Isolation of Efficient Xylooligosaccharides-Fermenting Probiotic Lactic Acid Bacteria from Ethnic Pickled Bamboo Shoot Products
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Chemicals
2.2. Microorganisms and Culture Conditions
2.3. Sampling Locations and Sample Collection
2.4. Enrichment and Isolation of Xylose-Fermenting LAB
2.5. Confirmation and Screening for XOSs-Fermenting LAB
2.6. Bacterial Identification
2.7. In Vitro Survival Test for Simulated Gastric Conditions
2.8. In Vitro Survival Test for Simulated Intestinal Conditions
2.9. Cell Surface Hydrophobicity Assay
2.10. Auto-Aggregation Assay
2.11. Fermentation of XOSs
2.11.1. Investigation of Antimicrobial Activity
2.11.2. Determination of Xylanase and β-Xylosidase Activities
2.11.3. Thin-Layer Chromatography
2.11.4. Determination of SCFAs
2.11.5. Antibiotic resistance
2.12. Hemolytic Activity Test
2.13. Analytical Methods
3. Results
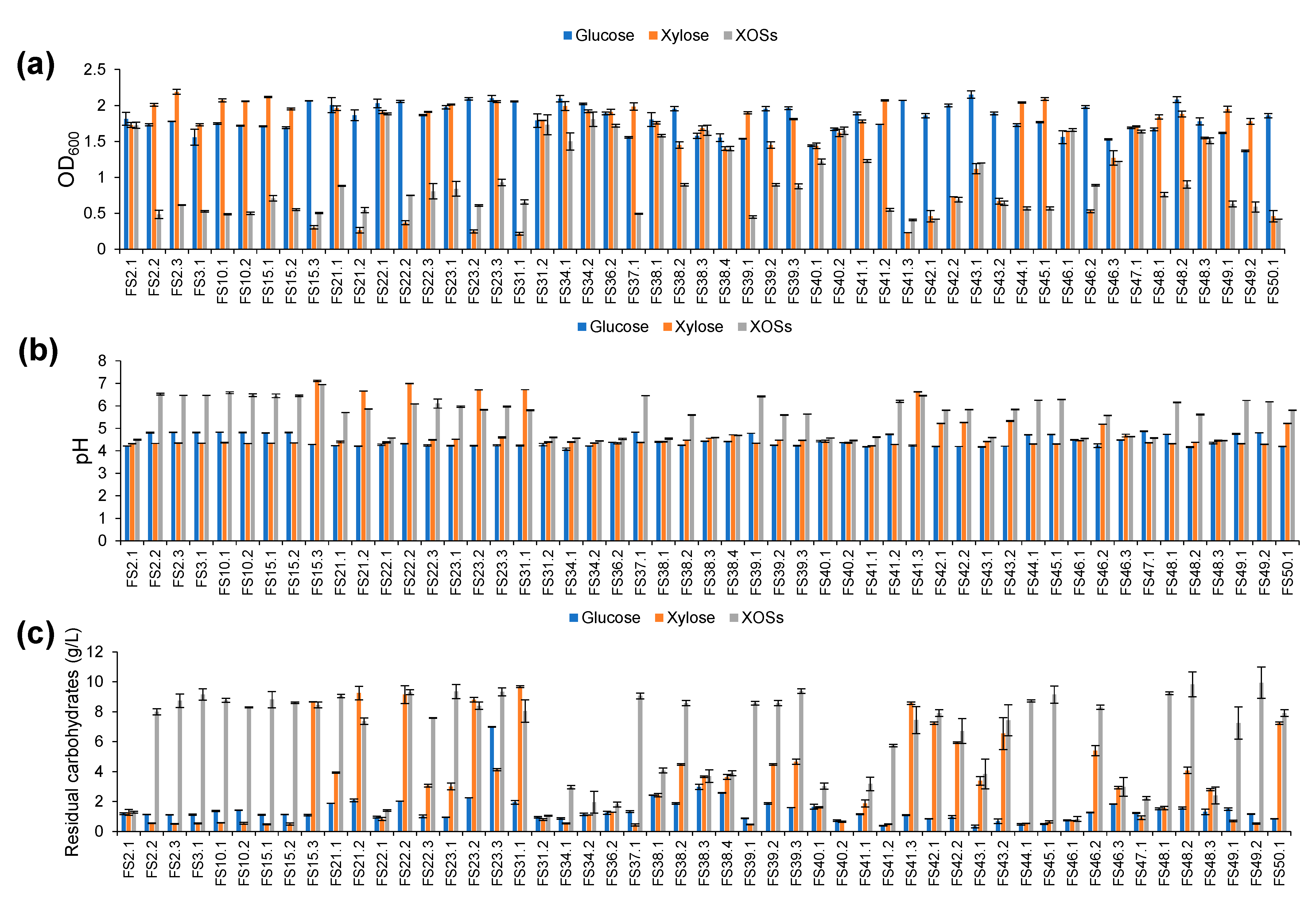
3.1. Isolation of XOSs-Fermenting LAB
3.2. Selection of XOSs-Fermenting LAB
3.3. Identification of XOSs-Fermenting LAB
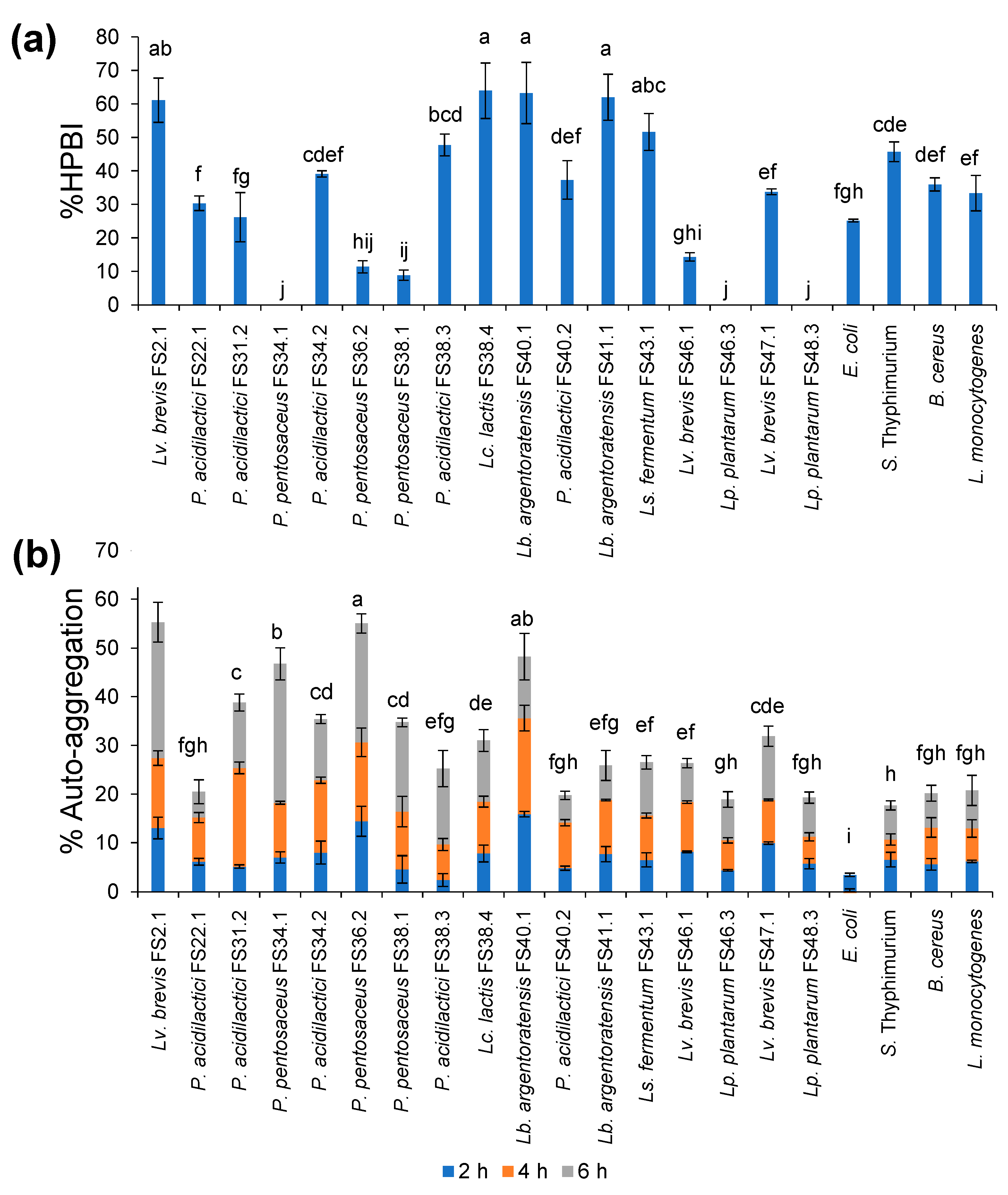
3.4. Tolerance of XOSs-Fermenting LAB under Simulated GIT Conditions
3.5. Cell Surface Hydrophobicity and Auto-Aggregation
3.6. Antimicrobial Activity
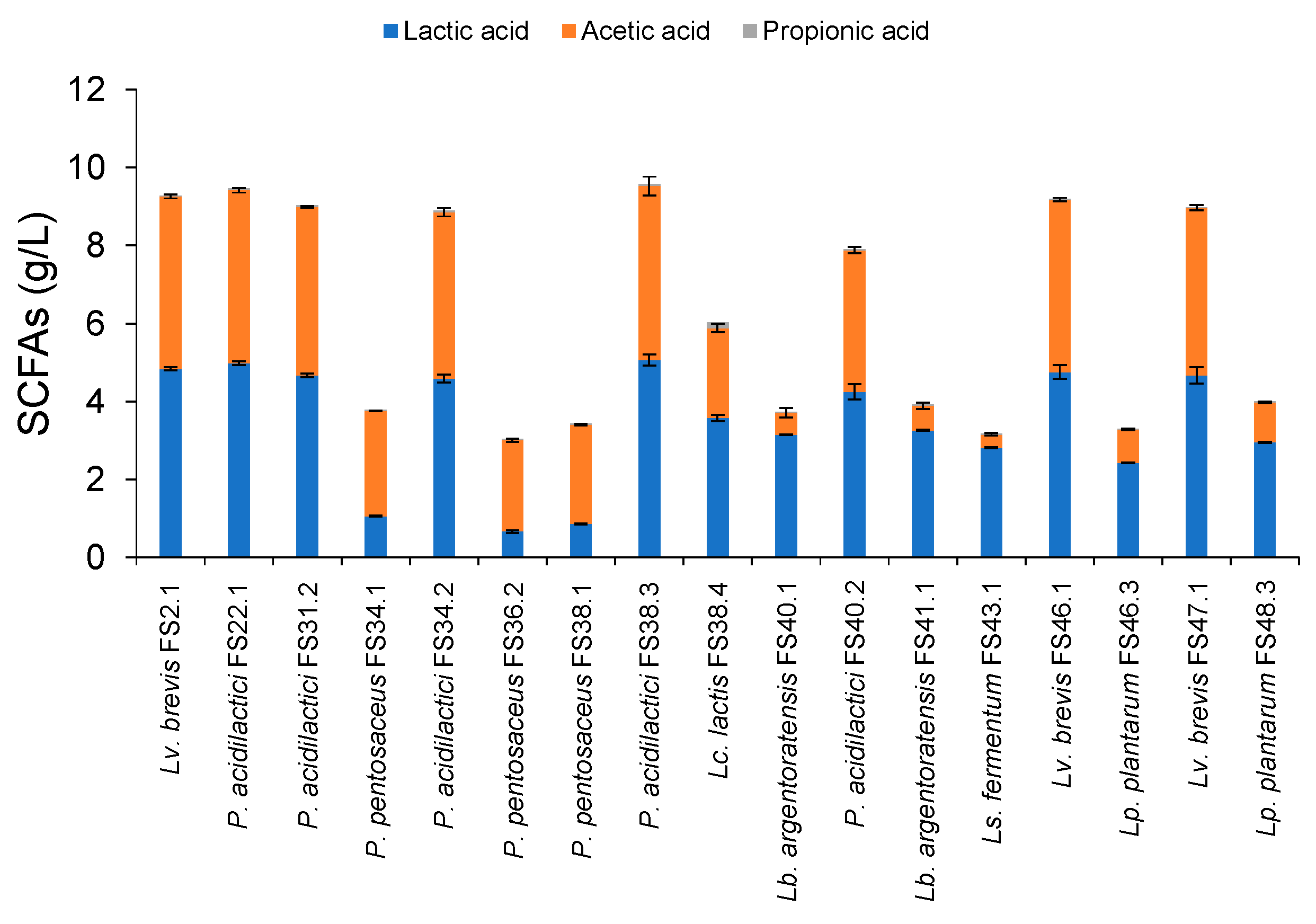
3.7. SCFAs Production
3.8. Antibiotic Susceptibility and Hemolytic Activity
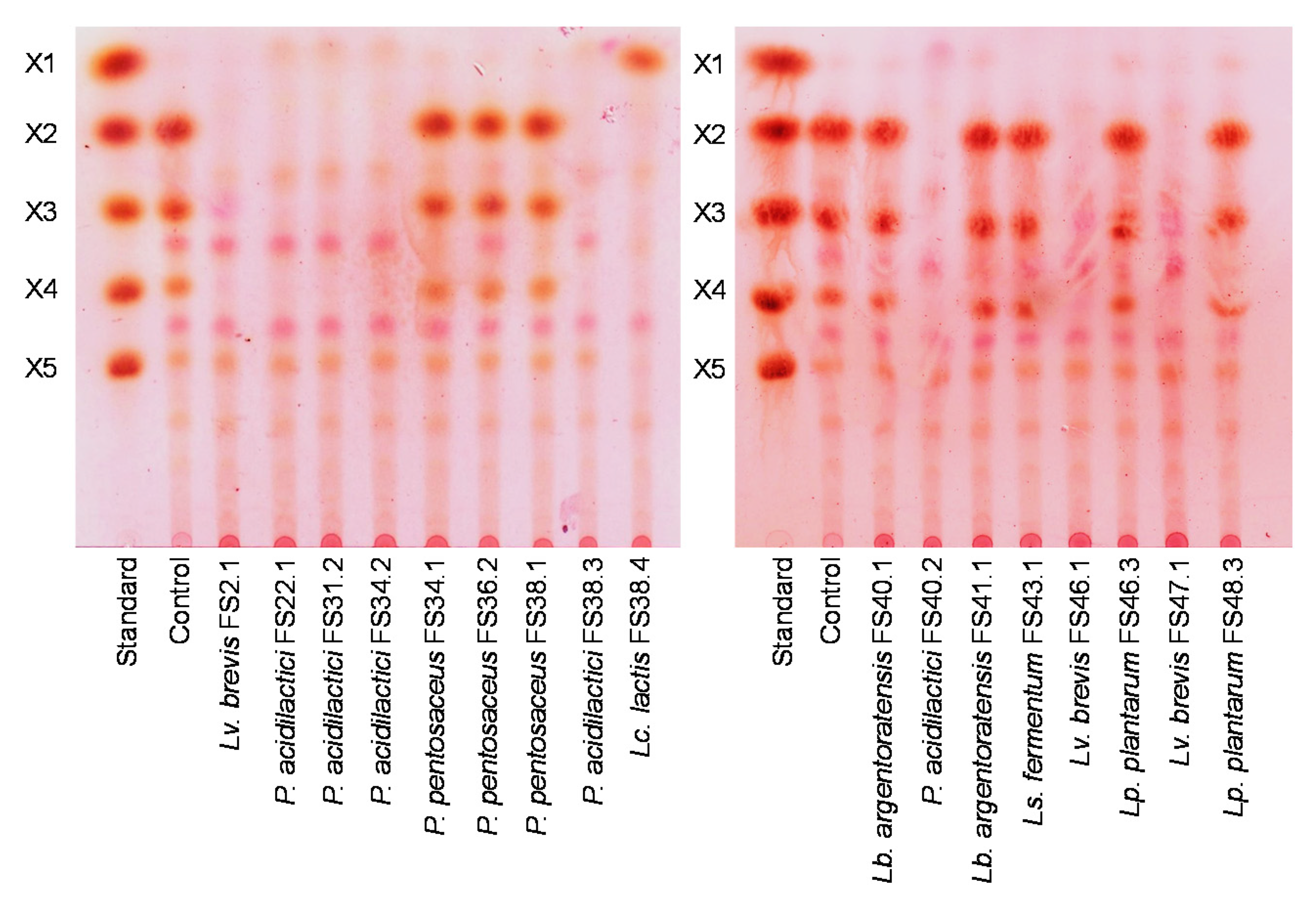
3.9. Detection of Xylanolytic Enzymes
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- de Freitas, C.; Terrone, C.C.; Masarin, F.; Carmona, E.C.; Brienzo, M. In vitro study of the effect of xylooligosaccharides obtained from banana pseudostem xylan by enzymatic hydrolysis on probiotic bacteria. Biocatal. Agric. Biotechnol. 2021, 33, 101973. [Google Scholar] [CrossRef]
- Rashid, R.; Sohail, M. Xylanolytic Bacillus species for xylooligosaccharides production: A critical review. Bioresour. Bioprocess 2021, 8, 16. [Google Scholar] [CrossRef]
- Li, Z.; Summanen, P.H.; Komoriya, T.; Finegold, S.M. In vitro study of the prebiotic xylooligosaccharide (XOS) on the growth of Bifidobacterium spp. and Lactobacillus spp. Int. J. Food Sci. Nutr. 2015, 66, 919–922. [Google Scholar] [CrossRef] [PubMed]
- Petrova, P.; Petrov, K. Chapter 7—Prebiotic–Probiotic relationship: The genetic fundamentals of polysaccharides conversion by Bifidobacterium and Lactobacillus Genera. In Food Bioconversion; Grumezescu, A.M., Holban, A.M., Eds.; Academic Press: Amsterdam, The Netherlands, 2017; pp. 237–278. [Google Scholar]
- Michlmayr, H.; Hell, J.; Lorenz, C.; Böhmdorfer, S.; Rosenau, T.; Kneifel, W. Arabinoxylan oligosaccharide hydrolysis by family 43 and 51 glycosidases from Lactobacillus brevis DSM 20054. Appl. Environ. Microbiol. 2013, 79, 6747–6754. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, K.; Wang, X.; Wang, J.; Zhang, J. Benefits from additives and xylanase during enzymatic hydrolysis of bamboo shoot and mature bamboo. Bioresour. Technol. 2015, 192, 424–431. [Google Scholar] [CrossRef] [PubMed]
- Kanpiengjai, A.; Rieantrakoonchai, W.; Pratanaphon, R.; Pathom-aree, W.; Lumyong, S.; Khanongnuch, C. High efficacy bioconversion of starch to lactic acid using an amylolytic lactic acid bacterium isolated from Thai indigenous fermented rice noodles. Food Sci. Biotechnol. 2014, 23, 1541–1550. [Google Scholar] [CrossRef]
- Dubois, M.; Gilles, K.; Hamilton, J.K.; Rebers, P.A.; Smith, F. A colorimetric method for the determination of sugars. Nature 1951, 168, 167. [Google Scholar] [CrossRef] [PubMed]
- Tamura, K.; Stecher, G.; Kumar, S. MEGA11: Molecular Evolutionary Genetics Analysis Version 11. Mol. Biol. Evol. 2021, 38, 3022–3027. [Google Scholar] [CrossRef]
- Kanpiengjai, A.; Khanongnuch, C.; Lumyong, S.; Kummasook, A.; Kittibunchakul, S. Characterization of Sporidiobolus ruineniae A45.2 cultivated in tannin substrate for use as a potential multifunctional probiotic yeast in aquaculture. J. Fungi 2020, 6, 378. [Google Scholar] [CrossRef]
- Unban, K.; Chaichana, W.; Baipong, S.; Abdullahi, A.D.; Kanpiengjai, A.; Shetty, K.; Khanongnuch, C. Probiotic and antioxidant properties of lactic acid bacteria isolated from indigenous fermented tea leaves (Miang) of north Thailand and promising application in synbiotic formulation. Fermentation 2021, 7, 195. [Google Scholar] [CrossRef]
- CLSI. CLSI Document M07-A9. Methods for Dilution Antimicrobial Susceptibility Tests for Bacteria That Grow Aerobically: Approved Standard, 19th ed.; CLSI: Wayne, PA, USA, 2012. [Google Scholar]
- Liu, D.D.; Gu, C.T. Proposal to reclassify Lactobacillus zhaodongensis, Lactobacillus zeae, Lactobacillus argentoratensis and Lactobacillus buchneri subsp. silagei as Lacticaseibacillus zhaodongensis comb. nov., Lacticaseibacillus zeae comb. nov., Lactiplantibacillus argentoratensis comb. nov. and Lentilactobacillus buchneri subsp. silagei comb. nov., respectively and Apilactobacillus kosoi as a later heterotypic synonym of Apilactobacillus micheneri. Int. J. Syst. Evol. Microbiol. 2020, 70, 6414–6417. [Google Scholar] [PubMed]
- Swain, M.R.; Anandharaj, M.; Ray, R.C.; Parveen Rani, R. Fermented fruits and vegetables of Asia: A potential source of probiotics. Biotechnol. Res. Int. 2014, 2014, 250424. [Google Scholar] [CrossRef]
- Jeyaram, K.; Romi, W.; Singh, T.A.; Devi, A.R.; Devi, S.S. Bacterial species associated with traditional starter cultures used for fermented bamboo shoot production in Manipur state of India. Int. J. Food Microbiol. 2010, 143, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Nongdam, P.; Tikendra, L. The nutritional facts of bamboo shoots and their usage as important traditional foods of northeast India. Int. Sch. Res. Not. 2014, 2014, 679073. [Google Scholar] [CrossRef] [PubMed]
- Yongsawas, R.; Inta, A.; Kampuansai, J.; Pandith, H.; Suwannarach, N.; Lamyong, S.; Chantawannakul, P.; Chitov, T.; Disayathanoowat, T. Bacterial communities in Lanna Phak-Gard-Dong (pickled mustard green) from three different ethnolinguistic groups in northern Thailand. Biology 2022, 11, 150. [Google Scholar] [CrossRef] [PubMed]
- Tamang, B.; Tamang, J.P.; Schillinger, U.; Franz, C.M.; Gores, M.; Holzapfel, W.H. Phenotypic and genotypic identification of lactic acid bacteria isolated from ethnic fermented bamboo tender shoots of North East India. Int. J. Food Microbiol. 2008, 121, 35–40. [Google Scholar] [CrossRef]
- Li, W.; Ren, M.; Duo, L.; Li, J.; Wang, S.; Sun, Y.; Li, M.; Ren, W.; Hou, Q.; Yu, J.; et al. Fermentation characteristics of Lactococcus lactis subsp. lactis isolated from naturally fermented dairy products and screening of potential starter isolates. Front. Microbiol. 2020, 11, 1477–1483. [Google Scholar] [CrossRef]
- Kelly, W.J.; Davey, G.P.; Ward, L.J. Characterization of lactococci isolated from minimally processed fresh fruit and vegetables. Int. J. Food Microbiol. 1998, 45, 85–92. [Google Scholar] [CrossRef]
- Tanaka, K.; Komiyama, A.; Sonomoto, K.; Ishizaki, A.; Hall, S.J.; Stanbury, P.F. Two different pathways for D-xylose metabolism and the effect of xylose concentration on the yield coefficient of L-lactate in mixed-acid fermentation by the lactic acid bacterium Lactococcus lactis IO-1. Appl. Microbiol. Biotechnol. 2002, 60, 160–167. [Google Scholar]
- Abbasiliasi, S.; Tan, J.S.; Ibrahim, T.A.T.; Ramanan, R.N.; Vakhshiteh, F.; Mustafa, S.; Ling, T.C.; Rahim, R.A.; Ariff, A.B. Isolation of Pediococcus acidilactici Kp10 with ability to secrete bacteriocin-like inhibitory substance from milk products for applications in food industry. BMC Microbiol. 2012, 12, 260. [Google Scholar] [CrossRef] [Green Version]
- Ahn, H.; Kim, J.; Kim, W.J. Isolation and characterization of bacteriocin-producing Pediococcus acidilactici HW01 from malt and its potential to control beer spoilage lactic acid bacteria. Food Control 2017, 80, 59–66. [Google Scholar] [CrossRef]
- Kim, M.; Choi, E.; Kim, J.; Ahn, H.; Han, H.; Kim, W.J. Effect of bacteriocin-producing Pediococcus acidilactici K10 on beer fermentation. J. Inst. Brew. 2016, 122, 422–429. [Google Scholar] [CrossRef] [Green Version]
- Xia, X.; Ran, C.; Ye, X.; Li, G.; Kan, J.; Zheng, J. Monitoring of the bacterial communities of bamboo shoots (Dendrocalamus latiflorus) during pickling process. Int. J. Food Sci. Technol. 2017, 52, 1101–1110. [Google Scholar] [CrossRef]
- Gut, A.M.; Vasiljevic, T.; Yeager, T.; Donkor, O.N. Characterization of yeasts isolated from traditional kefir grains for potential probiotic properties. J. Funct. Foods 2019, 58, 56–66. [Google Scholar] [CrossRef]
- Kavitha, M.; Raja, M.; Perumal, P. Evaluation of probiotic potential of Bacillus spp. isolated from the digestive tract of freshwater fish Labeo calbasu. Aquac. Rep. 2018, 11, 59–69. [Google Scholar] [CrossRef]
- Krausova, G.; Hyrslova, I.; Hynstova, I. In vitro evaluation of adhesion capacity, hydrophobicity, and auto-aggregation of newly isolated potential probiotic strains. Fermentation 2019, 5, 100. [Google Scholar] [CrossRef] [Green Version]
- Chen, C.C.; Lai, C.C.; Huang, H.L.; Huang, W.Y.; Toh, H.S.; Weng, T.C.; Chuang, Y.C.; Lu, Y.C.; Tang, H.J. Antimicrobial activity of Lactobacillus species against carbapenem-resistant Enterobacteriaceae. Front. Microbiol. 2019, 10, 789. [Google Scholar] [CrossRef]
- Jiang, S.; Cai, L.; Lv, L.; Li, L. Pediococcus pentosaceus, a future additive or probiotic candidate. Microb. Cell Fact. 2021, 20, 45. [Google Scholar] [CrossRef]
- Neut, C.; Mahieux, S.; Dubreuil, L.J. Antibiotic susceptibility of probiotic strains: Is it reasonable to combine probiotics with antibiotics? Med. Mal. Infect. 2017, 47, 477–483. [Google Scholar] [CrossRef]
- Cave, R.; Cole, J.; Mkrtchyan, H.V. Surveillance and prevalence of antimicrobial resistant bacteria from public settings within urban built environments: Challenges and opportunities for hygiene and infection control. Environ. Int. 2021, 157, 106836. [Google Scholar] [CrossRef]
- Sanders, M.E.; Akkermans, L.M.; Haller, D.; Hammerman, C.; Heimbach, J.; Hörmannsperger, G.; Huys, G.; Levy, D.D.; Lutgendorff, F.; Mack, D.; et al. Safety assessment of probiotics for human use. Gut Microbes 2010, 1, 164–185. [Google Scholar] [CrossRef] [PubMed]
- Iliev, I.; Vasileva, T.; Bivolarski, V.; Momchilova, A.; Ivanova, I. Metabolic profiling of xylooligosaccharides by Lactobacilli. Polymers 2020, 12, 2387. [Google Scholar] [CrossRef] [PubMed]
- Lei, Z.; Wu, Y.; Nie, W.; Yin, D.; Yin, X.; Guo, Y.; Aggrey, S.E.; Yuan, J. Transcriptomic analysis of xylan oligosaccharide utilization systems in Pediococcus acidilactici strain BCC-1. J. Agric. Food Chem. 2018, 66, 4725–4733. [Google Scholar] [CrossRef] [PubMed]
- Han, D.; Yan, Q.; Liu, J.; Jiang, Z.; Yang, S. Transcriptomic analysis of Pediococcus pentosaceus reveals carbohydrate metabolic dynamics under lactic acid stress. Front. Microbiol. 2021, 12, 736411. [Google Scholar] [CrossRef]
- Gilad, O.; Jacobsen, S.; Stuer-Lauridsen, B.; Pedersen, M.B.; Garrigues, C.; Svensson, B. Combined transcriptome and proteome analysis of Bifidobacterium animalis subsp. lactis BB-12 grown on xylo-oligosaccharides and a model of their utilization. Appl. Environ. Microbiol. 2010, 76, 7285–7291. [Google Scholar] [CrossRef] [Green Version]
- Jordan, D.B.; Wagschal, K. Properties and applications of microbial beta-D-xylosidases featuring the catalytically efficient enzyme from Selenomonas ruminantium. Appl. Microbiol. Biotechnol. 2010, 86, 1647–1658. [Google Scholar] [CrossRef]

| No. | Sample Code | Subdistrict, District, Province | Location | Date Achieved | No. of Isolates | Isolate Code |
|---|---|---|---|---|---|---|
| 1 | FS2 | Mae Faek Mai, San Sai, Chiang Mai | 18.9950, 98.9778 | 8 June 2020 | 3 | FS2.1, FS2.2, FS2.3 |
| 2 | FS3 | Suthep, Muang, Chiang Mai | 18.7904, 98.9595 | 9 June 2020 | 1 | FS3.1 |
| 3 | FS10 | Rim Tai, Mae Rim, Chiang Mai | 18.9133, 98.9445 | 16 June 2020 | 2 | FS10.1, FS10.2 |
| 4 | FS15 | Pa Daet, Pa Daet, Chiang Rai | 19.4998, 99.9905 | 20 June 2020 | 3 | FS15.1, FS15.2, FS15.3 |
| 5 | FS21 | Pong Tam, Chai Prakarn, Chiang Mai | 19.7312, 99.1029 | 11 August 2020 | 2 | FS21.1, FS21.2 |
| 6 | FS22 | Mae Sa, Mae Rim, Chiang Mai | 18.8924, 98.6798 | 12 August 2020 | 3 | FS22.1, FS22.2, FS22.3 |
| 7 | FS23 | Mae Sa, Mae Rim, Chiang Mai | 18.8924, 98.6798 | 16 August 2020 | 3 | FS23.1, FS23.2, FS23.3 |
| 8 | FS31 | Ki Lek, Mae Taeng, Chiang Mai | 19.0781, 98.9405 | 16 August 2020 | 2 | FS31.1, FS31.2 |
| 9 | FS34 | San Phi Suea, Mueang, Chiang Mai | 18.8371, 98.9872 | 23 August 2020 | 2 | FS34.1, FS34.2 |
| 10 | FS36 | Mae Taeng, Mae Taeng, Chiang Mai | 19.0945, 98.9012 | 24 August 2020 | 1 | FS36.2 |
| 11 | FS37 | San Phak Wan, Hang Dong, Chiang Mai | 18.7227, 98.9575 | 25 August 2020 | 1 | FS37.1 |
| 12 | FS38 | Nong Kwai, Hang Dong, Chiang Mai | 18.7381, 98.9312 | 31 August 2020 | 4 | FS38.1, FS38.2, FS38.3, FS38.4 |
| 13 | FS39 | Han Kaeo, Hang Dong, Chiang Mai | 18.6535, 98.9035 | 31 August 2020 | 3 | FS39.1, FS39.2, FS39.3 |
| 14 | FS40 | Hang Dong, Hang Dong, Chiang Mai | 18.6894, 98.9036 | 5 September 2020 | 2 | FS40.1, FS40.2 |
| 15 | FS41 | Rong Chang, Pa Daet, Chiang Rai | 19.4948, 99.9414 | 5 September 2020 | 3 | FS41.1, FS41.2, FS41.3 |
| 16 | FS42 | Pa Ngae, Pa Daet, Chiang Rai | 19.5545, 99.9463 | 5 September 2020 | 2 | FS42.1, FS42.2 |
| 17 | FS43 | Pa Daet, Pa Daet, Chiang Rai | 19.5116, 99.9925 | 5 September 2020 | 2 | FS43.1, FS43.2 |
| 18 | FS44 | Tha Wang Thong, Muang, Phayao | 19.1768, 99.8932 | 5 September 2020 | 1 | FS44.1 |
| 19 | FS45 | Wang Nuer, Wang Nuer, Lampang | 19.1355, 99.6323 | 5 September 2020 | 1 | FS45.1 |
| 20 | FS46 | Wang Nuer, Wang Nuer, Lampang | 19.1355, 99.6323 | 5 September 2020 | 3 | FS46.1, FS46.2, FS46.3 |
| 21 | FS47 | Rong Kwang, Rong Kwang, Phrae | 18.3371, 100.3165 | 20 September 2020 | 1 | FS47.1 |
| 22 | FS48 | Rong Kwang, Rong Kwang, Phrae | 18.3371, 100.3165 | 20 September 2020 | 3 | FS48.1, FS48.2, FS48.3 |
| 23 | FS49 | Klang Wiang, Wiang Sa, Nan | 18.5626, 100.7466 | 20 September 2020 | 2 | FS49.1, FS49.2 |
| 24 | FS50 | Klang Wiang, Wiang Sa, Nan | 18.5626, 100.7466 | 20 September 2020 | 1 | FS50.1 |
| Isolates | Nucleotides | % Similarity | Identification Results | Accession Number |
|---|---|---|---|---|
| FS2.1 | 1430 | 99.86 | Levilactobacillus brevis | OM899733 |
| FS22.1 | 1454 | 99.79 | Pediococcus acidilactici | OM899734 |
| FS31.2 | 1447 | 99.79 | Pediococcus acidilactici | OM899735 |
| FS34.1 | 1450 | 100.00 | Pediococcus pentosaceus | OM899736 |
| FS34.2 | 1460 | 99.79 | Pediococcus acidilactici | OM899737 |
| FS36.2 | 1440 | 100.00 | Pediococcus pentosaceus | OM899738 |
| FS38.1 | 1440 | 100.00 | Pediococcus pentosaceus | OM899739 |
| FS38.3 | 1440 | 99.79 | Pediococcus acidilactici | OM899740 |
| FS38.4 | 1420 | 99.93 | Lactococcus lactis | OM899741 |
| FS40.1 | 1428 | 99.93 | Lactobacillus argentoratensis | OM899742 |
| FS40.2 | 1440 | 99.79 | Pediococcus acidilactici | OM899743 |
| FS41.1 | 1440 | 99.86 | Lactobacillus argentoratensis | OM899744 |
| FS43.1 | 1440 | 99.93 | Limosilactobacillus fermentum | OM899745 |
| FS46.1 | 1430 | 99.79 | Levilactobacillus brevis | OM899746 |
| FS46.3 | 1424 | 99.93 | Lactiplantibacillus plantarum | OM899747 |
| FS47.1 | 1430 | 99.86 | Levilactobacillus brevis | OM899748 |
| FS48.3 | 1428 | 99.93 | Lactiplantibacillus plantarum | OM899749 |
| Strains | Gastric Conditions | Intestinal Conditions | ||||
|---|---|---|---|---|---|---|
| 0 h | 1 h | 2 h | 0 h | 4 h | 8 h | |
| Lv. brevis FS2.1 | 100.0 ± 0.15 | 93.0 ± 1.70 | 74.5 ± 0.94 | 100.0 ± 0.98 | 103.1 ± 0.43 | 109.1 ± 0.38 |
| P. acidilactici FS22.1 | 100.0 ± 0.19 | 85.5 ± 1.23 | 47.2 ± 2.16 | 100.0 ± 0.41 | 101.3 ± 0.07 | 104.7 ± 0.18 |
| P. acidilactici FS31.2 | 100.0 ± 0.13 | 76.8 ± 0.51 | 46.8 ± 2.14 | 100.0 ± 0.37 | 102.5 ± 0.23 | 108.0 ± 0.66 |
| P. pentosaceus FS34.1 | 100.0 ± 0.11 | 85.3 ± 1.78 | 43.0 ± 2.41 | 100.0 ± 0.22 | 101.6 ± 0.22 | 101.9 ± 0.23 |
| P. acidilactici FS34.2 | 100.0 ± 0.20 | 84.6 ± 0.37 | 37.9 ± 4.12 | 100.0 ± 0.09 | 101.2 ± 0.08 | 106.1 ± 0.67 |
| P. pentosaceus FS36.2 | 100.0 ± 0.30 | 87.3 ± 0.65 | 43.9 ± 2.46 | 100.0 ± 0.47 | 100.6 ± 0.19 | 101.4 ± 0.20 |
| P. pentosaceus FS38.1 | 100.0 ± 0.53 | 82.3 ± 2.21 | 53.0 ± 2.21 | 100.0 ± 0.42 | 102.3 ± 0.50 | 103.0 ± 0.35 |
| P. acidilactici FS38.3 | 100.0 ± 0.16 | 91.8 ± 0.10 | 78.6 ± 0.48 | 100.0 ± 0.29 | 103.0 ± 0.51 | 108.6 ± 0.33 |
| Lc. lactis FS38.4 | 100.0 ± 1.34 | 0.0 ± 0.00 | 0.0 ± 0.00 | 100.0 ± 0.93 | 102.9 ± 0.11 | 103.7 ± 0.08 |
| L. argentoratensis FS40.1 | 100.0 ± 1.15 | 77.4 ± 2.08 | 73.6 ± 0.27 | 100.0 ± 0.47 | 102.7 ± 0.79 | 108.4 ± 0.27 |
| P. acidilactici FS40.2 | 100.0 ± 0.15 | 79.5 ± 1.55 | 40.8 ± 1.23 | 100.0 ± 0.48 | 102.7 ± 0.28 | 106.8 ± 0.28 |
| L. argentoratensis FS41.1 | 100.0 ± 0.17 | 53.4 ± 2.36 | 39.0 ± 0.00 | 100.0 ± 0.05 | 104.4 ± 0.20 | 105.6 ± 0.38 |
| Ls. fermentum FS43.1 | 100.0 ± 2.29 | 93.8 ± 0.50 | 91.6 ± 0.92 | 100.0 ± 0.95 | 97.9 ± 0.79 | 97.0 ± 1.54 |
| Lv. brevis FS46.1 | 100.0 ± 0.48 | 94.2 ± 0.77 | 90.0 ± 0.45 | 100.0 ± 0.91 | 101.4 ± 0.37 | 106.3 ± 0.11 |
| Lp. plantarum FS46.3 | 100.0 ± 0.89 | 95.6 ± 0.13 | 98.3 ± 0.90 | 100.0 ± 0.77 | 105.3 ± 0.14 | 105.4 ± 0.18 |
| Lv. brevis FS47.1 | 100.0 ± 1.17 | 97.5 ± 0.05 | 84.3 ± 0.77 | 100.0 ± 0.08 | 101.0 ± 0.80 | 102.4 ± 0.19 |
| Lp. plantarum FS48.3 | 100.0 ± 0.79 | 94.3 ± 0.16 | 94.0 ± 0.42 | 100.0 ± 0.41 | 108.6 ± 0.55 | 112.5 ± 0.68 |
| Strains | E. coli | B. cereus | S. Thyphimurium | L. monocytogenes |
|---|---|---|---|---|
| Lv. brevis FS2.1 | 0.2 ± 0.0 | 0.4 ± 0.0 | 0.4 ± 0.0 | 1.0 ± 0.1 |
| P. acidilactici FS22.1 | 0.7 ± 0.0 | 0.8 ± 0.0 | 0.3 ± 0.0 | 0.3 ± 0.0 |
| P. acidilactici FS31.2 | 0.6 ± 0.1 | 0.6 ± 0.3 | 0.5 ± 0.1 | 0.5 ± 0.1 |
| P. pentosaceus FS34.1 | 0.8 ± 0.1 | 1.0 ± 0.1 | 1.0 ± 0.3 | 1.0 ± 0.1 |
| P. acidilactici FS34.2 | 0.6 ± 0.0 | 0.7 ± 0.0 | 0.4 ± 0.1 | 0.4 ± 0.0 |
| P. pentosaceus FS36.2 | 0.4 ± 0.1 | 1.1 ± 0.0 | 0.7 ± 0.4 | 0.7 ± 0.1 |
| P. pentosaceus FS38.1 | 0.4 ± 0.1 | 0.9 ± 0.0 | 1.5 ± 0.2 | 0.5 ± 0.1 |
| P. acidilactici FS38.3 | 0.2 ± 0.1 | 1.0 ± 0.1 | 1.3 ± 0.4 | 0.3 ± 0.0 |
| Lc. lactis FS38.4 | 0.5 ± 0.1 | 1.0 ± 0.1 | 0.8 ± 0.1 | 0.4 ± 0.1 |
| L. argentorantensis FS40.1 | 0.6 ± 0.1 | 1.0 ± 0.1 | 0.6 ± 0.1 | 0.5 ± 0.1 |
| P. acidilactici FS40.2 | 0.6 ± 0.1 | 0.7 ± 0.1 | 0.9 ± 0.1 | 0.8 ± 0.1 |
| L. argentorantensis FS41.1 | 0.8 ± 0.1 | 0.3 ± 0.1 | 0.5 ± 0.1 | 0.8 ± 0.1 |
| Ls. fermentum FS43.1 | 0.7 ± 0.0 | 0.5 ± 0.1 | 0.6 ± 0.1 | 0.7 ± 0.1 |
| Lv. brevis FS46.1 | 0.9 ± 0.0 | 0.7 ± 0.1 | 0.9 ± 0.0 | 0.9 ± 0.0 |
| Lp. plantarum FS46.3 | 0.6 ± 0.1 | 0.6 ± 0.1 | 0.9 ± 0.0 | 0.8 ± 0.1 |
| Lv. brevis FS47.1 | 0.6 ± 0.0 | 0.7 ± 0.1 | 0.9 ± 0.0 | 0.9 ± 0.0 |
| Lp. plantarum FS48.3 | 1.0 ± 0.1 | 1.0 ± 0.2 | 1.0 ± 0.1 | 1.1 ± 0.1 |
| Strains | Amp | Chl | Ery | Gen | Van | Kan | Tet | Cli |
|---|---|---|---|---|---|---|---|---|
| Lv. brevis FS2.1 | S | S | S | S | R | S | S | S |
| P. acidilactici FS22.1 | S | S | S | I | R | R | S | S |
| P. acidilactici FS31.2 | S | S | S | I | R | R | S | S |
| P. pentosaceus FS34.1 | S | S | S | I | R | R | S | S |
| P. acidilactici FS34.2 | S | S | S | S | R | R | S | S |
| P. pentosaceus FS36.2 | S | S | S | S | R | R | S | S |
| P. pentosaceus FS38.1 | S | S | S | S | R | R | S | S |
| P. acidilactici FS38.3 | S | S | S | I | R | R | S | S |
| L. argentoratensis FS40.1 | S | S | S | S | R | R | S | S |
| P. acidilactici FS40.2 | S | S | S | I | R | R | S | S |
| L. argentoratensis FS41.1 | S | S | S | S | R | R | S | S |
| Ls. fermentum FS43.1 | S | S | S | S | R | R | S | S |
| Lc. brevis FS46.1 | S | S | S | S | R | S | S | S |
| Lp. plantarum FS46.3 | S | S | S | S | R | R | S | S |
| Lv. brevis FS47.1 | S | S | S | S | R | S | S | I |
| Lp. plantarum FS48.3 | S | S | S | S | R | R | S | S |
| Strains | β-Xylosidase (mU/mL) | Xylanase (mU/mL) | ||||
|---|---|---|---|---|---|---|
| Extracellular | Cell-Associated | Intracellular | Extracellular | Cell-Associated | Intracellular | |
| Lv. brevis FS2.1 | ND | 29.4 ± 3.1 | ND | 320.8 ± 17.3 | ND | ND |
| P. acidilactici FS22.1 | ND | 79.2 ± 1.8 | 9.3 ± 0.1 | 280.9 ± 76.6 | ND | ND |
| P. acidilactici FS31.2 | ND | 110.2 ± 0.5 | 12.5 ± 0.7 | 450.5 ± 7.2 | ND | ND |
| P. pentosaceus FS34.1 | ND | ND | ND | 871.4 ± 10.1 | ND | ND |
| P. acidilactici FS34.2 | ND | 122 ± 7.2 | 9.8 ± 0.4 | 278.9 ± 67.9 | ND | ND |
| P. pentosaceus FS36.2 | ND | ND | ND | 545.5 ± 52.0 | ND | ND |
| P. pentosaceus FS38.1 | ND | ND | ND | 476.1± 89.6 | ND | ND |
| P. acidilactici FS38.3 | ND | 106.5 ± 3.5 | 5.8 ± 0.3 | 256.4 ± 7.2 | ND | ND |
| Lc. lactis FS38.4 | ND | 103.6 ± 3.8 | 31.2 ± 0.1 | 524.1 ± 70.8 | ND | ND |
| Lb. argentoratensis FS40.1 | ND | ND | ND | 2187.2 ± 198.1 | ND | ND |
| P. acidilactici FS40.2 | ND | 151.5 ± 28.4 | 6.6 ± 0.6 | 422.9 ± 23.1 | ND | ND |
| Lb. argentoratensis FS41.1 | ND | ND | ND | 673.2 ± 79.5 | ND | ND |
| Ls. fermentum FS43.1 | ND | ND | 5.1 ± 0.1 | 602.7 ± 23.1 | ND | ND |
| Lv. brevis FS46.1 | ND | 54.0 ± 3.0 | ND | 279.9 ± 28.9 | ND | ND |
| Lp. plantarum FS46.3 | ND | ND | 20.3 ± 0.9 | 561.9 ± 46.2 | ND | ND |
| Lv. brevis FS47.1 | ND | 67.9 ± 7.4 | 24.5 ± 1.1 | 446.4 ± 85.2 | ND | ND |
| Lp. plantarum FS48.3 | ND | ND | ND | 578.2 ± 60.7 | ND | ND |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kanpiengjai, A.; Nuntikaew, P.; Wongsanittayarak, J.; Leangnim, N.; Khanongnuch, C. Isolation of Efficient Xylooligosaccharides-Fermenting Probiotic Lactic Acid Bacteria from Ethnic Pickled Bamboo Shoot Products. Biology 2022, 11, 638. https://doi.org/10.3390/biology11050638
Kanpiengjai A, Nuntikaew P, Wongsanittayarak J, Leangnim N, Khanongnuch C. Isolation of Efficient Xylooligosaccharides-Fermenting Probiotic Lactic Acid Bacteria from Ethnic Pickled Bamboo Shoot Products. Biology. 2022; 11(5):638. https://doi.org/10.3390/biology11050638
Chicago/Turabian StyleKanpiengjai, Apinun, Pongsakorn Nuntikaew, Jirat Wongsanittayarak, Nalapat Leangnim, and Chartchai Khanongnuch. 2022. "Isolation of Efficient Xylooligosaccharides-Fermenting Probiotic Lactic Acid Bacteria from Ethnic Pickled Bamboo Shoot Products" Biology 11, no. 5: 638. https://doi.org/10.3390/biology11050638
APA StyleKanpiengjai, A., Nuntikaew, P., Wongsanittayarak, J., Leangnim, N., & Khanongnuch, C. (2022). Isolation of Efficient Xylooligosaccharides-Fermenting Probiotic Lactic Acid Bacteria from Ethnic Pickled Bamboo Shoot Products. Biology, 11(5), 638. https://doi.org/10.3390/biology11050638






