N-WASP Attenuates Cell Proliferation and Migration through ERK2-Dependent Enhanced Expression of TXNIP
Abstract
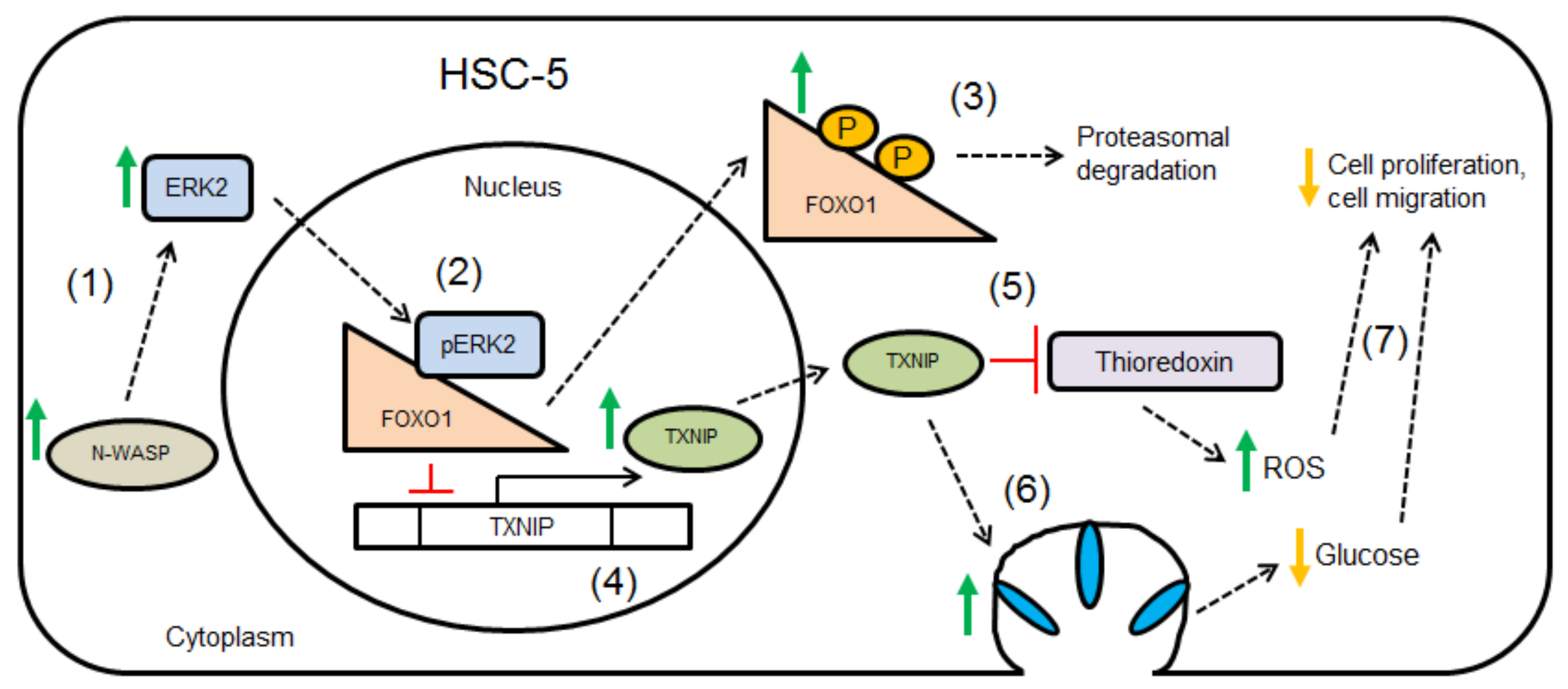
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Cell Culture and Generation of Stable Sublines by Lentiviral Transduction
2.2. Cell Proliferation Assay
2.3. Wound Healing Assay
2.4. Western Blot Analysis
2.5. Immunofluorescence
2.6. KinexTM Antibody Microarray Analysis
2.7. Real-Time PCR
2.8. Statistical Analysis
3. Results
3.1. Expression of N-WASP Is Reduced in SCC Compared to Matched Perilesional Controls
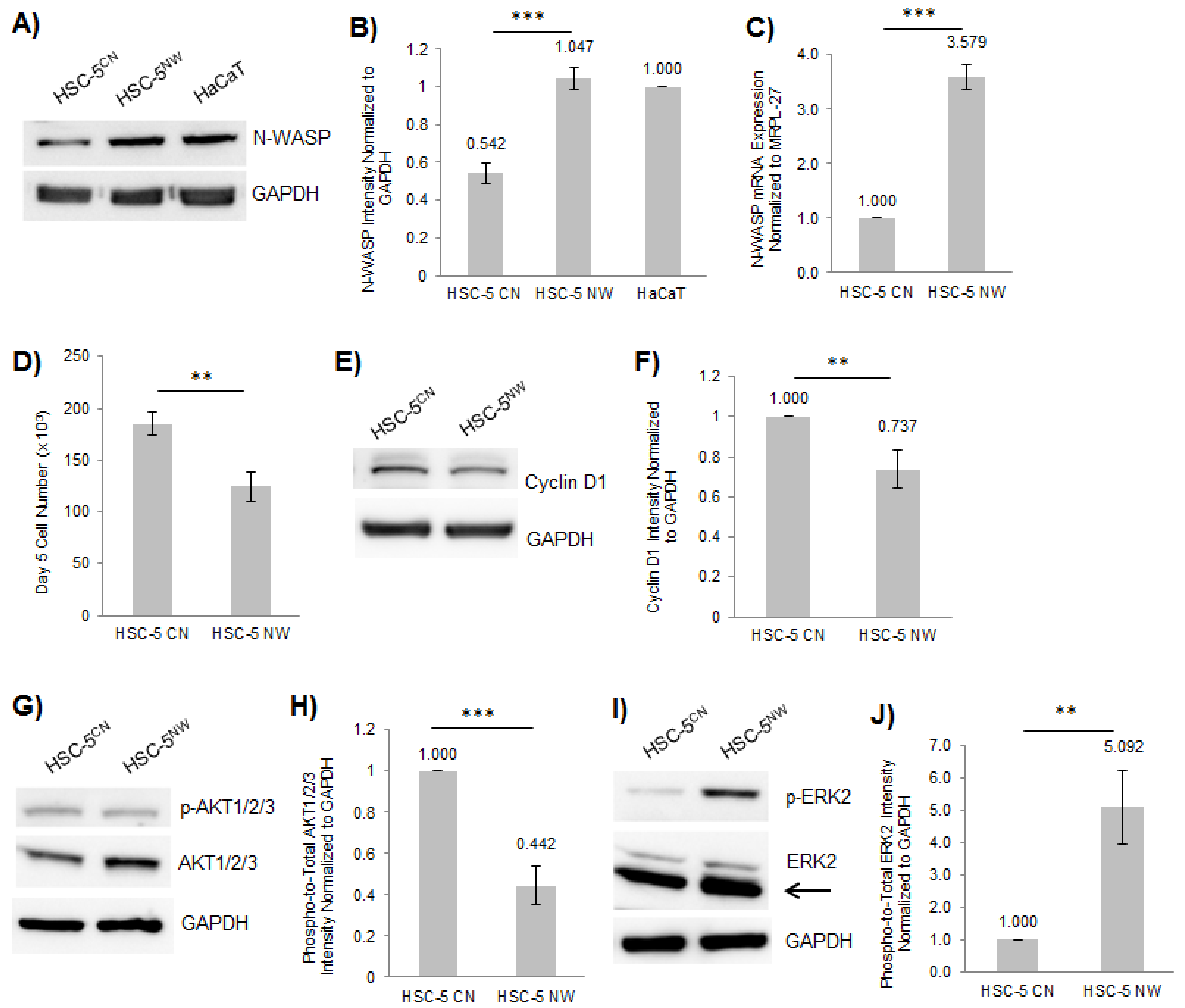
3.2. Elevated Expression of N-WASP in HSC-5 Cells Reduced Cell Proliferation and Cell Migration
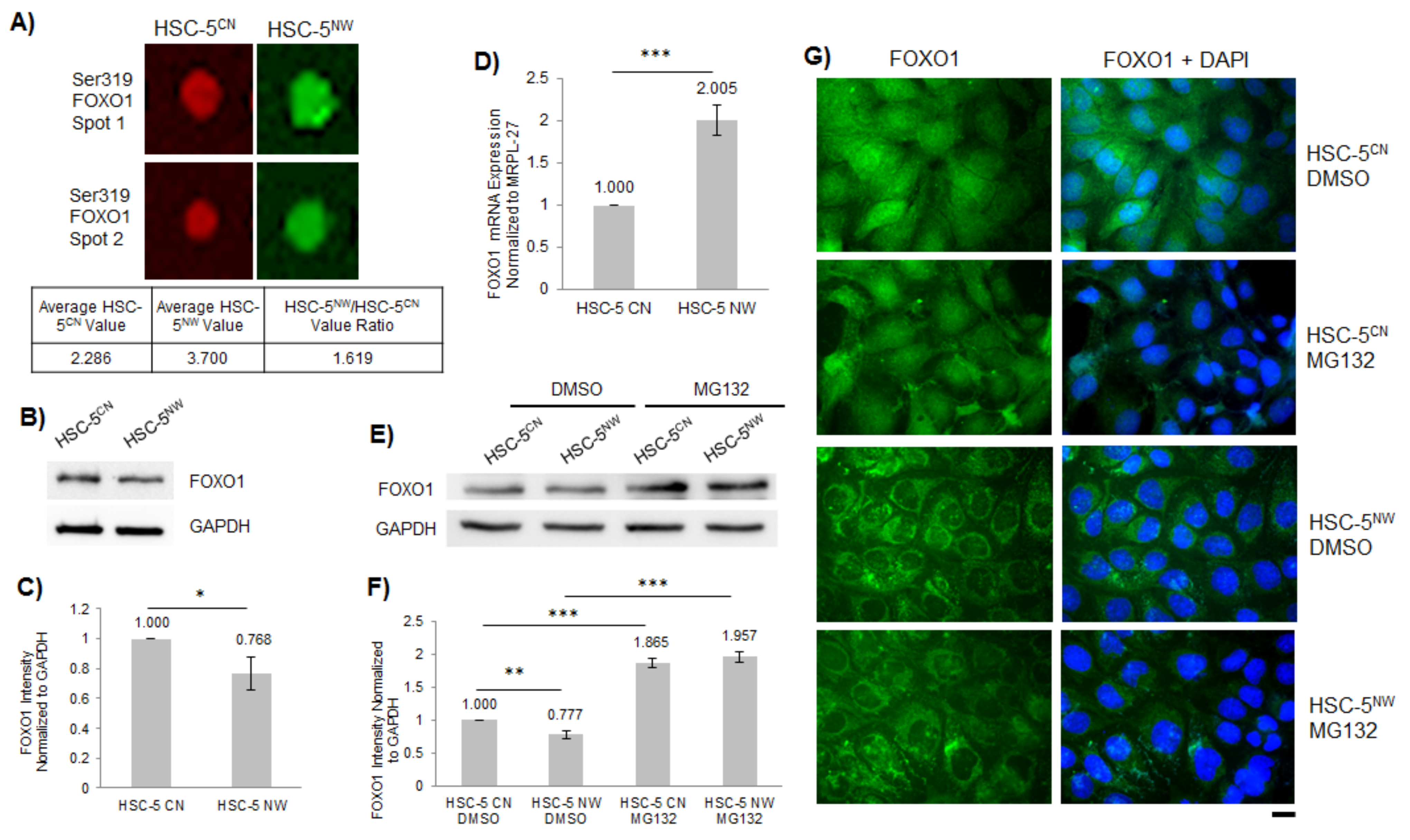
3.3. FOXO1 Expression Is Reduced in HSC-5NW Cells Compared to HSC-5CN Cells
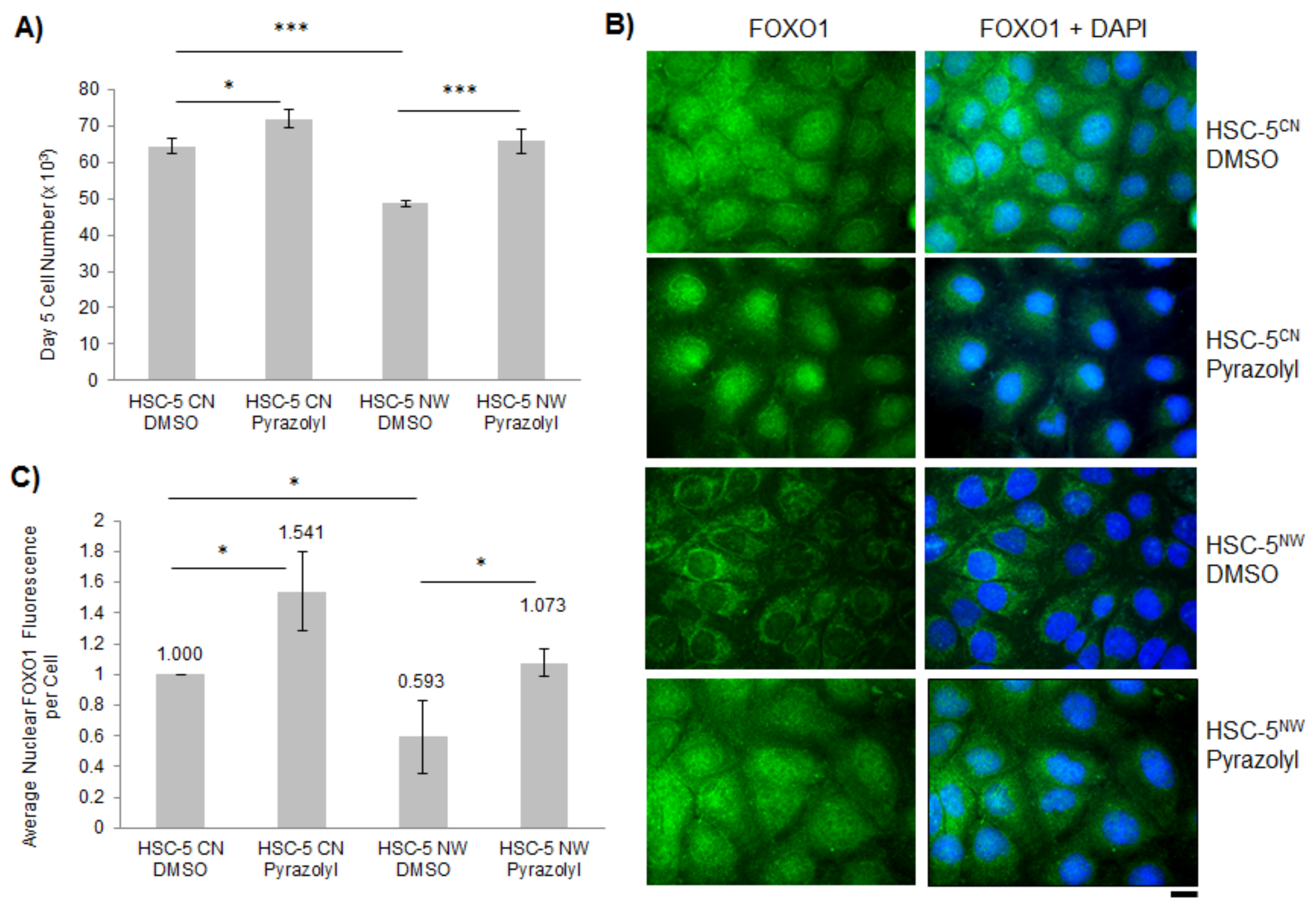
3.4. Inhibition of ERK2 in HSC-5NW Cells Enhanced Cell Proliferation, Nuclear Localization of FOXO1 and Cell Migration
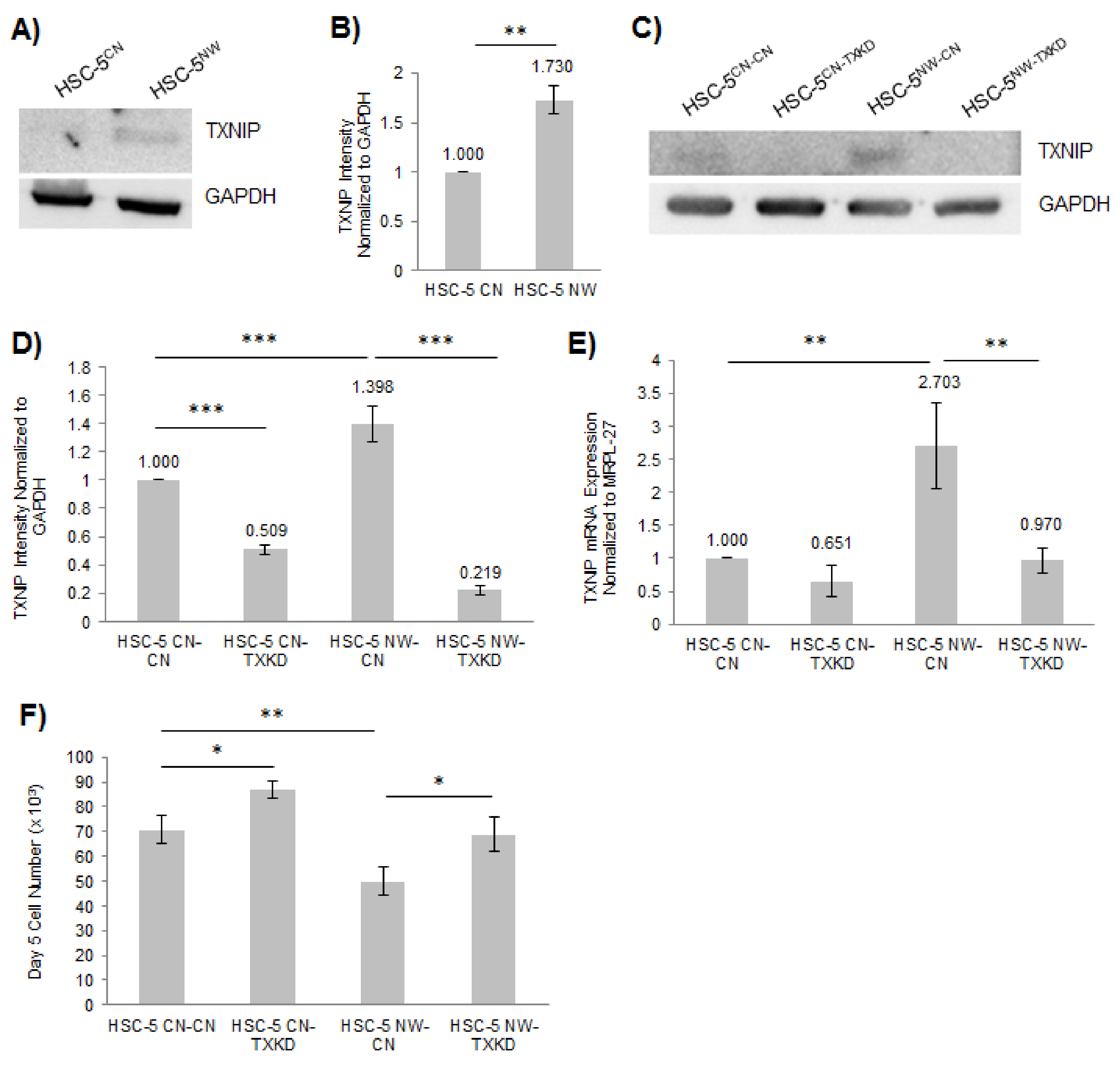
3.5. Knockdown of TXNIP in HSC-5NW Cells Restored Cell Proliferation and Migration
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Proksch, E.; Brandner, J.M.; Jensen, J.M. The skin: An indispensable barrier. Exp. Dermatol. 2008, 17, 1063–1072. [Google Scholar] [CrossRef] [PubMed]
- Bandarchi, B.; Ma, L.; Navab, R.; Seth, A.; Rasty, G. From melanocyte to metastatic malignant melanoma. Dermatol. Res. Pract. 2010, 2010, 583748. [Google Scholar] [CrossRef] [PubMed]
- Cakir, B.O.; Adamson, P.; Cingi, C. Epidemiology and economic burden of nonmelanoma skin cancer. Facial Plast. Surg. Clin. N. Am. 2012, 20, 419–422. [Google Scholar] [CrossRef] [PubMed]
- Janes, S.M.; Watt, F.M. New roles for integrins in squamous-cell carcinoma. Nat. Rev. Cancer 2006, 6, 175–183. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.A.; Tai, C.Y.; Mok, L.P.; Mosser, E.A.; Schuman, E.M. Calcium-dependent dynamics of cadherin interactions at cell-cell junctions. Proc. Natl. Acad. Sci. USA 2011, 108, 9857–9862. [Google Scholar] [CrossRef]
- Fletcher, D.A.; Mullins, R.D. Cell mechanics and the cytoskeleton. Nature 2010, 463, 485–492. [Google Scholar] [CrossRef] [PubMed]
- Lodish, H.; Berk, A.; Kaiser, C.A.; Krieger, M.; Bretscher, A.; Ploegh, H.; Amon, A.; Martin, K.C. Molecular Cell Biology; Macmillan: New York, NY, USA, 2016. [Google Scholar]
- Egile, C.; Loisel, T.P.; Laurent, V.; Li, R.; Pantaloni, D.; Sansonetti, P.J.; Carlier, M.F. Activation of the CDC42 effector N-WASP by the Shigella flexneri IcsA protein promotes actin nucleation by Arp2/3 complex and bacterial actin-based motility. J. Cell Biol. 1999, 146, 1319–1332. [Google Scholar] [CrossRef]
- Chesarone, M.A.; Goode, B.L. Actin nucleation and elongation factors: Mechanisms and interplay. Curr. Opin. Cell Biol. 2009, 21, 28–37. [Google Scholar] [CrossRef]
- Lee, S.H.; Dominguez, R. Regulation of actin cytoskeleton dynamics in cells. Mol. Cells 2010, 29, 311–325. [Google Scholar] [CrossRef]
- Takenawa, T.; Miki, H. WASP and WAVE family proteins: Key molecules for rapid rearrangement of cortical actin filaments and cell movement. J. Cell Sci. 2001, 114, 1801–1809. [Google Scholar] [CrossRef]
- Martin, T.A.; Pereira, G.; Watkins, G.; Mansel, R.E.; Jiang, W.G. N-WASP is a putative tumour suppressor in breast cancer cells, in vitro and in vivo, and is associated with clinical outcome in patients with breast cancer. Clin. Exp. Metastasis 2008, 25, 97–108. [Google Scholar] [CrossRef] [PubMed]
- Martin, T.A.; Toms, A.-M.; Davies, L.M.; Cheng, S.; Jiang, W.G. The clinical and biological implications of N-WASP expression in human colorectal cancer. Transl. Gastrointest. Cancer 2011, 1, 10–20. [Google Scholar]
- Frugtniet, B.A.; Martin, T.A.; Zhang, L.; Jiang, W.G. Neural Wiskott-Aldrich syndrome protein (nWASP) is implicated in human lung cancer invasion. BMC Cancer 2017, 17, 224. [Google Scholar] [CrossRef]
- Kalailingam, P.; Tan, H.B.; Jain, N.; Sng, M.K.; Chan, J.S.K.; Tan, N.S.; Thanabalu, T. Conditional knock out of N-WASP in keratinocytes causes skin barrier defects and atopic dermatitis-like inflammation. Sci. Rep. 2017, 7, 7311. [Google Scholar] [CrossRef]
- Karin, M. Too many transcription factors: Positive and negative interactions. New Biol. 1990, 2, 126–131. [Google Scholar]
- Lee, T.I.; Young, R.A. Transcription of eukaryotic protein-coding genes. Annu. Rev. Genet. 2000, 34, 77–137. [Google Scholar] [CrossRef] [PubMed]
- Nebert, D.W. Transcription factors and cancer: An overview. Toxicology 2002, 181, 131–141. [Google Scholar] [CrossRef]
- Tuteja, G.; Kaestner, K.H. SnapShot: Forkhead transcription factors I. Cell 2007, 130, 1160. [Google Scholar] [CrossRef]
- Lam, E.W.; Brosens, J.J.; Gomes, A.R.; Koo, C.Y. Forkhead box proteins: Tuning forks for transcriptional harmony. Nat. Rev. Cancer 2013, 13, 482–495. [Google Scholar] [CrossRef]
- Golson, M.L.; Kaestner, K.H. Fox transcription factors: From development to disease. Development 2016, 143, 4558–4570. [Google Scholar] [CrossRef]
- Asada, S.; Daitoku, H.; Matsuzaki, H.; Saito, T.; Sudo, T.; Mukai, H.; Iwashita, S.; Kako, K.; Kishi, T.; Kasuya, Y.; et al. Mitogen-activated protein kinases, Erk and p38, phosphorylate and regulate Foxo1. Cell. Signal. 2007, 19, 519–527. [Google Scholar] [CrossRef] [PubMed]
- Tsitsipatis, D.; Klotz, L.O.; Steinbrenner, H. Multifaceted functions of the forkhead box transcription factors FoxO1 and FoxO3 in skin. Biochim. Biophys. Acta 2017, 1861, 1057–1064. [Google Scholar] [CrossRef] [PubMed]
- Gunschmann, C.; Stachelscheid, H.; Akyuz, M.D.; Schmitz, A.; Missero, C.; Bruning, J.C.; Niessen, C.M. Insulin/IGF-1 controls epidermal morphogenesis via regulation of FoxO-mediated p63 inhibition. Dev. Cell 2013, 26, 176–187. [Google Scholar] [CrossRef] [PubMed]
- Watanabe, R.; Nakamura, H.; Masutani, H.; Yodoi, J. Anti-oxidative, anti-cancer and anti-inflammatory actions by thioredoxin 1 and thioredoxin-binding protein-2. Pharmacol. Ther. 2010, 127, 261–270. [Google Scholar] [CrossRef] [PubMed]
- Liu, B.; Zhou, W.; Chen, X.; Xu, F.; Chen, Y.; Liu, J.; Zhang, Q.; Bao, S.; Chen, N.; Li, M.; et al. Dihydromyricetin induces mouse hepatoma Hepal-6 cell apoptosis via the transforming growth factor-beta pathway. Mol. Med. Rep. 2015, 11, 1609–1614. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Nishiyama, A.; Matsui, M.; Iwata, S.; Hirota, K.; Masutani, H.; Nakamura, H.; Takagi, Y.; Sono, H.; Gon, Y.; Yodoi, J. Identification of thioredoxin-binding protein-2/vitamin D(3) up-regulated protein 1 as a negative regulator of thioredoxin function and expression. J. Biol. Chem. 1999, 274, 21645–21650. [Google Scholar] [CrossRef]
- Chen, K.S.; DeLuca, H.F. Isolation and characterization of a novel cDNA from HL-60 cells treated with 1,25-dihydroxyvitamin D-3. Biochim. Biophys. Acta 1994, 1219, 26–32. [Google Scholar] [CrossRef]
- Patwari, P.; Chutkow, W.A.; Cummings, K.; Verstraeten, V.L.; Lammerding, J.; Schreiter, E.R.; Lee, R.T. Thioredoxin-independent regulation of metabolism by the alpha-arrestin proteins. J. Biol. Chem. 2009, 284, 24996–25003. [Google Scholar] [CrossRef]
- Wu, N.; Zheng, B.; Shaywitz, A.; Dagon, Y.; Tower, C.; Bellinger, G.; Shen, C.H.; Wen, J.; Asara, J.; McGraw, T.E.; et al. AMPK-dependent degradation of TXNIP upon energy stress leads to enhanced glucose uptake via GLUT1. Mol. Cell 2013, 49, 1167–1175. [Google Scholar] [CrossRef]
- Shin, D.J.; Joshi, P.; Hong, S.H.; Mosure, K.; Shin, D.G.; Osborne, T.F. Genome-wide analysis of FoxO1 binding in hepatic chromatin: Potential involvement of FoxO1 in linking retinoid signaling to hepatic gluconeogenesis. Nucleic Acids Res. 2012, 40, 11499–11509. [Google Scholar] [CrossRef]
- Saxena, G.; Chen, J.; Shalev, A. Intracellular shuttling and mitochondrial function of thioredoxin-interacting protein. J. Biol. Chem. 2010, 285, 3997–4005. [Google Scholar] [CrossRef] [PubMed]
- Junn, E.; Han, S.H.; Im, J.Y.; Yang, Y.; Cho, E.W.; Um, H.D.; Kim, D.K.; Lee, K.W.; Han, P.L.; Rhee, S.G.; et al. Vitamin D3 up-regulated protein 1 mediates oxidative stress via suppressing the thioredoxin function. J. Immunol. 2000, 164, 6287–6295. [Google Scholar] [CrossRef] [PubMed]
- Su, C.; Shi, A.; Cao, G.; Tao, T.; Chen, R.; Hu, Z.; Shen, Z.; Tao, H.; Cao, B.; Hu, D.; et al. Fenofibrate suppressed proliferation and migration of human neuroblastoma cells via oxidative stress dependent of TXNIP upregulation. Biochem. Biophys. Res. Commun. 2015, 460, 983–988. [Google Scholar] [CrossRef] [PubMed]
- Zhou, J.; Yu, Q.; Chng, W.J. TXNIP (VDUP-1, TBP-2): A major redox regulator commonly suppressed in cancer by epigenetic mechanisms. Int. J. Biochem. Cell Biol. 2011, 43, 1668–1673. [Google Scholar] [CrossRef] [PubMed]
- Sancak, Y.; Peterson, T.R.; Shaul, Y.D.; Lindquist, R.A.; Thoreen, C.C.; Bar-Peled, L.; Sabatini, D.M. The Rag GTPases bind raptor and mediate amino acid signaling to mTORC1. Science 2008, 320, 1496–1501. [Google Scholar] [CrossRef]
- Schneider, C.A.; Rasband, W.S.; Eliceiri, K.W. NIH Image to ImageJ: 25 years of image analysis. Nat. Methods 2012, 9, 671–675. [Google Scholar] [CrossRef]
- Hozumi, Y.; Kondo, S.; Shimoura, T.; Aso, K. Human squamous cell carcinoma from skin: Establishment and characterization of a new cell line (HSC-5). J. Dermatol. 1990, 17, 143–148. [Google Scholar] [CrossRef]
- Boukamp, P.; Petrussevska, R.T.; Breitkreutz, D.; Hornung, J.; Markham, A.; Fusenig, N.E. Normal keratinization in a spontaneously immortalized aneuploid human keratinocyte cell line. J. Cell Biol. 1988, 106, 761–771. [Google Scholar] [CrossRef]
- Mamay, C.L.; Schauer, I.E.; Rice, P.L.; McDoniels-Silvers, A.; Dwyer-Nield, L.D.; You, M.; Sclafani, R.A.; Malkinson, A.M. Cyclin D1 as a proliferative marker regulating retinoblastoma phosphorylation in mouse lung epithelial cells. Cancer Lett. 2001, 168, 165–172. [Google Scholar] [CrossRef]
- Lawlor, M.A.; Alessi, D.R. PKB/Akt: A key mediator of cell proliferation, survival and insulin responses? J. Cell Sci. 2001, 114, 2903–2910. [Google Scholar] [CrossRef]
- Kovacs, E.M.; Verma, S.; Ali, R.G.; Ratheesh, A.; Hamilton, N.A.; Akhmanova, A.; Yap, A.S. N-WASP regulates the epithelial junctional actin cytoskeleton through a non-canonical post-nucleation pathway. Nat. Cell Biol. 2011, 13, 934–943. [Google Scholar] [CrossRef] [PubMed]
- Misra, A.; Lim, R.P.; Wu, Z.; Thanabalu, T. N-WASP plays a critical role in fibroblast adhesion and spreading. Biochem. Biophys. Res. Commun. 2007, 364, 908–912. [Google Scholar] [CrossRef] [PubMed]
- Pierre, S.; Bats, A.S.; Coumoul, X. Understanding SOS (Son of Sevenless). Biochem. Pharmacol. 2011, 82, 1049–1056. [Google Scholar] [CrossRef] [PubMed]
- Lefloch, R.; Pouyssegur, J.; Lenormand, P. Single and combined silencing of ERK1 and ERK2 reveals their positive contribution to growth signaling depending on their expression levels. Mol. Cell. Biol. 2008, 28, 511–527. [Google Scholar] [CrossRef]
- Higgs, H.N.; Pollard, T.D. Activation by Cdc42 and PIP(2) of Wiskott-Aldrich syndrome protein (WASp) stimulates actin nucleation by Arp2/3 complex. J. Cell Biol. 2000, 150, 1311–1320. [Google Scholar] [CrossRef]
- Morris, H.T.; Fort, L.; Spence, H.J.; Patel, R.; Vincent, D.F.; Park, J.H.; Snapper, S.B.; Carey, F.A.; Sansom, O.J.; Machesky, L.M. Loss of N-WASP drives early progression in an Apc model of intestinal tumourigenesis. J. Pathol. 2018, 245, 337–348. [Google Scholar] [CrossRef]
- Waldhart, A.N.; Dykstra, H.; Peck, A.S.; Boguslawski, E.A.; Madaj, Z.B.; Wen, J.; Veldkamp, K.; Hollowell, M.; Zheng, B.; Cantley, L.C.; et al. Phosphorylation of TXNIP by AKT Mediates Acute Influx of Glucose in Response to Insulin. Cell Rep. 2017, 19, 2005–2013. [Google Scholar] [CrossRef]
- Lyubimova, A.; Garber, J.J.; Upadhyay, G.; Sharov, A.; Anastasoaie, F.; Yajnik, V.; Cotsarelis, G.; Dotto, G.P.; Botchkarev, V.; Snapper, S.B. Neural Wiskott-Aldrich syndrome protein modulates Wnt signaling and is required for hair follicle cycling in mice. J. Clin. Investig. 2010, 120, 446–456. [Google Scholar] [CrossRef][Green Version]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chung, Y.J.; Salvi, A.; Kalailingam, P.; Alnawaz, M.; Tan, S.H.; Pan, J.Y.; Tan, N.S.; Thanabalu, T. N-WASP Attenuates Cell Proliferation and Migration through ERK2-Dependent Enhanced Expression of TXNIP. Biology 2022, 11, 582. https://doi.org/10.3390/biology11040582
Chung YJ, Salvi A, Kalailingam P, Alnawaz M, Tan SH, Pan JY, Tan NS, Thanabalu T. N-WASP Attenuates Cell Proliferation and Migration through ERK2-Dependent Enhanced Expression of TXNIP. Biology. 2022; 11(4):582. https://doi.org/10.3390/biology11040582
Chicago/Turabian StyleChung, Yat Joong, Amrita Salvi, Pazhanichamy Kalailingam, Myra Alnawaz, Suat Hoon Tan, Jiun Yit Pan, Nguan Soon Tan, and Thirumaran Thanabalu. 2022. "N-WASP Attenuates Cell Proliferation and Migration through ERK2-Dependent Enhanced Expression of TXNIP" Biology 11, no. 4: 582. https://doi.org/10.3390/biology11040582
APA StyleChung, Y. J., Salvi, A., Kalailingam, P., Alnawaz, M., Tan, S. H., Pan, J. Y., Tan, N. S., & Thanabalu, T. (2022). N-WASP Attenuates Cell Proliferation and Migration through ERK2-Dependent Enhanced Expression of TXNIP. Biology, 11(4), 582. https://doi.org/10.3390/biology11040582







