] [PubMed]
- Chang, M.C. Fertilizing capacity of spermatozoa deposited into the fallopian tubes. Nature 1951, 168, 697–698. [Google Scholar] [CrossRef]
- Austin, C.R. The capacitation of the mammalian sperm. Nature 1952, 170, 326. [Google Scholar] [CrossRef] [PubMed]
- Yanagimachi, R. Fertility of mammalian spermatozoa: Its development and relativity. Zygote 1994, 2, 371–372. [Google Scholar] [CrossRef] [PubMed]
- Xu, Y.; Han, Q.; Ma, C.; Wang, Y.; Zhang, P.; Li, C.; Cheng, X.; Xu, H. Comparative Proteomics and Phosphoproteomics Analysis Reveal the Possible Breed Difference in Yorkshire and Duroc Boar Spermatozoa. Front. Cell Dev. Biol. 2021, 9, 652809. [Google Scholar] [CrossRef]

| Biological Process | Study | Type of Samples | Sperm Preparation | Phosphoproteomic Method |
|---|---|---|---|---|
| Spermatogenesis | Castillo et al. 2019 | Testicular tissue | LC-MS/MS | |
| Sperm motility | Chan et al. 2009 | Normozoospermic vs. asthenozoospermic spermatozoa | Percoll fractionation | 2DE-MALDI-TOF MS |
| Parte et al. 2012 | Normozoospermic vs. asthenozoospermic spermatozoa | Washing | Nano UPLC-MS | |
| Martin-Hidalgo et al. 2020 | High-mobility vs. low-mobility sperm subpopulations | PureSperm fractionation | Nano HPLC-MS/MS Triple TOF | |
| Sperm capacitation | Ficarro et al. 2003 | Capacitated vs. non-capacitated spermatozoa | Percoll fractionation | 2DE-anti-phosphotyrosine Immunoblots MS/MS |
| Wang et al. 2015 | Capacitated vs. non-capacitated spermatozoa | Percoll fractionation | LC-MS/MS |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Serrano, R.; Garcia-Marin, L.J.; Bragado, M.J. Sperm Phosphoproteome: Unraveling Male Infertility. Biology 2022, 11, 659. https://doi.org/10.3390/biology11050659
Serrano R, Garcia-Marin LJ, Bragado MJ. Sperm Phosphoproteome: Unraveling Male Infertility. Biology. 2022; 11(5):659. https://doi.org/10.3390/biology11050659
Chicago/Turabian StyleSerrano, Rebeca, Luis J. Garcia-Marin, and Maria J. Bragado. 2022. "Sperm Phosphoproteome: Unraveling Male Infertility" Biology 11, no. 5: 659. https://doi.org/10.3390/biology11050659
APA StyleSerrano, R., Garcia-Marin, L. J., & Bragado, M. J. (2022). Sperm Phosphoproteome: Unraveling Male Infertility. Biology, 11(5), 659. https://doi.org/10.3390/biology11050659






