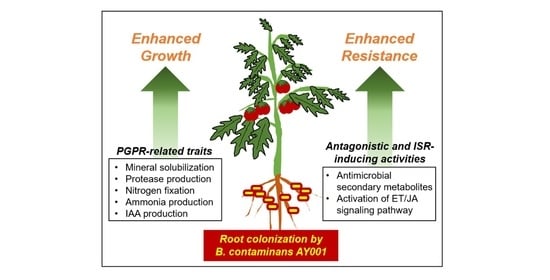
Biological Control Activity of Plant Growth Promoting Rhizobacteria Burkholderia contaminans AY001 against Tomato Fusarium Wilt and Bacterial Speck Diseases
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Isolation and Screening of Antagonistic Bacteria
2.2. Molecular Identification of AY001
2.3. Dual Culture and Culture Filtrate Assays
2.4. Zinc and Phosphate Solubilization
2.5. Protease Activity
2.6. Siderophore Production
2.7. Ammonia (NH3) Production
2.8. Indole-3-Acetic Acid (IAA) Production
2.9. Nitrogen Fixation
2.10. Plant Growth Condition and Growth of FOL and AY001
2.11. AY001 Treatment and FOL Inoculation
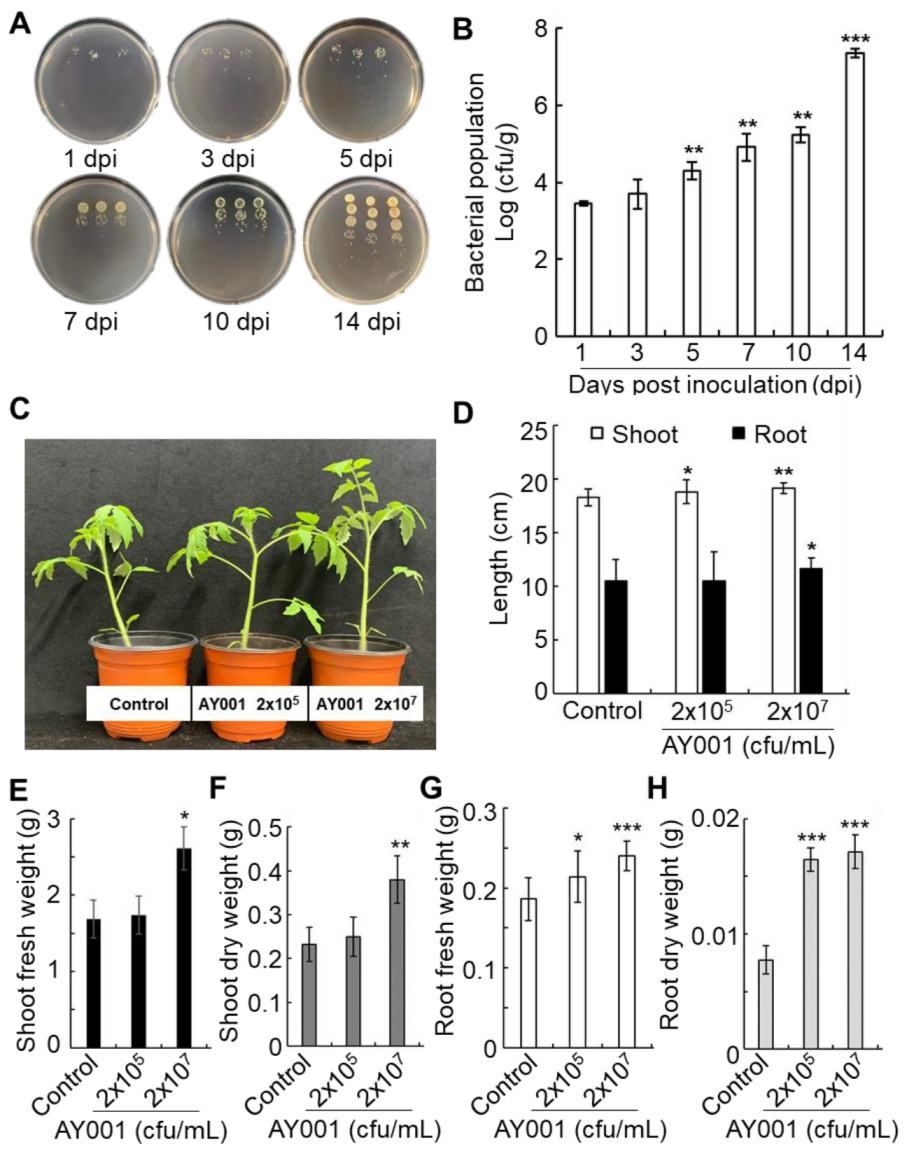
2.12. Root Colonization Assay
2.13. Pst DC3000 Inoculation and Bacterial Growth in Tomato Plants
2.14. Real-Time qRT-PCR Analysis of Marker Gene Expression
2.15. Gas Chromatography-Mass Spectrometry (GC-MS) Analysis
3. Results
3.1. Isolation and Identification of Burkholderia Conataminans AY001
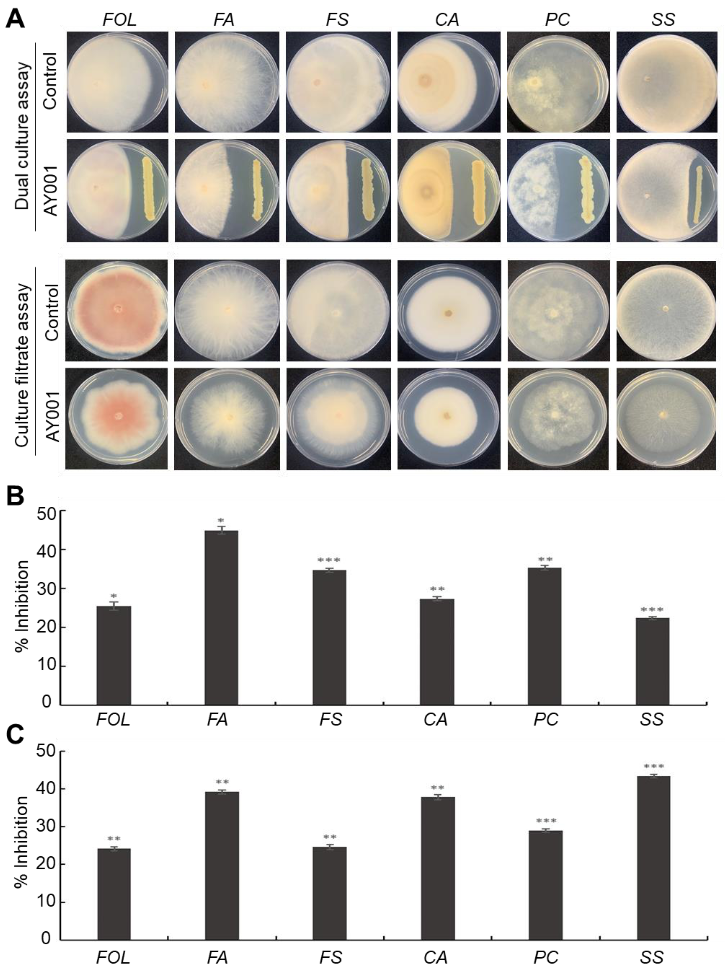
3.2. In Vitro Antagonistic Activity Assay
3.3. Plant Growth Promoting Rhizobacteria (PGPR)-Related Traits of AY001
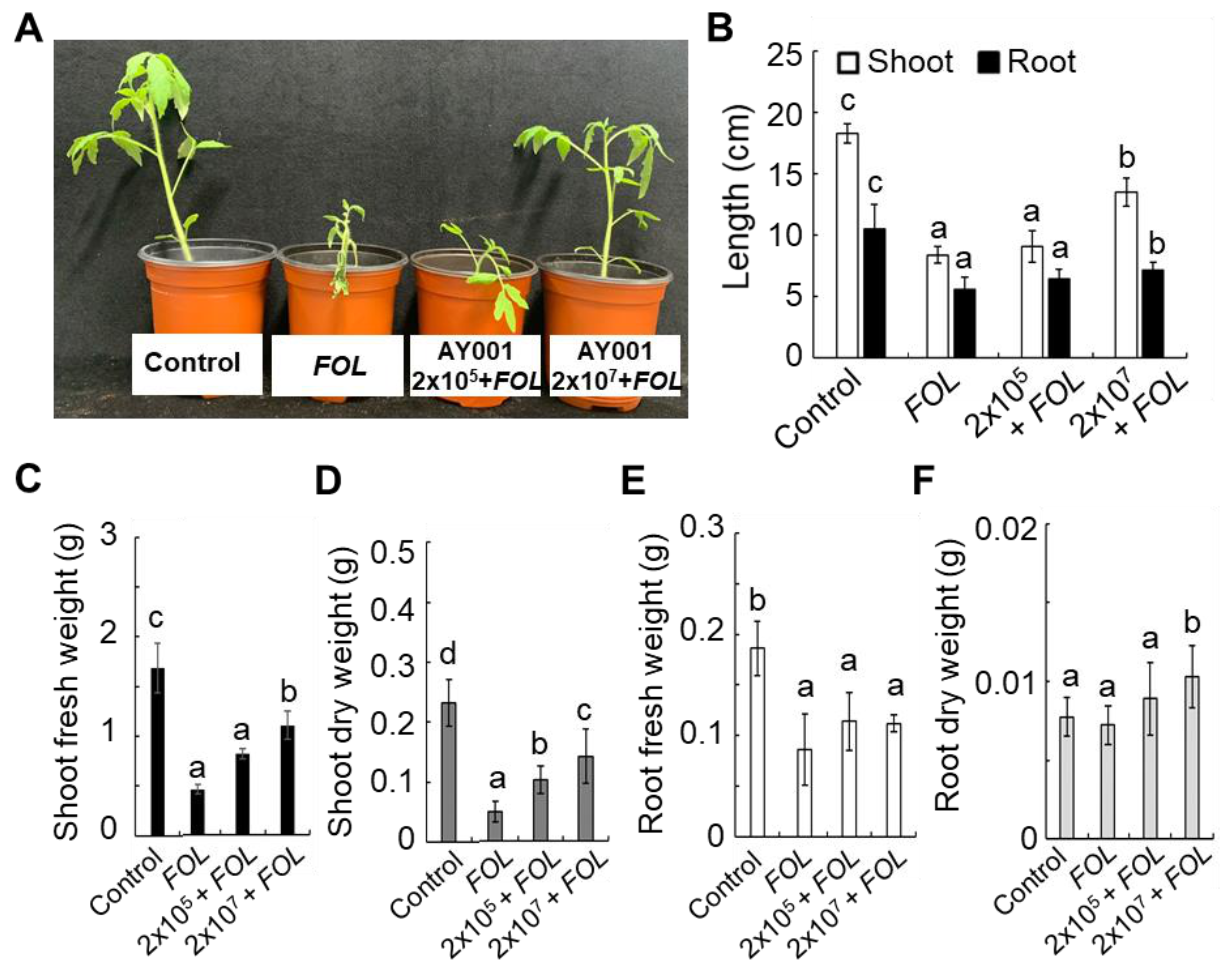
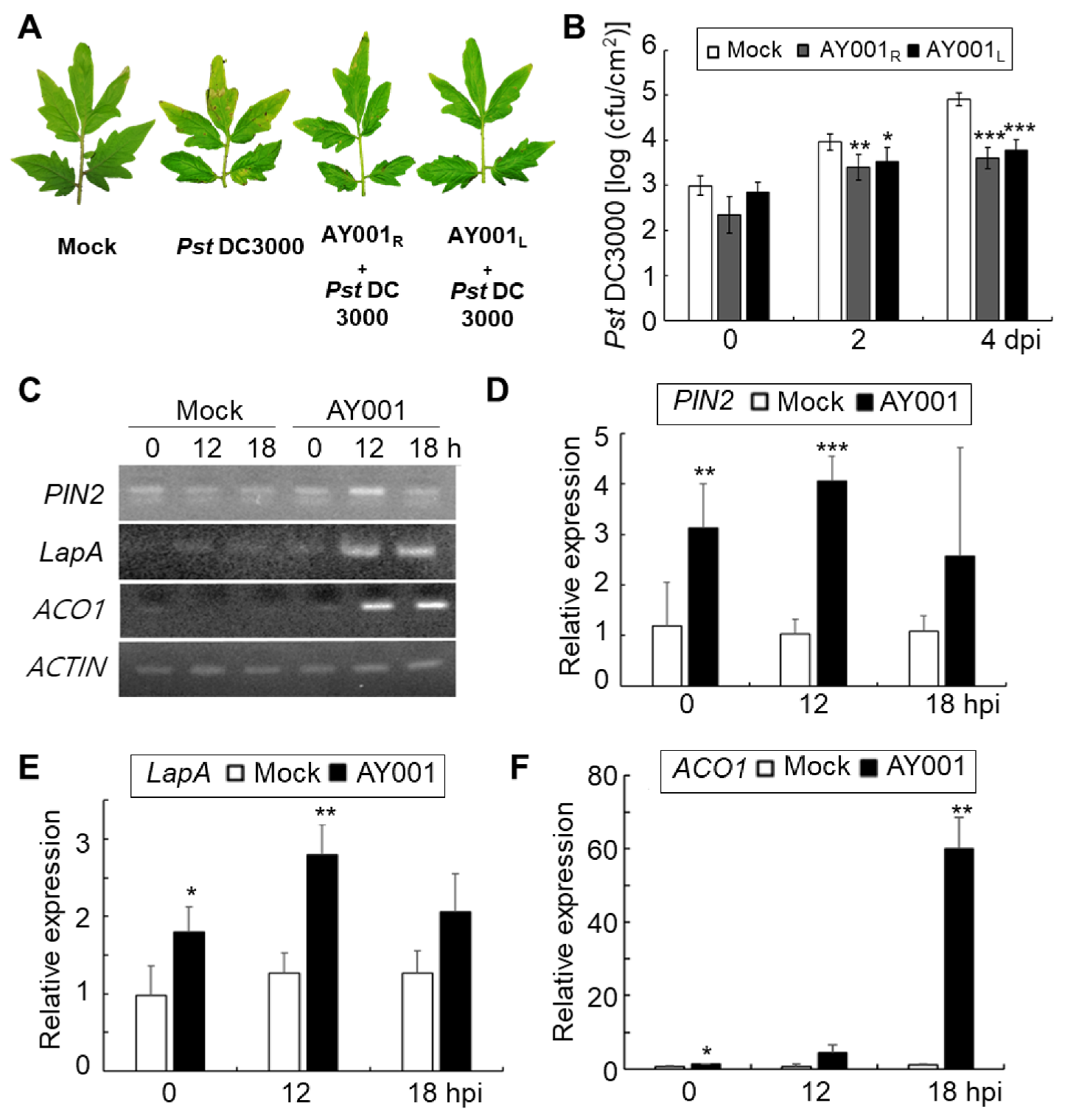
3.4. PGPR and Biocontrol Activities of AY001 in Tomato Plants
3.5. ISR-Inducing Activity of AY001 in Tomato Plants
3.6. Gas Chromatography-Mass Spectrometry (GC-MS) Analysis
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Stout, M.J.; Kurabchew, H.; Leite, G.L.D. Host-Plant Resistance in Tomato. In Sustainable Management of Arthropod Pests of Tomato; Wakil, W., Brust, G.E., Perring, T.M., Eds.; Academic Press: New York, NY, USA, 2018; pp. 217–236. [Google Scholar]
- Singh, V.K.; Singh, H.B.; Upadhyay, R.S. Role of fusaric acid in the development of ‘Fusarium wilt’ symptoms in tomato: Physiological, biochemical and proteomic perspectives. Plant Physiol. Biochem. 2017, 118, 320–332. [Google Scholar] [CrossRef] [PubMed]
- Bodah, E.T. Root rot diseases in plants: A review of common causal agents and management strategies. Agri. Res. Technol. Open Access J. 2017, 5, 555661. [Google Scholar] [CrossRef]
- Weller, D.M.; Raaijmakers, J.M.; Gardener, B.B.M.; Thomashow, L.S. Microbial populations responsible for specific soil suppressiveness to plant pathogens. Annu. Rev. Phytopathol. 2002, 40, 309–348. [Google Scholar] [CrossRef] [PubMed]
- Simonetti, E.; Roberts, I.N.; Monteccha, M.S.; Gutierrez-Boem, F.H.; Gomez, F.M.; Ruiz, A. A novel Burkholderia ambifaria strain able to degrade the mycotoxin fusaric acid and to inhibit Fusarium spp. growth. Microbiol. Res. 2018, 206, 50–59. [Google Scholar] [CrossRef]
- Abro, M.A.; Sun, X.; Li, X.; Jatoi, G.H.; Guo, L. Biocontrol Potential of Fungal Endophytes against Fusarium oxysporum f. sp. cucumerinum Causing Wilt in Cucumber. Plant Pathol. J. 2019, 35, 598–608. [Google Scholar] [CrossRef]
- Choi, H.W.; Ahsan, S.M. Biocontrol Activity of Aspergillus terreus ANU-301 against Two Distinct Plant Diseases, Tomato Fusarium Wilt and Potato Soft Rot. Plant Pathol. J. 2022, 38, 33–45. [Google Scholar] [CrossRef]
- Ting, A.S.Y.; Mah, S.W.; Tee, C.S. Evaluating the feasibility of induced host resistance by endophytic isolate Penicillium citrinum BTF08 as a control mechanism for Fusarium wilt in banana plantlets. Biol. Control 2012, 61, 155–159. [Google Scholar] [CrossRef]
- Ahemad, M.; Kibret, M. Mechanisms and applications of plant growth promoting rhizobacteria: Current perspective. J. King Saud Univ.-Sci. 2014, 26, 1–20. [Google Scholar] [CrossRef]
- Swarnalakshmi, K.; Yadav, V.; Tyagi, D.; Dhar, D.W.; Kannepalli, A.; Kumar, S. Significance of Plant Growth Promoting Rhizobacteria in Grain Legumes: Growth Promotion and Crop Production. Plants 2020, 9, 1596. [Google Scholar] [CrossRef]
- Wei, G.; Kloepper, J.W.; Tuzun, S. Induction of systemic resistance of cucumber to Colletotrichum orbiculare by select strains of plant growth promoting rhizobacteria. Phytopathology 1991, 81, 1508–1512. [Google Scholar] [CrossRef]
- Choudhary, D.K.; Prakash, A.; Johri, B. Induced systemic resistance (ISR) in plants: Mechanism of action. Indian J. Microbiol. 2007, 47, 289–297. [Google Scholar] [CrossRef]
- Gerber, I.B.; Zeidler, D.; Durner, J.; Dubery, I.A. Early perception responses of Nicotiana tabacum cells in response to lipopolysaccharides from Bulkholderia cepacia. Planta 2004, 218, 647–657. [Google Scholar] [CrossRef]
- Choudhary, D.K.; Johri, B.N. Interactions of Bacillus spp. and plants-With special reference to induced systemic resistance (ISR). Microbiol. Res. 2009, 164, 493–513. [Google Scholar] [CrossRef]
- Bordiec, S.; Paquis, S.; Lacroix, H.; Dhondt-Cordelier, S.; Barka, E.A.; Kauffmann, S.; Jeandet, P.; Mazeyrat-Gourbeyre, F.; Clément, C.; Baillieul, F.; et al. Comparative analysis of defence responses induced by the endophytic plant growth-promoting rhizobacterium Burkholderia phytofirmans strain PsJN and the non-host bacterium Pseudomonas syringae pv. pisi in grapevine cell suspensions. J. Exp. Bot. 2011, 62, 595–603. [Google Scholar] [CrossRef]
- Coenye, T.; Vandamme, P. Diversity and significance of Burkholderia species occupying diverse ecological niches. Environ. Microbiol. 2003, 5, 719–729. [Google Scholar] [CrossRef]
- Compant, S.; Nowak, J.; Coenye, T.; Clément, C.; Barka, E.A. Diversity and occurrence of Burkholderia spp. in the natural environment. FEMS Microbiol. Rev. 2008, 32, 607–626. [Google Scholar] [CrossRef]
- Esmaeel, Q.; Jacquard, C.; Sanchez, L.; Clément, C.; Barka, E.A. The mode of action of plant associated Burkholderia against grey mould disease in grapevine revealed through traits and genomic analyses. Sci. Rep. 2020, 10, 1–14. [Google Scholar] [CrossRef]
- Deng, P.; Wang, X.; Baird, S.M.; Showmaker, K.C.; Smith, L.; Peterson, D.G.; Lu, S. Comparative genome-wide analysis reveals that Burkholderia contaminans MS14 possesses multiple antimicrobial biosynthesis genes but not major genetic loci required for pathogenesis. MicrobiologyOpen 2016, 5, 353–369. [Google Scholar] [CrossRef]
- Nunvar, J.; Kalferstova, L.; Bloodworth, R.A.M.; Kolář, M.; Degrossi, J.; Lubovich, S.; Cardona, S.; Drevinek, P. Understanding the Pathogenicity of Burkholderia contaminans, an Emerging Pathogen in Cystic Fibrosis. PLoS ONE 2016, 11, e0160975. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular Evolutionary Genetics Analysis across computing platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef]
- Fasim, F.; Ahmed, N.; Parsons, R.; Gadd, G.M. Solubilization of zinc salts by a bacterium isolated from the air environment of a tannery. FEMS Microbiol. Lett. 2002, 213, 1–6. [Google Scholar] [CrossRef]
- Nautiyal, C.S. An efficient microbiological growth medium for screening phosphate solubilizing microorganisms. FEMS Microbiol. Lett. 2006, 170, 265–270. [Google Scholar] [CrossRef]
- Virpiranta, H.; Banasik, M.; Taskila, S.; Leiviskä, T.; Halttu, M.; Sotaniemi, V.-H.; Tanskanen, J. Isolation of Efficient Metal-Binding Bacteria from Boreal Peat Soils and Development of Microbial Biosorbents for Improved Nickel Scavenging. Water 2020, 12, 2000. [Google Scholar] [CrossRef]
- Goswami, D.; Parmar, S.; Vaghela, H.; Dhandhukia, P.; Thakker, J.N. Describing Paenibacillus mucilaginosus strain N3 as an efficient plant growth promoting rhizobacteria (PGPR). Cogent. Food Agric. 2015, 1, 1000714. [Google Scholar] [CrossRef]
- Gordon, S.A.; Weber, R.P. Colorimetric estimation of indole acetic acid. Plant Physiol. 1951, 26, 192–195. [Google Scholar] [CrossRef] [PubMed]
- Estrada-De Los Santos, P.; Bustillos-Cristales, R.; Caballero-Mellado, J. Burkholderia, a Genus Rich in Plant-Associated Nitrogen Fixers with Wide Environmental and Geographic Distribution. Appl. Environ. Microbiol. 2001, 67, 2790–2798. [Google Scholar] [CrossRef] [PubMed]
- Hossain, M.M.; Sultana, F.; Kubota, M.; Hyakumachi, M. Differential inducible defense mechanisms against bacterial speck pathogen in Arabidopsis thaliana by plant-growth-promoting-fungus Penicillium sp. GP16-2 and its cell free filtrate. Plant Soil 2008, 304, 227–239. [Google Scholar] [CrossRef]
- Steindler, L.; Bertani, I.; De Sordi, L.; Schwager, S.; Eberl, L.; Venturi, V. LasI/R and RhlI/R quorum sensing in a strain of Pseudomonas aeruginosa beneficial to plants. Appl. Environ. Microbiol. 2009, 75, 5131–5140. [Google Scholar] [CrossRef]
- Parke, J.L.; Gurian-Sherman, D. Diversity of the Burkholderia cepacia complex and implications for risk assessment of biological control strains. Annu. Rev. Phytopathol. 2001, 39, 225–258. [Google Scholar] [CrossRef]
- Sharma, S.B.; Sayyed, R.Z.; Trivedi, M.H.; Gobi, T.A. Phosphate solubilizing microbes: Sustainable approach for managing phosphorus deficiency in agricultural soils. SpringerPlus 2013, 2, 587. [Google Scholar] [CrossRef]
- Estrada-De Los Santos, P.; Rojas-Rojas, F.U.; Tapia-García, E.Y.; Vásquez-Murrieta, M.S.; Hirsch, A.M. To split or not to split: An opinion on dividing the genus Burkholderia. Ann. Microbiol. 2016, 66, 1303–1314. [Google Scholar] [CrossRef]
- Tagele, S.B.; Kim, S.W.; Lee, H.G.; Kim, H.S.; Lee, Y.S. Effectiveness of multi-trait Burkholderia contaminans KNU17BI1 in growth promotion and management of banded leaf and sheath blight in maize seedling. Microbiol. Res. 2018, 214, 8–18. [Google Scholar] [CrossRef]
- Liu, W.H.; Chen, F.F.; Wang, C.E.; Fu, H.H.; Fang, X.Q.; Ye, J.R.; Shi, J.Y. Indole-3-Acetic Acid in Burkholderia pyrrocinia JK-SH007: Enzymatic Identification of the Indole-3-Acetamide Synthesis Pathway. Front. Microbiol. 2019, 10, 2559. [Google Scholar] [CrossRef]
- Ren, J.H.; Ye, J.R.; Liu, H.; Xu, X.L.; Wu, X.Q. Isolation and characterization of a new Burkholderia pyrrocinia strain JK-SH007 as a potential biocontrol agent. World J. Microbiol. Biotechnol. 2011, 27, 2203–2215. [Google Scholar] [CrossRef]
- Wang, X.; Shi, J.; Wang, R. Effect of Burkholderia contaminans on Postharvest Diseases and Induced Resistance of Strawberry Fruits. Plant Pathol. J. 2018, 34, 403–411. [Google Scholar] [CrossRef]
- Jung, B.K.; Hong, S.-J.; Park, G.-S.; Kim, M.-C.; Shin, J.-H. Isolation of Burkholderia cepacian JBK9 with plant growth-promoting activity while producing pyrrolnitrin antagonistic to plant fungal diseases. Appl. Biol. Chem. 2018, 61, 173–180. [Google Scholar] [CrossRef]
- Pieterse, C.M.; Zamioudis, C.; Berendsen, R.L.; Weller, D.M.; Van Wees, S.C.M.; Bakker, P.A.H.M. Induced systemic resistance by beneficial microbes. Annu. Rev. Phytopathol. 2014, 52, 347–375. [Google Scholar] [CrossRef]
- Klessig, D.F.; Choi, H.W.; Dempsey, D.M.A. Systemic acquired resistance and salicylic acid: Past, present, and future. Mol. Plant Microbe Interact. 2018, 31, 871–888. [Google Scholar] [CrossRef]
- Koo, Y.M.; Heo, A.Y.; Choi, H.W. Salicylic Acid as a Safe Plant Protector and Growth Regulator. Plant Pathol. J. 2020, 36, 1–10. [Google Scholar] [CrossRef]
- Farmer, E.E.; Ryan, C.A. Octadecanoid precursors of jasmonic acid activate the synthesis of wound-inducible proteinase inhibitors. Plant Cell 1992, 4, 129–134. [Google Scholar] [CrossRef]
- Song, Y.; Chen, D.; Lu, K.; Sun, Z.; Zeng, R. Enhanced tomato disease resistance primed by arbuscular mycorrhizal fungus. Front. Plant Sci. 2015, 6, 786. [Google Scholar] [CrossRef]
- Fowler, J.H.; Narváez-Vásquez, J.; Aromdee, D.N.; Pautot, V.; Holzer, F.M.; Wallinga, L.L. Leucine Aminopeptidase Regulates Defense and Wound Signaling in Tomato Downstream of Jasmonic Acid. Plant Cell 2009, 21, 1239–1251. [Google Scholar] [CrossRef]
- Dimopoulou, A.; Theologidis, I.; Liebmann, B.; Kalantidis, K.; Vassilakos, N.; Skandalis, N. Bacillus amyloliquefaciens MBI600 differentially induces tomato defense signaling pathways depending on plant part and dose of application. Sci. Rep. 2019, 9, 19120. [Google Scholar] [CrossRef]
- Houben, M.; Van de Poel, B. 1-Aminocyclopropane-1-Carboxylic Acid Oxidase (ACO): The Enzyme That Makes the Plant Hormone Ethylene. Front. Plant Sci. 2019, 10, 695. [Google Scholar] [CrossRef]
- Klaus, J.R.; Coulon, P.M.L.; Koirala, P.; Seyedsayamdost, M.R.; Déziel, E.; Chandler, J.R. Secondary metabolites from the Burkholderia pseudomallei complex: Structure, ecology, and evolution. J. Ind. Microbiol. Biotechnol. 2020, 47, 877–887. [Google Scholar] [CrossRef]
- Elshafie, H.S.; Camele, I. An Overview of Metabolic Activity, Beneficial and Pathogenic Aspects of Burkholderia Spp. Metabolites 2021, 11, 321. [Google Scholar] [CrossRef]
- Ahuactzin-Pérez, M.; Tlecuitl-Beristain, S.; García-Dávila, J.; Santacruz-Juárez, E.; González-Pérez, M.; Gutiérrez-Ruíz, M.C.; Sánchez, C. A novel biodegradation pathway of the endocrine-disruptor di(2-ethyl hexyl) phthalate by Pleurotus ostreatus based on quantum chemical investigation. Ecotoxicol. Environ. Saf. 2018, 147, 494–499. [Google Scholar] [CrossRef]
- Habib, M.R.; Karim, M.R. Antimicrobial and cytotoxic activity of di-(2-ethylhexyl) phthalate and anhydrosophoradiol-3-acetate isolated from Calotropis gigantea (Linn.) Flower. Mycobiology 2009, 37, 31–36. [Google Scholar] [CrossRef] [PubMed]
- Nshimiyimana, J.B.; Khadka, S.; Zou, P.; Adhikari, S.; Proshad, R.; Thapa, A.; Xiong, L. Study on biodegradation kinetics of di-2-ethylhexyl phthalate by newly isolated halotolerant Ochrobactrum anthropic strain L1-W. BMC Res. Notes 2020, 13, 252. [Google Scholar] [CrossRef] [PubMed]
- Dilika, F.; Bremner, P.D.; Meyer, J.J. Antibacterial activity of linoleic and oleic acids isolated from Helichrysum pedunculatum: A plant used during circumcision rites. Fitoterapia 2000, 71, 450–452. [Google Scholar] [CrossRef]
- Kachroo, A.; Fu, D.; Havens, W.; Navarre, D.; Kachroo, P.; Ghabrial, S.A. An oleic acid-mediated pathway induces constitutive defense signaling and enhanced resistance to multiple pathogens in soybean. Mol. Plant Microbe. Interact. 2008, 21, 564–575. [Google Scholar] [CrossRef]
- Wang, E.; Liu, X.; Si, Z.; Li, X.; Bi, J.; Dong, W.; Chen, M.; Wang, S.; Zhang, J.; Song, A.; et al. Volatile Organic Compounds from Rice Rhizosphere Bacteria Inhibit Growth of the Pathogen Rhizoctonia solani. Agriculture 2021, 11, 368. [Google Scholar] [CrossRef]
- Zaman, N.R.; Chowdhury, U.F.; Reza, R.N.; Chowdhury, F.T.; Sarker, M.; Hossain, M.M.; Akbor, A.; Amin, A.; Islam, M.R.; Khan, H. Plant growth promoting endophyte Burkholderia contaminans NZ antagonizes phytopathogen Macrophomina phaseolina through melanin synthesis and pyrrolnitrin inhibition. PLoS ONE 2021, 16, e0257863. [Google Scholar] [CrossRef]
- Park, H.B.; Lee, B.; Kloepper, J.W.; Ryu, C. One shot-two pathogens blocked: Exposure of Arabidopsis to hexadecane, a long chain volatile organic compound, confers induced resistance against both Pectobacterium carotovorum and Pseudomonas syringae. Plant Signal. Behav. 2013, 8, e24619. [Google Scholar] [CrossRef]
- Kannabiran, K. Bioactivity of Pyrrolo[1,2-a]pyrazine-1,4-dione,hexahydro-3-(phenylmethyl)- Extracted from Streptomyces sp. VITPK9 Isolated from the Salt Spring Habitat of Manipur, India. Asian J. Pharm. 2016, 10, 265–270. [Google Scholar] [CrossRef]
- Putra, M.Y.; Karim, F. Antibacterial and Antioxidant Activity-Guided Isolation Studies on Fusarium sp. Associated with the Ascidian Botryllus schlosseri. In AIP Conference Proceedings; AIP Publishing LLC: Melville, NY, USA, 2020; Volume 2243, p. 020019. [Google Scholar] [CrossRef]
- Al-Rubaye, A.F.; Kadhim, M.J.; Hameed, I.H. Characterization of Antifungal Secondary Metabolites Produced by Klebsiella pneumoniae and Screening of its Chemical CompoundsUsing GC-MS. Int. J. Curr. Pharm. 2017, 8, 141–148. [Google Scholar] [CrossRef][Green Version]
- Milling, A.; Babujee, L.; Allen, C. Ralstonia solanacearum Extracellular Polysaccharide Is a Specific Elicitor of Defense Responses in Wilt-Resistant Tomato Plants. PLoS ONE 2011, 6, e15853. [Google Scholar] [CrossRef]
- Li, X.; Huang, L.; Zhang, Y.; Ouyang, Z.; Hong, Y.; Zhang, H.; Li, D.; Song, F. Tomato SR/CAMTA transcription factors SlSR1 and SlSR3L negatively regulate disease resistance response and SlSR1L positively modulates drought stress tolerance. BMC Plant Biol. 2014, 14, 286. [Google Scholar] [CrossRef]
- Jia, C.; Zhang, L.; Liu, L.; Wang, J.; Li, C.; Wang, Q. Multiple phytohormone signalling pathways modulate susceptibility of tomato plants to Alternaria alternata f. sp. lycopersici. J. Exp. Bot. 2013, 64, 637–650. [Google Scholar] [CrossRef]


| Peak No. | Compound | Structure | Formula | Molecular Weight | Retention Time (min) |
|---|---|---|---|---|---|
| 28 | Di(2-ethylhexyl) phthalate | 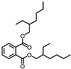 | C24H38O4 | 390.277 | 24.695 |
| 23 | Octadec-9-enoic acid | 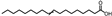 | C18H34O2 | 282.256 | 21.344 |
| 6 | 2-Ethyl-1-hexanol |  | C8H18O | 130.136 | 7.834 |
| 26 | (Z)-9-Octadecenamide | 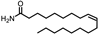 | C18H35NO | 281.272 | 23.318 |
| 17 | Hexadecane |  | C16H34 | 226.266 | 15.807 |
| Peak No. | Compound | Structure | Formula | Molecular Weight | Retention Time (min) |
|---|---|---|---|---|---|
| 43 | Pyrrolo[1,2-a]pyrazine-1,4-dione, hexahydro-3-(phenylmethyl) | 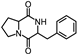 | C14H16N2O2 | 244.121 | 23.552 |
| 33 | Pyrrolo[1,2-a]pyrazine-1,4-dione, hexahydro-3-(2-methylpropyl)- | 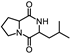 | C11H18N2O2 | 210.137 | 19.418 |
| 32 | L-Proline, N-pivaloyl-, ethyl ester | 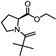 | C12H21NO3 | 227.152 | 19.25 |
| 31 | 2,4(1H,3H)-Pyrimidinedione, 1,3,6-trimethyl- | 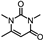 | C7H10N2O2 | 154.074 | 18.255 |
| 21 | N-Benzyl-N-ethyl-p-isopropylbenzamide | 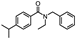 | C19H23NO | 281.178 | 14.102 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Heo, A.Y.; Koo, Y.M.; Choi, H.W. Biological Control Activity of Plant Growth Promoting Rhizobacteria Burkholderia contaminans AY001 against Tomato Fusarium Wilt and Bacterial Speck Diseases. Biology 2022, 11, 619. https://doi.org/10.3390/biology11040619
Heo AY, Koo YM, Choi HW. Biological Control Activity of Plant Growth Promoting Rhizobacteria Burkholderia contaminans AY001 against Tomato Fusarium Wilt and Bacterial Speck Diseases. Biology. 2022; 11(4):619. https://doi.org/10.3390/biology11040619
Chicago/Turabian StyleHeo, A Yeong, Young Mo Koo, and Hyong Woo Choi. 2022. "Biological Control Activity of Plant Growth Promoting Rhizobacteria Burkholderia contaminans AY001 against Tomato Fusarium Wilt and Bacterial Speck Diseases" Biology 11, no. 4: 619. https://doi.org/10.3390/biology11040619
APA StyleHeo, A. Y., Koo, Y. M., & Choi, H. W. (2022). Biological Control Activity of Plant Growth Promoting Rhizobacteria Burkholderia contaminans AY001 against Tomato Fusarium Wilt and Bacterial Speck Diseases. Biology, 11(4), 619. https://doi.org/10.3390/biology11040619