An Insight into Abiotic Stress and Influx Tolerance Mechanisms in Plants to Cope in Saline Environments
Abstract
Simple Summary
Abstract
Contents:
- ▪
- An overview of abiotic stress; effects of salinity stress on crop growth, development, and yield
- ▪
- Salinity—a major limiting factor in the ecosystem and an inhibitor of plant growth
- ▪
- Alkaline Salinity (high pH)
- ▪
- Classifications of plants; Glycophytes (salt-sensitive plants) and halophytes (salt-resistant plants) are two salinities (salt-tolerant plants)
- ▪
- Impact of salinity on photosynthesis and stomatal conductance
- ▪
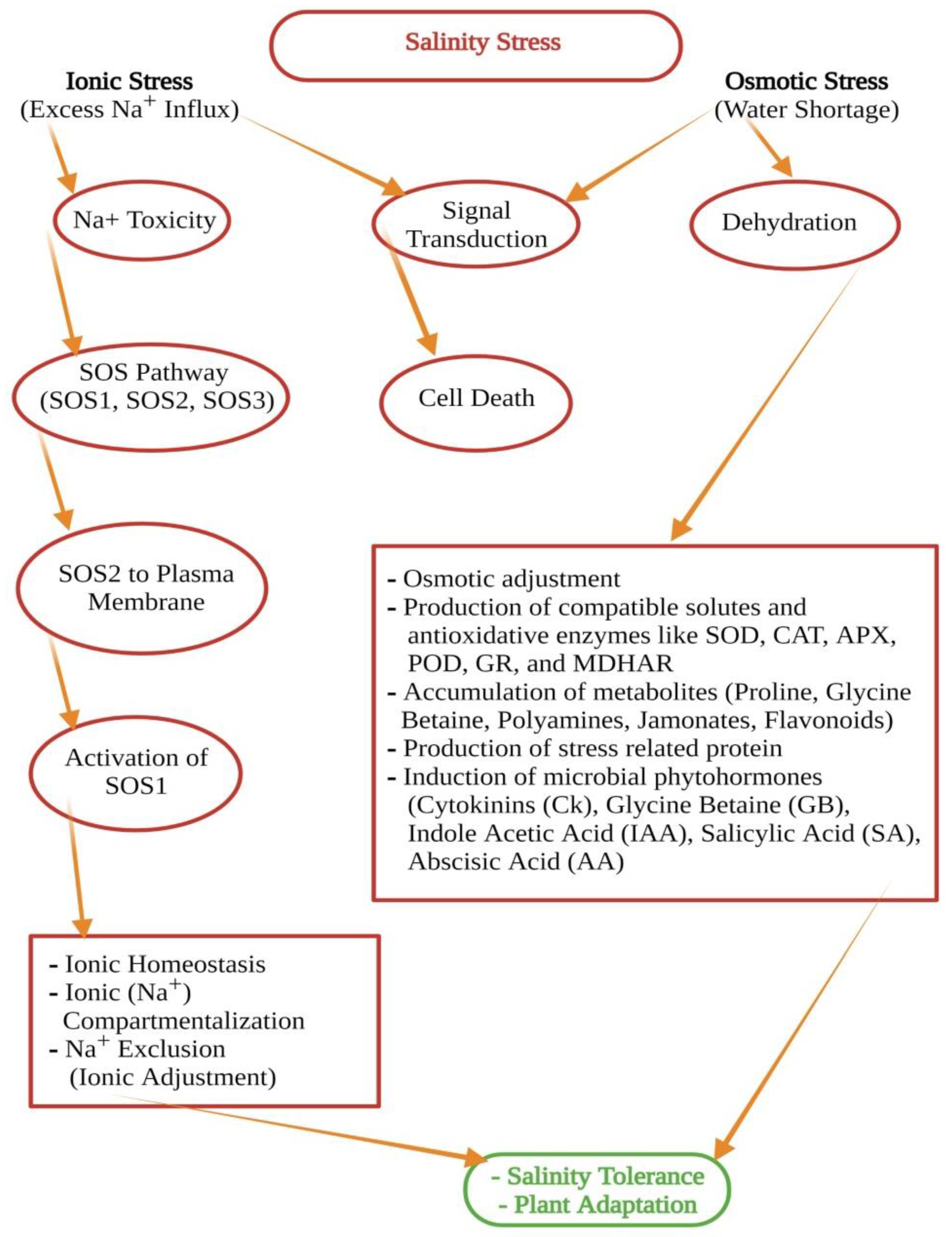
- Causes of soil salinity; signal transduction and ionic homeostasis under salt stress (leading to osmotic, ionic, and oxidative stresses)
- ▪
- SOS pathway (salt overly sensitive pathway)—sensing salt stress in plants
- ▪
- Channels involved in Na+ ion regulation—HKT (High-affinity K+ channel), NSCC (Non-selective cation channel), AKT1 (Arabidopsis K+ Transporter1), NORC (nonselective outward-rectifying conductance), VIC (Voltage-independent channel)
- ▪
- Metabolic profiling; osmolyte production in plants under salt stress; proline, glycine betaine, abscisic acid, jasmonates, flavonoids; plants and rhizosphere microbial activities in response to various stress conditions.
- ▪
- Conclusions and future perspectives.
1. Content Description
1.1. An Overview of Abiotic Stress; Soil Salinization—A Major Environmental Constraint and Plant Growth Inhibitor
1.2. Alkaline Salinity
1.3. Classification of Plants Based on Salinity: Glycophytes (Salt-Sensitive Plants) and Halophytes (Salt-Tolerant Plants)
1.4. Impact of Salinity on Photosynthesis and Stomatal Conductance
1.5. Signal Transduction and Ionic Homeostasis; Osmotic and Ionic Stress
1.6. SOS Pathway (Salt Overly Sensitive Pathway)—Sensing Salt Stress in Plants
1.7. Different Channels Involved in Na+ Regulation
1.8. Metabolic Profiling; Osmolyte Production in Plants during Salt Stress
1.9. Salinity Stress and Proline (A Crucial and Multifunctional Amino Acid That Can Affect Plant Growth as Well as Stress Responses)
1.10. Salinity Stress and Glycine Betaine (An Effective Protectant against Abiotic Stresses in Plants)
1.11. Salinity Stress and Abscisic Acid (A Ubiquitous Plant-Stress Hormone)
1.12. Jasmonates (Lipid-Derived Plant Stress Hormones)
1.13. Polyphenols/Flavonoids (Important Secondary Metabolites and Bio-Compounds in Plants)
2. Conclusions and Future Perspectives
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Shrivastava, P.; Kumar, R. Soil Salinity: A Serious Environmental Issue and Plant Growth Promoting Bacteria as One of the Tools for Its Alleviation. Saudi J. Biol. Sci. 2015, 22, 123–131. [Google Scholar] [CrossRef] [PubMed]
- Alvarez Rogel, J.; Alcaraz Ariza, F.; Ortiz Silla, R. Soil Salinity and Moisture Gradients and Plant Zonation in Mediterranean Salt Marshes of Southeast Spain. Wetlands 2000, 20, 357–372. [Google Scholar] [CrossRef]
- Rubio, J.S.; García-Sánchez, F.; Rubio, F.; Martínez, V. Yield, Blossom-End Rot Incidence, and Fruit Quality in Pepper Plants under Moderate Salinity Are Affected by K+ and Ca2+ Fertilization. Sci. Hortic. 2009, 119, 79–87. [Google Scholar] [CrossRef]
- Sonowal, H.; Pal, P.B.; Shukla, K.; Ramana, K.V. Aspalatone Prevents VEGF-Induced Lipid Peroxidation, Migration, Tube Formation, and Dysfunction of Human Aortic Endothelial Cells. Oxid. Med. Cell. Longev. 2017, 2017, 2769347. [Google Scholar] [CrossRef] [PubMed]
- Bockheim, J.G.; Gennadiyev, A.N. The Role of Soil-Forming Processes in the Definition of Taxa in Soil Taxonomy and the World Soil Reference Base. Geoderma 2000, 95, 53–72. [Google Scholar] [CrossRef]
- Hanin, M.; Ebel, C.; Ngom, M.; Laplaze, L.; Masmoudi, K. New Insights on Plant Salt Tolerance Mechanisms and Their Potential Use for Breeding. Front. Plant Sci. 2016, 7, 1787. [Google Scholar] [CrossRef]
- Ondrasek, G.; Rengel, Z. Environmental Salinization Processes: Detection, Implications & Solutions. Sci. Total Environ. 2021, 754, 142432. [Google Scholar] [CrossRef]
- Munns, R.; Tester, M. Mechanisms of Salinity Tolerance. Annu. Rev. Plant Biol. 2008, 59, 651–681. [Google Scholar] [CrossRef]
- Ismail, A.; Takeda, S.; Nick, P. Life and Death under Salt Stress: Same Players, Different Timing? J. Exp. Bot. 2014, 65, 2963–2979. [Google Scholar] [CrossRef]
- Flowers, T.J.; Colmer, T.D. Salinity Tolerance in Halo-Phytes. New Phytol. 2008, 179, 945–963. [Google Scholar] [CrossRef]
- Khan, N.; Bano, A.; Babar, M.A. The Stimulatory Effects of Plant Growth Promoting Rhizobacteria and Plant Growth Regulators on Wheat Physiology Grown in Sandy Soil. Arch. Microbiol. 2019, 201, 769–785. [Google Scholar] [CrossRef] [PubMed]
- Al-Farsi, S.M.; Nawaz, A.; Anees-Ur-Rehman; Nadaf, S.K.; Al-Sadi, A.M.; Siddique, K.H.M.; Farooq, M. Effects, Tolerance Mechanisms and Management of Salt Stress in Lucerne (Medicago sativa). Crop Pasture Sci. 2020, 71, 411–428. [Google Scholar] [CrossRef]
- Eynard, A.; Lal, R.; Wiebe, K. Crop Response in Salt-Affected Soils. J. Sustain. Agric. 2005, 27, 5–50. [Google Scholar] [CrossRef]
- Fang, S.; Hou, X.; Liang, X. Response Mechanisms of Plants Under Saline-Alkali Stress. Front. Plant Sci. 2021, 12, 1–20. [Google Scholar] [CrossRef] [PubMed]
- Liu, L.; Wang, Y.; Gai, Z.; Liu, D.; Wu, P.; Wang, B.; Zou, C.; Li, C.; Yang, F. Responses of Soil Microorganisms and Enzymatic Activities to Alkaline Stress in Sugar Beet Rhizosphere. Polish J. Environ. Stud. 2020, 29, 739–748. [Google Scholar] [CrossRef]
- Yu, S.; Yu, L.; Hou, Y.; Zhang, Y.; Guo, W.; Xue, Y. Contrasting Effects of NaCl and NaHCO3 Stresses on Seed Germination, Seedling Growth, Photosynthesis, and Osmoregulators of the Common Bean (Phaseolus vulgaris L.). Agronomy 2019, 9, 409. [Google Scholar] [CrossRef]
- Zou, C.; Wang, Y.; Wang, B.; Liu, D.; Liu, L.; Gai, Z.; Li, C. Long Non-Coding RNAs in the Alkaline Stress Response in Sugar Beet (Beta vulgaris L.). BMC Plant Biol. 2020, 20, 227. [Google Scholar] [CrossRef]
- Geng, G.; Wang, G.; Stevanato, P.; Lv, C.; Wang, Q.; Yu, L.; Wang, Y. Physiological and Proteomic Analysis of Different Molecular Mechanisms of Sugar Beet Response to Acidic and Alkaline PH Environment. Front. Plant Sci. 2021, 12, 682799. [Google Scholar] [CrossRef] [PubMed]
- Yolcu, S.; Alavilli, H.; Ganesh, P.; Asif, M.; Kumar, M.; Song, K. An Insight into the Abiotic Stress Responses of Cultivated Beets (Beta vulgaris L.). Plants 2022, 11, 12. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Tang, X.; Shao, C.; Shao, H.; Wang, H. Molecular Cloning and Bioinformatics Analysis of a New Plasma Membrane Na+/H+ Antiporter Gene from the Halophyte Kosteletzkya virginica. Sci. World J. 2014, 2014, 141675. [Google Scholar] [CrossRef]
- Greenway, H.; Munns, R. Mechanisms of Salt Tolerance in Nonhalophytes. Annu. Rev. Plant Physiol. 1980, 31, 149–190. [Google Scholar] [CrossRef]
- Ahmad, R.; Jamil, S.; Shahzad, M.; Zörb, C.; Irshad, U.; Khan, N.; Younas, M.; Khan, S.A. Metabolic Profiling to Elucidate Genetic Elements Due to Salt Stress. Clean–Soil Air Water 2017, 45, 1600574. [Google Scholar] [CrossRef]
- Muchate, N.S.; Nikalje, G.C.; Rajurkar, N.S.; Suprasanna, P.; Nikam, T.D. Plant Salt Stress: Adaptive Responses, Tolerance Mechanism and Bioengineering for Salt Tolerance. Bot. Rev. 2016, 82, 371–406. [Google Scholar] [CrossRef]
- Meng, X.; Zhou, J.; Sui, N. Mechanisms of Salt Tolerance in Halophytes: Current Understanding and Recent Advances. Open Life Sci. 2018, 13, 149–154. [Google Scholar] [CrossRef] [PubMed]
- Ashraf, M.; Harris, P.J.C. Photosynthesis under Stressful Environments: An Overview. Photosynthetica 2013, 51, 163–190. [Google Scholar] [CrossRef]
- Shabala, S. Learning from Halophytes: Physiological Basis and Strategies to Improve Abiotic Stress Tolerance in Crops. Ann. Bot. 2013, 112, 1209–1221. [Google Scholar] [CrossRef]
- Shabala, S.; Bose, J.; Hedrich, R. Salt Bladders: Do They Matter? Trends Plant Sci. 2014, 19, 687–691. [Google Scholar] [CrossRef]
- Slama, I.; Abdelly, C.; Bouchereau, A.; Flowers, T.; Savouré, A. Diversity, Distribution and Roles of Osmoprotective Compounds Accumulated in Halophytes under Abiotic Stress. Ann. Bot. 2015, 115, 433–447. [Google Scholar] [CrossRef]
- Sevin, D.C.; Stählin, J.N.; Pollak, G.R.; Kuehne, A.; Sauer, U. Global Metabolic Responses to Salt Stress in Fifteen Species. PLoS ONE 2016, 11, e0148888. [Google Scholar] [CrossRef]
- Bohm, J.; Messerer, M.; Müller, H.M.; Scholz-Starke, J.; Gradogna, A.; Scherzer, S.; Maierhofer, T.; Bazihizina, N.; Zhang, H.; Stigloher, C.; et al. Understanding the Molecular Basis of Salt Sequestration in Epidermal Bladder Cells of Chenopodium Quinoa. Curr. Biol. 2018, 28, 3075–3085.e7. [Google Scholar] [CrossRef]
- Zou, C.; Chen, A.; Xiao, L.; Muller, H.M.; Ache, P.; Haberer, G.; Zhang, M.; Jia, W.; Deng, P.; Huang, R.; et al. A High-Quality Genome Assembly of Quinoa Provides Insights into the Molecular Basis of Salt Bladder-Based Salinity Tolerance and the Exceptional Nutritional Value. Cell Res. 2017, 27, 1327–1340. [Google Scholar] [CrossRef] [PubMed]
- Lonard, R.I.; Judd, F.W.; Summy, K.R.; Deyoe, H.; Stalter, R. The Biological Flora of Coastal Dunes and Wetlands: Avicennia germinans (L.) L. J. Coast. Res. 2017, 33, 191–207. [Google Scholar] [CrossRef]
- Shahid, M.A.; Balal, R.M.; Khan, N.; Rossi, L.; Rathinasabapathi, B.; Liu, G.; Khan, J.; Cámara-Zapata, J.M.; Martínez-Nicolas, J.J.; Garcia-Sanchez, F. Polyamines Provide New Insights into the Biochemical Basis of Cr-Tolerance in Kinnow Mandarin Grafted on Diploid and Double-Diploid Rootstocks. Environ. Exp. Bot. 2018, 156, 248–260. [Google Scholar] [CrossRef]
- Zorb, C.; Geilfus, C.M.; Dietz, K.J. Salinity and Crop Yield. Plant Biol. 1982, 21, 31–38. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Ma, H.; Chen, T.; Pen, J.; Yu, S.; Zhaoe, X. Morphological and Physiological Responses of Cotton (Gossypium hirsutum L.) Plants to Salinity. PLoS ONE 2014, 9, e112807. [Google Scholar] [CrossRef] [PubMed]
- Tuteja, N. Mechanisms of High Salinity Tolerance in Plants. Methods Enzymol. 2007, 428, 419–438. [Google Scholar] [CrossRef] [PubMed]
- Batista-Santos, P.; Duro, N.; Rodrigues, A.P.; Semedo, J.N.; Alves, P.; da Costa, M.; Graça, I.; Pais, I.P.; Scotti-Campos, P.; Lidon, F.C.; et al. Is Salt Stress Tolerance in Casuarina Glauca Sieb. Ex Spreng. Associated with Its Nitrogen-Fixing Root-Nodule Symbiosis? An Analysis at the Photosynthetic Level. Plant Physiol. Biochem. 2015, 96, 97–109. [Google Scholar] [CrossRef]
- Author, C.; Tawaha, A.M.; Othman, Y.; Al-Karaki, G.; Al-Tawaha, A.R.; Al-Horani, A. Variation in Germination and Ion Uptake in Barley Genotypes under Salinity Conditions. World J. Agric. Sci. 2006, 2, 11–15. [Google Scholar]
- Khan, M.A.; Rizvi, Y. Effect of Salinity, Temperature, and Growth Regulators on the Germination and Early Seedling Growth of Atriplex Griffithii Var. Stocksii. Can. J. Bot. 1994, 72, 475–479. [Google Scholar] [CrossRef]
- Kandil, A.A.; Shareif, E.; Gad, A.M. Effect of Salinity on Germination and Seeding Parameters of Forage Cowpea Seed. Res. J. Seed Sci. 2016, 10, 17–26. [Google Scholar] [CrossRef]
- Rajjou, L.; Duval, M.; Gallardo, K.; Catusse, J.; Bally, J.; Job, C.; Job, D. Seed Germination and Vigor. Annu. Rev. Plant Biol. 2012, 63, 507–533. [Google Scholar] [CrossRef] [PubMed]
- Meena, K.K.; Sorty, A.M.; Bitla, U.M.; Choudhary, K.; Gupta, P.; Pareek, A.; Singh, D.P.; Prabha, R.; Sahu, P.K.; Gupta, V.K.; et al. Abiotic Stress Responses and Microbe-Mediated Mitigation in Plants: The Omics Strategies. Front. Plant Sci. 2017, 8, 172. [Google Scholar] [CrossRef] [PubMed]
- Stepien, P.; Kłbus, G. Water Relations and Photosynthesis in Cucumis Sativus L. Leaves under Salt Stress. Biol. Plant. 2006, 50, 610–616. [Google Scholar] [CrossRef]
- Puniran-Hartley, N.; Hartley, J.; Shabala, L.; Shabala, S. Salinity-Induced Accumulation of Organic Osmolytes in Barley and Wheat Leaves Correlates with Increased Oxidative Stress Tolerance: In Planta Evidence for Cross-Tolerance. Plant Physiol. Biochem. 2014, 83, 32–39. [Google Scholar] [CrossRef] [PubMed]
- Khan, N.; Bano, A. Effects of Exogenously Applied Salicylic Acid and Putrescine Alone and in Combination with Rhizobacteria on the Phytoremediation of Heavy Metals and Chickpea Growth in Sandy Soil. Int. J. Phytoremediat. 2018, 20, 405–414. [Google Scholar] [CrossRef]
- Baxter, A.; Mittler, R.; Suzuki, N. ROS as Key Players in Plant Stress Signalling. J. Exp. Bot. 2014, 65, 1229–1240. [Google Scholar] [CrossRef]
- Tripathy, B.C.; Oelmüller, R. Reactive Oxygen Species Generation and Signaling in Plants. Plant Signal. Behav. 2012, 7, 1621–1633. [Google Scholar] [CrossRef]
- Floryszak-Wieczorek, J.; Górski, Z.; Arasimowicz-Jelonek, M. Reactive Oxygen Species: Metabolism, Oxidative Stress, and Signal Transduction. Eur. J. plant Pathol. 2011, 130, 373–399. [Google Scholar] [CrossRef]
- Ma, Y.; Wang, L.; Wang, J.; Zhong, Y.; Cheng, Z.M. Isolation and Expression Analysis of Salt Overly Sensitive Gene Family in Grapevine (Vitisvinifera) in Response to Salt and PEG Stress. PLoS ONE 2019, 14, e0212666. [Google Scholar] [CrossRef]
- Ding, D.; Liu, M.; Arif, M.; Yuan, Z.; Li, J.; Hu, X.; Li, C. Responses of Ecological Stoichiometric Characteristics of Carbon, Nitrogen, and Phosphorus to Periodic Submergence in Mega-Reservoir: Growth of Taxodium distichum and Taxodium ascendens. Plants 2021, 10, 2040. [Google Scholar] [CrossRef]
- Miller, G.; Suzuki, N.; Ciftci-Yilmaz, S.; Mittler, R. Reactive Oxygen Species Homeostasis and Signalling during Drought and Salinity Stresses. Plant Cell Environ. 2010, 33, 453–467. [Google Scholar] [CrossRef] [PubMed]
- Cheeseman, J.M. The Integration of Activity in Saline Environments: Problems and Perspectives. Funct. Plant Biol. 2013, 40, 759–774. [Google Scholar] [CrossRef] [PubMed]
- Bose, J.; Munns, R.; Shabala, S.; Gilliham, M.; Pogson, B.; Tyerman, S.D. Chloroplast Function and Ion Regulation in Plants Growing on Saline Soils: Lessons from Halophytes. J. Exp. Bot. 2017, 68, 3129–3143. [Google Scholar] [CrossRef] [PubMed]
- Van Zelm, E.; Zhang, Y.; Testerink, C. Salt Tolerance Mechanisms of Plants. Annu. Rev. Plant Biol. 2020, 71, 403–433. [Google Scholar] [CrossRef]
- Hasanuzzaman, M.; Bhuyan, M.H.M.B.; Nahar, K.; Hossain, M.S.; Al Mahmud, J.; Hossen, M.S.; Masud, A.A.C.; Moumita; Fujita, M. Potassium: A Vital Regulator of Plant Responses and Tolerance to Abiotic Stresses. Agronomy 2018, 8, 31. [Google Scholar] [CrossRef]
- Gao, Q.; Cai, Z. Comparative Physiological and Biochemical Mechanisms of Salt Tolerance in Five Contrasting Highland Quinoa Cultivars. BMC Plant Biol. 2019, 20, 70. [Google Scholar] [CrossRef]
- Choi, W.G.; Toyota, M.; Kim, S.H.; Hilleary, R.; Gilroy, S. Salt Stress-Induced Ca2+ Waves Are Associated with Rapid, Long-Distance Root-to-Shoot Signaling in Plants. Proc. Natl. Acad. Sci. USA 2014, 111, 6497–6502. [Google Scholar] [CrossRef]
- Duan, L.; Dietrich, D.; Ng, C.H.; Chan, Y.P.M.; Bhalerao, R.; Bennett, M.J.; Dinneny, J.R. Endodermal ABA Signaling Promotes Lateral Root Quiescence during Salt Stress in Arabidopsis Seedlings. Plant Cell 2013, 25, 324–341. [Google Scholar] [CrossRef]
- Geilfus, C.-M.; Mithöfer, A.; Ludwig-Müller, J.; Zörb, C.; Muehling, K.H. Chloride- Inducible Transient Apoplastic Alkalinizations Induce Stomata Closure by Controlling Abscisic Acid Distribution between Leaf Apoplast and Guard Cells in Salt- Stressed Vicia Faba. New Phytol. 2015, 208, 802–816. [Google Scholar] [CrossRef]
- Flowers, T.J.; Munns, R.; Colmer, T.D. Sodium Chloride Toxicity and the Cellular Basis of Salt Tolerance in Halophytes. Ann. Bot. 2015, 115, 419–431. [Google Scholar] [CrossRef]
- Nxele, X.; Klein, A.; Ndimba, B.K. Drought and Salinity Stress Alters ROS Accumulation, Water Retention, and Osmolyte Content in Sorghum Plants. S. Afr. J. Bot. 2017, 108, 261–266. [Google Scholar] [CrossRef]
- Golldack, D.; Li, C.; Mohan, H.; Probst, N. Tolerance to Drought and Salt Stress in Plants: Unraveling the Signaling Networks. Front. Plant Sci. 2014, 5, 151. [Google Scholar] [CrossRef] [PubMed]
- Naseem, H.; Ahsan, M.; Shahid, M.A.; Khan, N. Exopolysaccharides Producing Rhizobacteria and Their Role in Plant Growth and Drought Tolerance. J. Basic Microbiol. 2018, 58, 1009–1022. [Google Scholar] [CrossRef] [PubMed]
- Zekri, M.; Parsons, L.R. Calcium Influences Growth and Leaf Mineral Concentration of Citrus under Saline Conditions. HortScience 2019, 25, 784–786. [Google Scholar] [CrossRef]
- Kumari, A.; Das, P.; Parida, A.K.; Agarwal, P.K. Proteomics, Metabolomics, and Ionomics Perspectives of Salinity Tolerance in Halophytes. Front. Plant Sci. 2015, 6, 537. [Google Scholar] [CrossRef]
- Bose, J.; Rodrigo-Moreno, A.; Lai, D.; Xie, Y.; Shen, W.; Shabala, S. Rapid Regulation of the Plasma Membrane H+-ATPase Activity Is Essential to Salinity Tolerance in Two Halophyte Species, Atriplex Lentiformis and Chenopodium Quinoa. Ann. Bot. 2015, 115, 481–494. [Google Scholar] [CrossRef]
- Lin, H.; Yang, Y.; Quan, R.; Mendoza, I.; Wu, Y.; Du, W.; Zhao, S.; Schumaker, K.S.; Pardo, J.M.; Guo, Y. Phosphorylation of SOS3-like Calcium Binding Protein8 by SOS2 Protein Kinase Stabilizes Their Protein Complex and Regulates Salt Tolerance in Arabidopsis. Plant Cell 2009, 21, 1607–1619. [Google Scholar] [CrossRef]
- Ji, H.; Pardo, J.M.; Batelli, G.; Van Oosten, M.J.; Bressan, R.A.; Li, X. The Salt Overly Sensitive (SOS) Pathway: Established and Emerging Roles. Mol. Plant 2013, 6, 275–286. [Google Scholar] [CrossRef]
- Assaha, D.V.M.; Ueda, A.; Saneoka, H.; Al-Yahyai, R.; Yaish, M.W. The Role of Na+ and K+ Transporters in Salt Stress Adaptation in Glycophytes. Front. Physiol. 2017, 8, 509. [Google Scholar] [CrossRef]
- Quintero, F.J.; Ohta, M.; Shi, H.; Zhu, J.K.; Pardo, J.M. Reconstitution in Yeast of the Arabidopsis SOS Signaling Pathway for Na+ Homeostasis. Proc. Natl. Acad. Sci. USA 2002, 99, 9061–9066. [Google Scholar] [CrossRef]
- Oh, D.H.; Dassanayake, M.; Haas, J.S.; Kropornika, A.; Wright, C.; d’Urzo, M.P.; Hong, H.; Ali, S.; Hernandez, A.; Lambert, G.M.; et al. Genome Structures and Halophyte-Specific Gene Expression of the Extremophile Thellungiella Parvula in Comparison with Thellungiella Salsuginea (Thellungiella halophila) and Arabidopsis. Plant Physiol. 2010, 154, 1040–1052. [Google Scholar] [CrossRef] [PubMed]
- Qiu, Q.S.; Guo, Y.; Dietrich, M.A.; Schumaker, K.S.; Zhu, J.K. Regulation of SOS1, a Plasma Membrane Na+/H+ Exchanger in Arabidopsis Thaliana, by SOS2 and SOS3. Proc. Natl. Acad. Sci. USA 2002, 99, 8436–8441. [Google Scholar] [CrossRef] [PubMed]
- Guo, K.M.; Babourina, O.; Rengel, Z. Na+/H+ Antiporter Activity of the SOS1 Gene: Lifetime Imaging Analysis and Electrophysiological Studies on Arabidopsis Seedlings. Physiol. Plant. 2009, 137, 155–165. [Google Scholar] [CrossRef] [PubMed]
- Brindha, C.; Vasantha, S.; Raja, A.K.; Tayade, A.S. Characterization of the Salt Overly Sensitive Pathway Genes in Sugarcane under Salinity Stress. Physiol. Plant. 2020, 171, 677–687. [Google Scholar] [CrossRef] [PubMed]
- El Mahi, H.; Hormaeche, J.P.; De Luca, A.; Villalta, I.; Espartero, J.; Arjona, F.G.; Fernández, J.L.; Bundó, M.; Mendoza, I.; Mieulet, D.; et al. A Critical Role of Sodium Flux via the Plasma Membrane Na+/H+ Exchanger SOS1 in the Salt Tolerance of Rice. Plant Physiol. 2019, 180, 1046–1065. [Google Scholar] [CrossRef]
- Yadav, N.S.; Shukla, P.S.; Jha, A.; Agarwal, P.K.; Jha, B. The SbSOS1 Gene from the Extreme Halophyte Salicornia Brachiata Enhances Na+ Loading in Xylem and Confers Salt Tolerance in Transgenic Tobacco. BMC Plant Biol. 2012, 12, 188. [Google Scholar] [CrossRef] [PubMed]
- Katschnig, D.; Bliek, T.; Rozema, J.; Schat, H. Constitutive High-Level SOS1 Expression and Absence of HKT1;1 Expression in the Salt-Accumulating Halophyte Salicornia Dolichostachya. Plant Sci. 2015, 234, 144–154. [Google Scholar] [CrossRef]
- Rezaei Moshaei, M.; Nematzadeh, G.A.; Askari, H.; Mozaffari Nejad, A.S.; Pakdin, A. Quantitative Gene Expression Analysis of Some Sodium Ion Transporters under Salinity Stress in Aeluropus littoralis. Saudi J. Biol. Sci. 2014, 21, 394–399. [Google Scholar] [CrossRef]
- Jannesar, M.; Razavi, K.; Saboora, A. Effects of Salinity on Expression of the Salt Overly Sensitive Genes in Aeluropus lagopoides. Aust. J. Crop Sci. 2014, 8, 1–8. [Google Scholar]
- Xia, T.; Apse, M.P.; Aharon, G.S.; Blumwald, E. Identification and Characterization of a NaCl-Inducible Vacuolar Na+/H+ Antiporter in Beta Vulgaris. Physiol. Plant. 2002, 116, 206–212. [Google Scholar] [CrossRef]
- Yuan, Z.; Ni, X.; Arif, M.; Dong, Z.; Zhang, L.; Tan, X.; Li, C. Transcriptomic Analysis of the Photosynthetic, Respiration, and Aerenchyma Adaptation Strategies in Bermudagrass (Cynodon dactylon) under Different Submergence Stress. Int. J. Mol. Sci. 2021, 22, 7905. [Google Scholar] [CrossRef] [PubMed]
- Suarez, N.; Medina, E. Salinity Effects on Leaf Ion Composition and Salt Secretion Rate in Avicennia germinans (L.) L. Braz. J. Plant Physiol. 2008, 20, 131–140. [Google Scholar] [CrossRef]
- Akcin, A.; Yalcin, E. Effect of Salinity Stress on Chlorophyll, Carotenoid Content, and Proline in Salicornia Prostrata Pall. and Suaeda Prostrata Pall. Subsp. Prostrata (Amaranthaceae). Rev. Bras. Bot. 2016, 39, 101–106. [Google Scholar] [CrossRef]
- Chakraborty, K.; Sairam, R.K.; Bhattacharya, R.C. Differential Expression of Salt Overly Sensitive Pathway Genes Determines Salinity Stress Tolerance in Brassica Genotypes. Plant Physiol. Biochem. 2012, 51, 90–101. [Google Scholar] [CrossRef]
- Nutan, K.K.; Kumar, G.; Singla-Pareek, S.L.; Pareek, A. A Salt Overly Sensitive Pathway Member from Brassica Juncea BjSOS3 Can Functionally Complement ΔAtsos3 in Arabidopsis. Curr. Genom. 2017, 19, 60–69. [Google Scholar] [CrossRef] [PubMed]
- Martinez-Atienza, J.; Jiang, X.; Garciadeblas, B.; Mendoza, I.; Zhu, J.K.; Pardo, J.M.; Quintero, F.J. Conservation of the Salt Overly Sensitive Pathway in Rice. Plant Physiol. 2007, 143, 1001–1012. [Google Scholar] [CrossRef]
- Wu, J.; Zhang, X.; Nie, Y.; Jin, S.; Liang, S. Factors Affecting Somatic Embryogenesis and Plant Regeneration from a Range of Recalcitrant Genotypes of Chinese Cottons (Gossypium Hirsutum L.). Vitr. Cell. Dev. Biol. Plant 2004, 40, 371–375. [Google Scholar] [CrossRef]
- Sandhu, D.; Pudussery, M.V.; Kaundal, R.; Suarez, D.L.; Kaundal, A.; Sekhon, R.S. Molecular Characterization and Expression Analysis of the Na+/H+ Exchanger Gene Family in Medicago Truncatula. Funct. Integr. Genomics 2018, 18, 141–153. [Google Scholar] [CrossRef]
- De Souza Miranda, R.; Mesquita, R.O.; Costa, J.H.; Alvarez-Pizarro, J.C.; Prisco, J.T.; Gomes-Filho, E. Integrative Control between Proton Pumps and SOS1 Antiporters in Roots Is Crucial for Maintaining Low Na+ Accumulation and Salt Tolerance in Ammonium-Supplied Sorghum Bicolor. Plant Cell Physiol. 2017, 58, 522–536. [Google Scholar] [CrossRef]
- Zorb, C.; Noll, A.; Karl, S.; Leib, K.; Yan, F.; Schubert, S. Molecular Characterization of Na+/H+ Antiporters (ZmNHX) of Maize (Zea mays L.) and Their Expression under Salt Stress. J. Plant Physiol. 2005, 162, 55–66. [Google Scholar] [CrossRef]
- Kotula, L.; Garcia Caparros, P.; Zörb, C.; Colmer, T.D.; Flowers, T.J. Improving Crop Salt Tolerance Using Transgenic Approaches: An Update and Physiological Analysis. Plant Cell Environ. 2020, 43, 2932–2956. [Google Scholar] [CrossRef] [PubMed]
- Ma, D.M.; Xu, W.R.; Li, H.W.; Jin, F.X.; Guo, L.N.; Wang, J.; Dai, H.J.; Xu, X. Co-Expression of the Arabidopsis SOS Genes Enhances Salt Tolerance in Transgenic Tall Fescue (Festuca arundinacea Schreb.). Protoplasma 2014, 251, 219–231. [Google Scholar] [CrossRef] [PubMed]
- Hu, X.; Xie, T.; Arif, M.; Ding, D.; Li, J.; Yuan, Z.; Li, C. Response of Annual Herbaceous Plant Leaching and Decomposition to Periodic Submergence in Mega-Reservoirs: Changes in Litter Nutrients and Soil Properties for Restoration. Biology 2021, 10, 1141. [Google Scholar] [CrossRef] [PubMed]
- Platten, J.D.; Cotsaftis, O.; Berthomieu, P.; Zhu, J.-K.; Dennis, E.S.; Tester, M. Nomenclature for HKT Transporters, Key Determinants of Plant Salinity Tolerance. Trends Plant Sci. 2006, 11, 372–374. [Google Scholar] [CrossRef]
- Møller, I.S.; Gilliham, M.; Jha, D.; Mayo, G.M.; Roy, S.; Coates, J.; Haseloff, J.; Tester, M. Shoot Na+ Exclusion and Increased Salinity Tolerance engineered by Cell Type-Specific Alteration of Na+ Transport in Arabidopsis. Plant Cell 2009, 21, 2163–2178. [Google Scholar] [CrossRef]
- Tester, M.; Davenport, R. Na+ Tolerance and Na+ Transport in Higher Plants. Ann. Bot. 2003, 91, 503–527. [Google Scholar] [CrossRef]
- Hasegawa, P.M.; Rus, A.; Yokoi, S.; Sharkhuu, A.; Reddy, M.; Lee, B.H.; Matsumoto, T.K.; Koiwa, H.; Zhu, J.K.; Bressan, R.A. AtHKT1 Is a Salt Tolerance Determinant That Controls Na+ Entry into Plant Roots. Proc. Natl. Acad. Sci. USA 2001, 98, 14150–14155. [Google Scholar] [CrossRef]
- Asins, M.J.; Villalta, I.; Aly, M.M.; Olías, R.; Álvarez De Morales, P.; Huertas, R.; Li, J.; Jaime-Pérez, N.; Haro, R.; Raga, V.; et al. Two Closely Linked Tomato HKT Coding Genes Are Positional Candidates for the Major Tomato QTL Involved in Na+/K+ Homeostasis. Plant Cell Environ. 2013, 36, 1171–1191. [Google Scholar] [CrossRef]
- Ariyarathna, H.A.C.K.; Oldach, K.H.; Francki, M.G. A Comparative Gene Analysis with Rice Identified Orthologous Group II HKT Genes and Their Association with Na+ Concentration in Bread Wheat. BMC Plant Biol. 2016, 16, 21. [Google Scholar] [CrossRef]
- Munns, R.; James, R.A.; Xu, B.; Athman, A.; Conn, S.J.; Jordans, C.; Byrt, C.S.; Hare, R.A.; Tyerman, S.D.; Tester, M.; et al. Wheat Grain Yield on Saline Soils Is Improved by an Ancestral Na+ Transporter Gene. Nat. Biotechnol. 2012, 30, 360–364. [Google Scholar] [CrossRef]
- Hamamoto, S.; Horie, T.; Hauser, F.; Deinlein, U.; Schroeder, J.I.; Uozumi, N. HKT Transporters Mediate Salt Stress Resistance in Plants: From Structure and Function to the Field. Curr. Opin. Biotechnol. 2015, 32, 113–120. [Google Scholar] [CrossRef] [PubMed]
- Monti, L.L.; Bustamante, C.A.; Osorio, S.; Gabilondo, J.; Borsani, J.; Lauxmann, M.A.; Maulión, E.; Valentini, G.; Budde, C.O.; Fernie, A.R.; et al. Metabolic Profiling of a Range of Peach Fruit Varieties Reveals High Metabolic Diversity and Commonalities and Differences during Ripening. Food Chem. 2016, 190, 879–888. [Google Scholar] [CrossRef] [PubMed]
- Thouvenot, L.; Deleu, C.; Berardocco, S.; Haury, J.; Thiébaut, G. Characterization of the Salt Stress Vulnerability of Three Invasive Freshwater Plant Species Using a Metabolic Profiling Approach. J. Plant Physiol. 2015, 175, 113–121. [Google Scholar] [CrossRef] [PubMed]
- Sharma, A.; Kumar, D.; Kumar, S.; Rampuria, S.; Reddy, A.R.; Kirti, P.B. Ectopic Expression of an Atypical Hydrophobic Group 5 LEA Protein from Wild Peanut, Arachis Diogoi Confers Abiotic Stress Tolerance in Tobacco. PLoS ONE 2016, 11, e150609. [Google Scholar] [CrossRef]
- Wang, L.; Pan, D.; Lv, X.; Cheng, C.L.; Li, J.; Liang, W.; Xing, J.; Chen, W. A Multilevel Investigation to Discover Why Kandelia Candel Thrives in High Salinity. Plant Cell Environ. 2016, 39, 2486–2497. [Google Scholar] [CrossRef]
- Li, J.; Li, L.; Arif, M.; Ding, D.; Hu, X.; Zheng, J.; Li, C. Artificial Plantation Responses to Periodic Submergence in Massive Dam and Reservoir Riparian Zones: Changes in Soil Properties and Bacterial Community Characteristics. Biology 2021, 10, 819. [Google Scholar] [CrossRef]
- Liu, X.; Luo, Y.; Li, Z.; Wang, J.; Wei, G. Role of Exopolysaccharide in Salt Stress Resistance and Cell Motility of Mesorhizobium Alhagi CCNWXJ12–2T. Appl. Microbiol. Biotechnol. 2017, 101, 2967–2978. [Google Scholar] [CrossRef]
- Wang, L.M.; Zhang, L.D.; Chen, J.B.; Huang, D.F.; Zhang, Y.D. Physiological Analysis and Transcriptome Comparison of Two Muskmelon (Cucumis melo L.) Cultivars in Response to Salt Stress. Genet. Mol. Res. 2016, 15, 1–18. [Google Scholar] [CrossRef]
- Zhang, J.; Yang, D.; Li, M.; Shi, L. Metabolic Profiles Reveal Changes in Wild and Cultivated Soybean Seedling Leaves under Salt Stress. PLoS ONE 2016, 11, e0159622. [Google Scholar] [CrossRef]
- Wei, M.; Zhuang, Y.; Li, H.; Li, P.; Huo, H.; Shu, D.; Huang, W.; Wang, S. The Cloning and Characterization of Hypersensitive to Salt Stress Mutant, Affected in Quinolinate Synthase, Highlights the Involvement of NAD in Stress-Induced Accumulation of ABA and Proline. Plant J. 2020, 102, 85–98. [Google Scholar] [CrossRef]
- Suzuki, N.; Bassil, E.; Hamilton, J.S.; Inupakutika, M.A.; Zandalinas, S.I.; Tripathy, D.; Luo, Y.; Dion, E.; Fukui, G.; Kumazaki, A.; et al. ABA Is Required for Plant Acclimation to a Combination of Salt and Heat Stress. PLoS ONE 2016, 11, e0147625. [Google Scholar] [CrossRef]
- Yang, N.; Song, X.; Lu, X.; Chen, Q.; Liu, J.; Liu, Y.; Wang, H.; Zhang, Z.; Tang, Z. Comparative Study on Metabolites and Elements of Two Dominant Plant Communities in Saline-Alkali Grassland. Environ. Exp. Bot. 2021, 190, 104587. [Google Scholar] [CrossRef]
- Sharma, A.; Shahzad, B.; Kumar, V.; Kohli, S.K.; Sidhu, G.P.S.; Bali, A.S.; Handa, N.; Kapoor, D.; Bhardwaj, R.; Zheng, B. Phytohormones Regulate Accumulation of Osmolytes under Abiotic Stress. Biomolecules 2019, 9, 285. [Google Scholar] [CrossRef] [PubMed]
- Guo, Y.; Liu, Y.; Zhang, Y.; Liu, J.; Gul, Z.; Guo, X.R.; Abozeid, A.; Tang, Z.H. Effects of Exogenous Calcium on Adaptive Growth, Photosynthesis, Ion Homeostasis and Phenolics of Gleditsia Sinensis Lam. Plants under Salt Stress. Agriculture 2021, 11, 978. [Google Scholar] [CrossRef]
- Zhou, W.; Zhou, T.; Li, M.X.; Zhao, C.L.; Jia, N.; Wang, X.X.; Sun, Y.Z.; Li, G.L.; Xu, M.; Zhou, R.G.; et al. The Arabidopsis J-Protein AtDjB1 Facilitates Thermotolerance by Protecting Cells against Heat-Induced Oxidative Damage. New Phytol. 2012, 194, 364–378. [Google Scholar] [CrossRef] [PubMed]
- Yuan, T.T.; Xiang, Z.X.; Li, W.; Gao, X.; Lu, Y.T. Osmotic Stress Represses Root Growth by Modulating the Transcriptional Regulation of PIN-FORMED3. New Phytol. 2021, 232, 1661–1673. [Google Scholar] [CrossRef] [PubMed]
- Elhamid, E.M.A.; Sadak, M.S.; Tawfik, M.M. Alleviation of Adverse Effects of Salt Stress in Wheat Cultivars by Foliar Treatment with Antioxidant 2—Changes in Some Biochemical Aspects, Lipid Peroxidation, Antioxidant Enzymes and Amino Acid Contents. Agric. Sci. 2014, 5, 1269–1280. [Google Scholar] [CrossRef]
- Khan, N.; Zandi, P.; Ali, S.; Mehmood, A.; Shahid, M.A. Impact of Salicylic Acid and PGPR on the Drought Tolerance and Phytoremediation Potential of Helianthus Annus. Front. Microbiol. 2018, 9, 2507. [Google Scholar] [CrossRef]
- Yang, Y.; Guo, Y. Elucidating the Molecular Mechanisms Mediating Plant Salt-Stress Responses. New Phytol. 2018, 217, 523–539. [Google Scholar] [CrossRef]
- Meringer, M.V.; Villasuso, A.L.; Margutti, M.P.; Usorach, J.; Pasquare, S.J.; Giusto, N.M.; Machado, E.E.; Racagni, G.E. Saline and Osmotic Stresses Stimulate PLD/Diacylglycerol Kinase Activities and Increase the Level of Phosphatidic Acid and Proline in Barley Roots. Environ. Exp. Bot. 2016, 128, 69–78. [Google Scholar] [CrossRef]
- Hu, Y.; Xia, S.; Su, Y.; Wang, H.; Luo, W.; Su, S.; Xiao, L. Brassinolide Increases Potato Root Growth in Vitro in a Dose-Dependent Way and Alleviates Salinity Stress. Biom. Res. Int. 2016, 2016, 8231873. [Google Scholar] [CrossRef]
- He, X.; Wang, T.; Wu, K.; Wang, P.; Qi, Y.; Arif, M.; Wei, H. Responses of Swamp Cypress (Taxodium distichum) and Chinese Willow (Salix matsudana) Roots to Periodic Submergence in Mega-Reservoir: Changes in Organic Acid Concentration. Forests 2021, 12, 203. [Google Scholar] [CrossRef]
- Hare, P.D.; Cress, W.A.; Van Staden, J. Disruptive Effects of Exogenous Proline on Chloroplast and Mitochondrial Ultrastructure in Arabidopsis Leaves. S. Afr. J. Bot. 2002, 68, 393–396. [Google Scholar] [CrossRef]
- Chakraborty, K.; Bishi, S.K.; Goswami, N.; Singh, A.L.; Zala, P.V. Differential Fine-Regulation of Enzyme Driven ROS Detoxification Network Imparts Salt Tolerance in Contrasting Peanut Genotypes. Environ. Exp. Bot. 2016, 128, 79–90. [Google Scholar] [CrossRef]
- Borgo, L.; Marur, C.J.; Vieira, L.G.E. Effects of High Proline Accumulation on Chloroplast and Mitochondrial Ultrastructure and on Osmotic Adjustment in Tobacco Plants. Acta Sci. Agron. 2015, 37, 191. [Google Scholar] [CrossRef]
- Zheng, J.; Arif, M.; Zhang, S.; Yuan, Z.; Zhang, L.; Li, J.; Li, C. Dam inundation simplifies the plant community composition. Sci. Total Environ. 2021, 801, 149827. [Google Scholar] [CrossRef]
- Saibi, W.; Feki, K.; Ben Mahmoud, R.; Brini, F. Durum Wheat Dehydrin (DHN-5) Confers Salinity Tolerance to Transgenic Arabidopsis Plants through the Regulation of Proline Metabolism and ROS Scavenging System. Planta 2015, 242, 1187–1194. [Google Scholar] [CrossRef]
- Stavridou, E.; Hastings, A.; Webster, R.J.; Robson, P.R.H. The Impact of Soil Salinity on the Yield, Composition and Physiology of the Bioenergy Grass Miscanthus × Giganteus. GCB Bioenergy 2017, 9, 92–104. [Google Scholar] [CrossRef]
- Wahb-Allah, M.A.; Alsadon, A.A.; Sadder, M. Physiological, Chemical and Gene Expression Analyses under Salt Stress for Several Tomato Genotypes. Acta Hortic. 2016, 1145, 57–68. [Google Scholar] [CrossRef]
- Huang, Z.; Zhao, L.; Chen, D.; Liang, M.; Liu, Z.; Shao, H.; Long, X. Salt Stress Encourages Proline Accumulation by Regulating Proline Biosynthesis and Degradation in Jerusalem Artichoke Plantlets. PLoS ONE 2013, 8, e62085. [Google Scholar] [CrossRef]
- Long, R.; Li, M.; Zhang, T.; Kang, J.; Sun, Y.; Cong, L.; Gao, Y.; Liu, F.; Yang, Q. Comparative Proteomic Analysis Reveals Differential Root Proteins in Medicago Sativa and Medicago Truncatula in Response to Salt Stress. Front. Plant Sci. 2016, 7, 424. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Joseph, E.; Radhakrishnan, V.; Mohanan, K. A Study on the Accumulation of Proline-An Osmoprotectant Amino Acid under Salt Stress in Some Native Rice Cultivars of North Kerala, India. Univers. J. Agric. Res. 2015, 3, 15–22. [Google Scholar] [CrossRef]
- Ashraf, M. and Foolad, M.R. Roles of Glycinebetaine and Proline in Improving Plant Abiotic Stress Tolerance. Environ. Exp. Bot. 2007, 59, 206–216. [Google Scholar] [CrossRef]
- Kahlaoui, B.; Hachicha, M.; Misle, E.; Fidalgo, F.; Teixeira, J. Physiological and Biochemical Responses to the Exogenous Application of Proline of Tomato Plants Irrigated with Saline Water. J. Saudi Soc. Agric. Sci. 2018, 17, 17–23. [Google Scholar] [CrossRef]
- Nemoto, Y.; Sasakuma, T. Differential Stress Responses of Early Salt-Stress Responding Genes in Common Wheat. Phytochemistry 2002, 61, 129–133. [Google Scholar] [CrossRef]
- Khan, M.S.; Ahmad, D.; Khan, M.A. Utilization of Genes Encoding Osmoprotectants in Transgenic Plants for Enhanced Abiotic Stress Tolerance. Electron. J. Biotechnol. 2015, 18, 257–266. [Google Scholar] [CrossRef]
- Schertl, P.; Cabassa, C.; Saadallah, K.; Bordenave, M.; Savouré, A.; Braun, H.P. Biochemical Characterization of Proline Dehydrogenase in Arabidopsis Mitochondria. FEBS J. 2014, 281, 2794–2804. [Google Scholar] [CrossRef]
- Dar, M.I.; Naikoo, M.I.; Rehman, F.; Naushin, F.; Khan, F.A. Proline Accumulation in Plants: Roles in Stress Tolerance and Plant Development. Osmolytes Plants Acclim. to Chang. Environ. Emerg. Omi. Technol. 2015, 155–166. [Google Scholar] [CrossRef]
- Ali, B.; Hayat, S.; Ahmad, A. 28-Homobrassinolide Ameliorates the Saline Stress in Chickpea (Cicer arietinum L.). Environ. Exp. Bot. 2007, 59, 217–223. [Google Scholar] [CrossRef]
- Hasanuzzaman, M.; Alam, M.M.; Rahman, A.; Hasanuzzaman, M.; Nahar, K.; Fujita, M. Exogenous Proline and Glycine Betaine Mediated Upregulation of Antioxidant Defense and Glyoxalase Systems Provides Better Protection against Salt-Induced Oxidative Stress in Two Rice (Oryza sativa L.) Varieties. Biomed Res. Int. 2014, 2014, 757219. [Google Scholar] [CrossRef]
- Zeyner, A.; Romanowski, K.; Vernunft, A.; Harris, P.; Müller, A.M.; Wolf, C.; Kienzle, E. Effects of Different Oral Doses of Sodium Chloride on the Basal Acid-Base and Mineral Status of Exercising Horses Fed Low Amounts of Hay. PLoS ONE 2017, 12, e168325. [Google Scholar] [CrossRef]
- Gill, S.S.; Tuteja, N. Polyamines and Abiotic Stress Tolerance in Plants. Plant Signal. Behavior 2010, 5, 26–33. [Google Scholar] [CrossRef] [PubMed]
- Kumar, R.; Khurana, A.; Sharma, A.K. Role of Plant Hormones and Their Interplay in Development and Ripening of Fleshy Fruits. J. Exp. Bot. 2014, 65, 4561–4575. [Google Scholar] [CrossRef] [PubMed]
- Jaarsma, R.; de Vries, R.S.M.; de Boer, A.H. Effect of Salt Stress on Growth, Na+ Accumulation and Proline Metabolism in Potato (Solanum tuberosum) Cultivars. PLoS ONE 2013, 8, e60183. [Google Scholar] [CrossRef]
- Zhang, Y.; Yang, J.; Lu, S.; Cai, J.; Guo, Z. Overexpressing SgNCED1 in Tobacco Increases ABA Level, Antioxidant Enzyme Activities, and Stress Tolerance. J. Plant Growth Regul. 2008, 27, 151–158. [Google Scholar] [CrossRef]
- Park, H.J.; Kim, W.Y.; Yun, D.J. A New Insight of Salt Stress Signaling in Plant. Mol. Cells 2016, 39, 447–459. [Google Scholar] [CrossRef] [PubMed]
- Wutipraditkul, N.; Wongwean, P.; Buaboocha, T. Alleviation of Salt-Induced Oxidative Stress in Rice Seedlings by Proline and/or Glycinebetaine. Biol. Plant. 2015, 59, 547–553. [Google Scholar] [CrossRef]
- Hussain Wani, S.; Brajendra Singh, N.; Haribhushan, A.; Iqbal Mir, J. Compatible Solute Engineering in Plants for Abiotic Stress Tolerance-Role of Glycine Betaine. Curr. Genom. 2013, 14, 157–165. [Google Scholar] [CrossRef]
- Das, M.K.; Roychoudhury, A. ROS and Responses of Antioxidant as ROS-Scavengers during Environmental Stress in Plants. Front. Environ. Sci. 2014, 2, 800. [Google Scholar] [CrossRef]
- Chen, Z.; Arif, M.; Wang, C.; Chen, X.; Li, C. Effects of Hydrological Regime on Foliar Decomposition and Nutrient Release in the Riparian Zone of the Three Gorges Reservoir, China. Front. Plant Sci. 2021, 12, 661865. [Google Scholar] [CrossRef]
- Fita, A.; Rodríguez-Burruezo, A.; Boscaiu, M.; Prohens, J.; Vicente, O. Breeding and Domesticating Crops Adapted to Drought and Salinity: A New Paradigm for Increasing Food Production. Front. Plant Sci. 2015, 6, 978. [Google Scholar] [CrossRef] [PubMed]
- Hasthanasombut, S.; Ntui, V.; Supaibulwatana, K.; Mii, M.; Nakamura, I. Expression of Indica Rice OsBADH1 Gene under Salinity Stress in Transgenic Tobacco. Plant Biotechnol. Rep. 2010, 4, 75–83. [Google Scholar] [CrossRef]
- Lutts, S.; Lefèvre, I. How Can We Take Advantage of Halophyte Properties to Cope with Heavy Metal Toxicity in Salt-Affected Areas? Ann. Bot. 2015, 115, 509–528. [Google Scholar] [CrossRef] [PubMed]
- Nisar, M.; Khan, N.; Nausheen; Ahmad, Z.; Ghafoor, A. Genetic Diversity and Disease Response of Rust in Bread Wheat Collected from Waziristan Agency, Pakistan. Int. J. Biodivers. Conserv. 2011, 3, 10–18. [Google Scholar]
- Arif, M.; Zhang, S.; Jie, Z.; Charles, W.; Mzondi, P.S.; Li, C. Evaluating the Effects of Pressure Indicators on Riparian Zone Health Conditions in the Three Gorges Dam Reservoir, China. Forests 2020, 11, 214. [Google Scholar] [CrossRef]
- Hoque, M.A.; Banu, M.N.A.; Nakamura, Y.; Shimoishi, Y.; Murata, Y. Proline and Glycinebetaine Enhance Antioxidant Defense and Methylglyoxal Detoxification Systems and Reduce NaCl-Induced Damage in Cultured Tobacco Cells. J. Plant Physiol. 2008, 165, 813–824. [Google Scholar] [CrossRef]
- Osman, H.S. Enhancing Antioxidant–Yield Relationship of Pea Plant under Drought at Different Growth Stages by Exogenously Applied Glycine Betaine and Proline. Ann. Agric. Sci. 2015, 60, 389–402. [Google Scholar] [CrossRef]
- Khan, N.; Bano, A. Rhizobacteria and Abiotic Stress Management. In Plant Growth Promoting Rhizobacteria for Sustainable Stress Management; Springer: Singapore, 2019; pp. 65–80. [Google Scholar]
- Nawaz, K.; Ashraf, M. Exogenous Application of Glycinebetaine Modulates Activities of Antioxidants in Maize Plants Subjected to Salt Stress. J. Agron. Crop Sci. 2010, 196, 28–37. [Google Scholar] [CrossRef]
- Annunziata, M.G.; Ciarmiello, L.F.; Woodrow, P.; Maximova, E.; Fuggi, A.; Carillo, P. Durum Wheat Roots Adapt to Salinity Remodeling the Cellular Content of Nitrogen Metabolites and Sucrose. Front. Plant Sci. 2017, 7, 2035. [Google Scholar] [CrossRef]
- Woodrow, P.; Ciarmiello, L.F.; Annunziata, M.G.; Pacifico, S.; Iannuzzi, F.; Mirto, A.; D’Amelia, L.; Dell’Aversana, E.; Piccolella, S.; Fuggi, A.; et al. Durum Wheat Seedling Responses to Simultaneous High Light and Salinity Involve a Fine Reconfiguration of Amino Acids and Carbohydrate Metabolism. Physiol. Plant. 2017, 159, 290–312. [Google Scholar] [CrossRef]
- Abbas Al-Hamzawi, M.K. Effect of Calcium Nitrate, Potassium Nitrate and Anfaton on Growth and Storability of Plastic Houses Cucumber (Cucumis sativus L. Cv. Al-Hytham). Am. J. Plant Physiol. 2010, 5, 278–290. [Google Scholar] [CrossRef]
- Orlovsky, N.; Japakova, U.; Zhang, H.; Volis, S. Effect of Salinity on Seed Germination, Growth and Ion Content in Dimorphic Seeds of Salicornia europaea L. (Chenopodiaceae). Plant Divers. 2016, 38, 183–189. [Google Scholar] [CrossRef] [PubMed]
- Kong, X.; Luo, Z.; Dong, H.; Eneji, A.E.; Li, W. Effects of Non-Uniform Root Zone Salinity on Water Use, Na+ Recirculation, and Na+ and H+ Flux in Cotton. J. Exp. Bot. 2012, 63, 2105–2116. [Google Scholar] [CrossRef] [PubMed]
- Wu, H.; Shabala, L.; Liu, X.; Azzarello, E.; Zhou, M.; Pandolfi, C.; Chen, Z.H.; Bose, J.; Mancuso, S.; Shabala, S. Linking Salinity Stress Tolerance with Tissue-Specific Na+ Sequestration in Wheat Roots. Front. Plant Sci. 2015, 6, 71. [Google Scholar] [CrossRef]
- Athar, H.U.R.; Zafar, Z.U.; Ashraf, M. Glycinebetaine Improved Photosynthesis in Canola under Salt Stress: Evaluation of Chlorophyll Fluorescence Parameters as Potential Indicators. J. Agron. Crop Sci. 2015, 201, 428–442. [Google Scholar] [CrossRef]
- Luo, J.Y.; Zhang, S.; Peng, J.; Zhu, X.Z.; Lv, L.M.; Wang, C.Y.; Li, C.H.; Zhou, Z.G.; Cui, J.J. Effects of Soil Salinity on the Expression of Bt Toxin (Cry1Ac) and the Control Efficiency of Helicoverpa Armigera in Field-Grown Transgenic Bt Cotton. PLoS ONE 2017, 12, e170379. [Google Scholar] [CrossRef]
- Raza, M.A.S.; Shahid, A.M.; Saleem, M.F.; Khan, I.H.; Ahmad, S.; Ali, M.; Iqbal, R. Effects and Management Strategies to Mitigate Drought Stress in Oilseed Rape (Brassica napus L.): A Review Sausros Sukelto Streso Mažinimo Būdai Aliejinių Rapsų Pasėliuose: Apžvalga. Zemdirb. Agric. 2017, 104, 85–94. [Google Scholar] [CrossRef]
- Akram, R.; Natasha; Fahad, S.; Hashmi, M.Z.; Wahid, A.; Adnan, M.; Mubeen, M.; Khan, N.; Rehmani, M.I.A.; Awais, M.; et al. Trends of Electronic Waste Pollution and Its Impact on the Global Environment and Ecosystem. Environ. Sci. Pollut. Res. 2019, 26, 16923–16938. [Google Scholar] [CrossRef]
- Bhuiyan, M.S.I.; Maynard, G.; Raman, A.; Hodgkins, D.; Mitchell, D.; Nicol, H. Salt Effects on Proline and Glycine Betaine Levels and Photosynthetic Performance in Melilotus Siculus, Tecticornia Pergranulata and Thinopyrum Ponticum Measured in Simulated Saline Conditions. Funct. Plant Biol. 2016, 43, 254–265. [Google Scholar] [CrossRef]
- Dolatabadi, N.; Toorchi, M. Rapeseed (Brassica napus L.) Genotypes Response to NaCl Salinity. J. Biodivers. Environ. Sci. 2017, 10, 265–270. [Google Scholar] [CrossRef]
- Yang, R.; Yang, T.; Zhang, H.; Qi, Y.; Xing, Y.; Zhang, N.; Li, R.; Weeda, S.; Ren, S.; Ouyang, B.; et al. Hormone Profiling and Transcription Analysis Reveal a Major Role of ABA in Tomato Salt Tolerance. Plant Physiol. Biochem. 2014, 77, 23–34. [Google Scholar] [CrossRef] [PubMed]
- Solis, J.; Baisakh, N.; Brandt, S.R.; Villordon, A.; La Bonte, D. Transcriptome Profiling of Beach Morning Glory (Ipomoea imperati) under Salinity and Its Comparative Analysis with Sweetpotato. PLoS ONE 2016, 11, e147398. [Google Scholar] [CrossRef] [PubMed]
- Zhou, H.; Qian, J.; Zhao, M.-D.; Li, F.; Tong, W.; Li, L.; Fang, R.J.; Zhao, W.G.; Kim, H.J. Cloning and Sequence Analysis of the Δ1-Pyrroline-5-Carboxylate Synthase Gene (MP5CS) from Mulberry (Morus alba) and Patterns of MP5CS Gene Expression under Abiotic Stress Conditions. J. Hortic. Sci. Biotechnol. 2016, 91, 102–110. [Google Scholar] [CrossRef]
- Zorb, C.; Geilfus, C.M.; Mühling, K.H.; Ludwig-Müller, J. The Influence of Salt Stress on ABA and Auxin Concentrations in Two Maize Cultivars Differing in Salt Resistance. J. Plant Physiol. 2013, 170, 220–224. [Google Scholar] [CrossRef] [PubMed]
- Ahmad, P.; Rasool, S.; Gul, A.; Sheikh, S.A.; Akram, N.A.; Ashraf, M.; Kazi, A.M.; Gucel, S. Jasmonates: Multifunctional Roles in Stress Tolerance. Front. Plant Sci. 2016, 7, 813. [Google Scholar] [CrossRef]
- Sun, J.Q.; Jiang, H.L.; Li, C.Y. Systemin/Jasmonate-Mediated Systemic Defense Signaling in Tomato. Mol. Plant 2011, 4, 607–615. [Google Scholar] [CrossRef] [PubMed]
- Manan, A.; Ayyub, C.M.; Pervez, M.A.; Ahmad, R. Methyl Jasmonate Brings about Resistance against Salinity Stressed Tomato Plants by Altering Biochemical and Physiological Processes. Pak. J. Agric. Sci. 2016, 53, 35–41. [Google Scholar] [CrossRef]
- Hazman, M.; Hause, B.; Eiche, E.; Nick, P.; Riemann, M. Increased Tolerance to Salt Stress in OPDA-Deficient Rice Allene Oxide Cyclase Mutants Is Linked to an Increased ROS-Scavenging Activity. J. Exp. Bot. 2015, 66, 3339–3352. [Google Scholar] [CrossRef]
- Agati, G.; Biricolti, S.; Guidi, L.; Ferrini, F.; Fini, A.; Tattini, M. The Biosynthesis of Flavonoids Is Enhanced Similarly by UV Radiation and Root Zone Salinity in L. Vulgare Leaves. J. Plant Physiol. 2011, 168, 204–212. [Google Scholar] [CrossRef]
- Gavin, N.M.; Durako, M.J. Localization and Antioxidant Capacity of Flavonoids in Halophila Johnsonii in Response to Experimental Light and Salinity Variation. J. Exp. Mar. Bio. Ecol. 2012, 416–417, 32–40. [Google Scholar] [CrossRef]
- Khan, S.A.; Chibon, P.Y.; De Vos, R.C.H.; Schipper, B.A.; Walraven, E.; Beekwilder, J.; Van Dijk, T.; Finkers, R.; Visser, R.G.F.; Van De Weg, E.W.; et al. Genetic Analysis of Metabolites in Apple Fruits Indicates an MQTL Hotspot for Phenolic Compounds on Linkage Group 16. J. Exp. Bot. 2012, 63, 2895–2908. [Google Scholar] [CrossRef] [PubMed]
- Gavin, N.M.; Durako, M.J. Localization and Antioxidant Capacity of Flavonoids from Intertidal and Subtidal Halophila Johnsonii and Halophila Decipiens. Aquat. Bot. 2011, 95, 242–247. [Google Scholar] [CrossRef]
- Hashem, A.; Tabassum, B.; Fathi Abd Allah, E. Bacillus subtilis: A plant-growth promoting rhizobacterium that also impacts biotic stress. Saudi J. Biol. Sci. 2019, 26, 1291–1297. [Google Scholar] [CrossRef] [PubMed]
- Mokabel, S.; Olama, Z.; Ali, S. The Role of Plant Growth Promoting Rhizosphere Microbiome as Alternative Biofertilizer in Boosting Solanum melongena L. Adaptation to Salinity Stress. Plants 2022, 11, 659. [Google Scholar] [CrossRef] [PubMed]
- Zhao, S.; Zhang, Q.; Liu, M.; Zhou, H.; Ma, C.; Wang, P. Regulation of Plant Responses to Salt Stress. Int. J. Mol. Sci. 2021, 22, 4609. [Google Scholar] [CrossRef] [PubMed]
- Geng, G.; Lv, C.; Stevanato, P.; Li, R.; Liu, H.; Yu, L.; Wang, Y. Transcriptome Analysis of Salt-Sensitive and Tolerant Genotypes Reveals Salt-Tolerance Metabolic Pathways in Sugar Beet. Int. J. Mol. Sci. 2019, 20, 5910. [Google Scholar] [CrossRef]
- Song, Q.; Joshi, M.; Joshi, V. Transcriptomic Analysis of Short-Term Salt Stress Response in Watermelon Seedlings. Int. J. Mol. Sci. 2020, 21, 6036. [Google Scholar] [CrossRef]
- Xiong, Y.; Yan, H.; Liang, H.; Zhang, Y.; Guo, B.; Niu, M.; Jian, S.; Ren, H.; Zhang, X.; Li, Y.; et al. RNA-Seq Analysis of Clerodendrum inerme (L.) Roots in Response to Salt Stress. BMC Genom. 2019, 20, 724. [Google Scholar] [CrossRef]
- Ponce, K.S.; Meng, L.; Guo, L.; Leng, Y.; Ye, G. Advances in Sensing, Response and Regulation Mechanism of Salt Tolerance in Rice. Int. J. Mol. Sci. 2021, 22, 2254. [Google Scholar] [CrossRef]
- Zörb, C.; Schmitt, S.; Mühling, K.H. Proteomic Changes in Maize Roots after Short-Term Adjustment to Saline Growth Conditions. Proteomics 2010, 10, 4441–4449. [Google Scholar] [CrossRef]
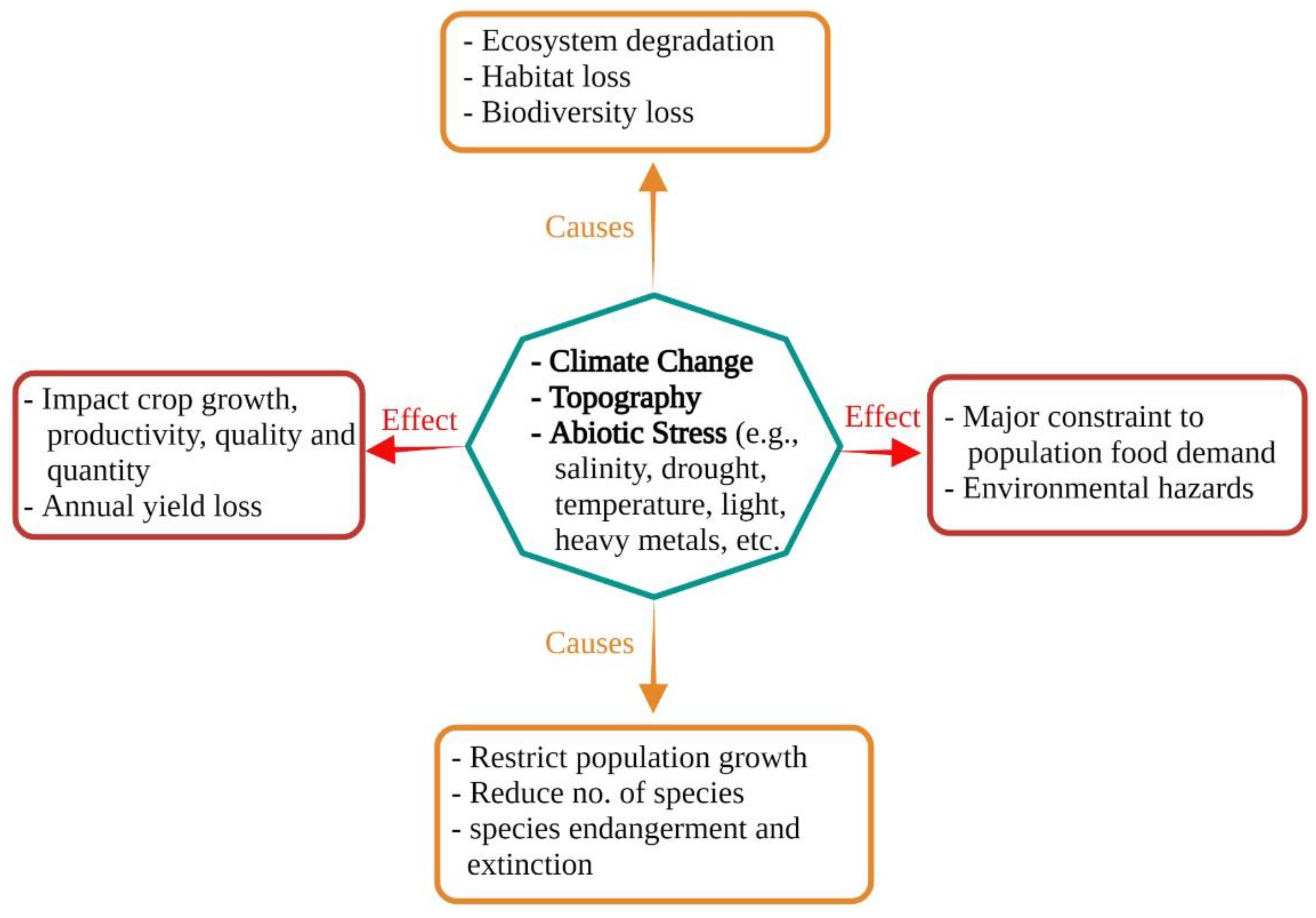
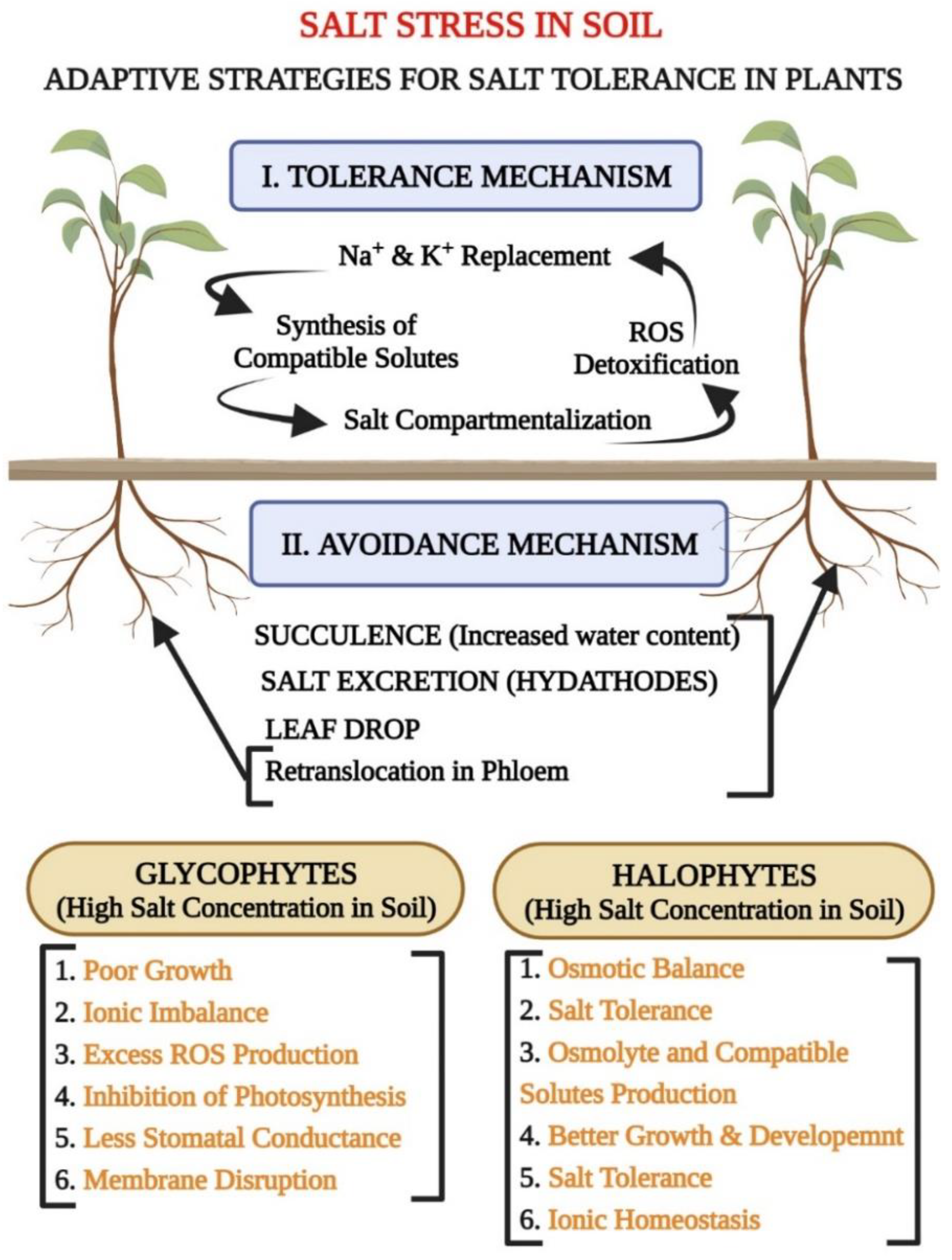
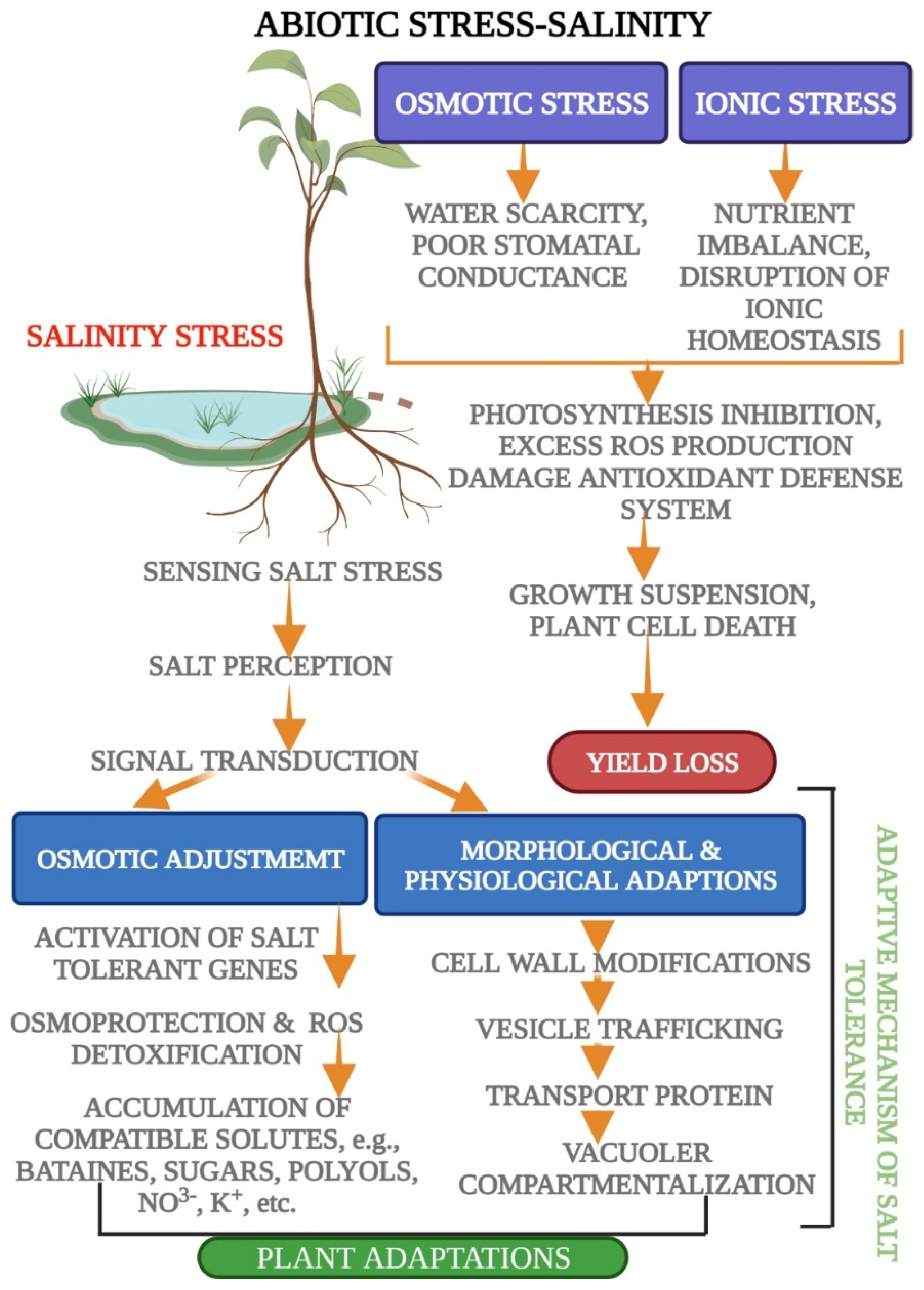
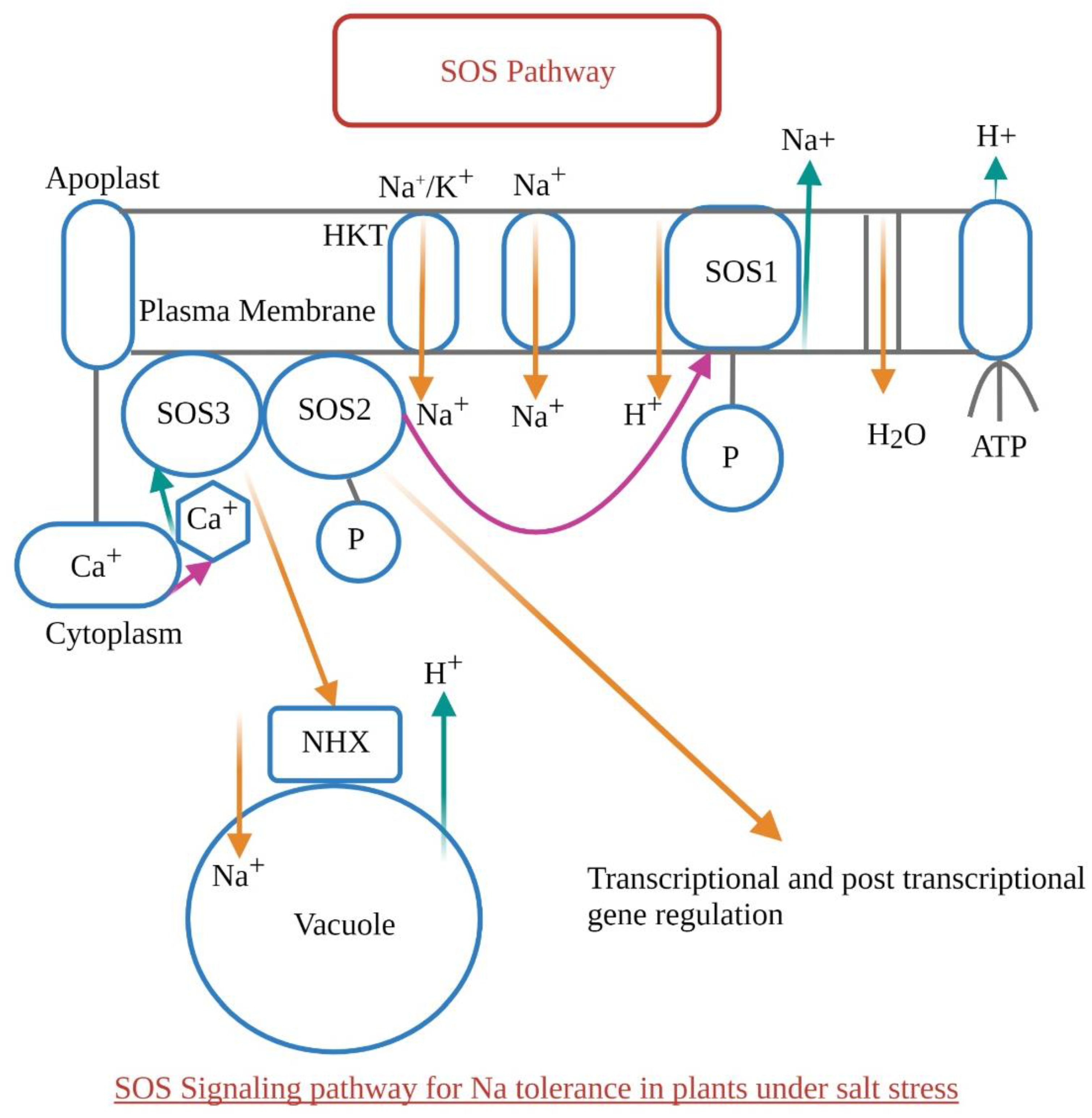
| Type of Abiotic Stress | Secondary Metabolites/Osmolytes Production | References |
|---|---|---|
| Salt stress | Proline, Glycine Betaine (GB), Flavonoids, Jasmonates (JA), Abscisic acid (ABA) | [110,111,112,113,114] |
| Drought stress | Proline, Glycine Betaine (GB), Polyamines | [3] |
| Heat stress | Abscisic acid (ABA), Glycine Betaine (GB), Proline, Polyols | [111,113,115] |
| Osmotic stress | Glycine Betaine (GB), Polyamines | [57,58,116] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gul, Z.; Tang, Z.-H.; Arif, M.; Ye, Z. An Insight into Abiotic Stress and Influx Tolerance Mechanisms in Plants to Cope in Saline Environments. Biology 2022, 11, 597. https://doi.org/10.3390/biology11040597
Gul Z, Tang Z-H, Arif M, Ye Z. An Insight into Abiotic Stress and Influx Tolerance Mechanisms in Plants to Cope in Saline Environments. Biology. 2022; 11(4):597. https://doi.org/10.3390/biology11040597
Chicago/Turabian StyleGul, Zarmina, Zhong-Hua Tang, Muhammad Arif, and Zhang Ye. 2022. "An Insight into Abiotic Stress and Influx Tolerance Mechanisms in Plants to Cope in Saline Environments" Biology 11, no. 4: 597. https://doi.org/10.3390/biology11040597
APA StyleGul, Z., Tang, Z.-H., Arif, M., & Ye, Z. (2022). An Insight into Abiotic Stress and Influx Tolerance Mechanisms in Plants to Cope in Saline Environments. Biology, 11(4), 597. https://doi.org/10.3390/biology11040597








