Promising Novel Method of Acetylation Modification for Regulating Fatty Acid Metabolism in Brassica napus L.
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Materials
2.2. Methods
2.2.1. Acetylation Modification
Protein Extraction
Trypsin Digestion
Tandem Mass Tag (TMT) Labeling
Liquid Chromatography Coupled with Tandem Mass Spectrometry (LC-MS/MS) Analysis
Database Search
2.2.2. Analysis and Verification of Sequencing Results
Western Blot Analysis
Gene Expression Validation by Real-Time PCR (qPCR) Analysis
2.2.3. Gene Function Verification
Cloning of Fatty Acid Metabolism-Related Genes and Introduction of Base-Directed Mutations
Overexpression Vector Construction
Functional Verification of Arabidopsis
3. Results
3.1. Acetylation Modification Results
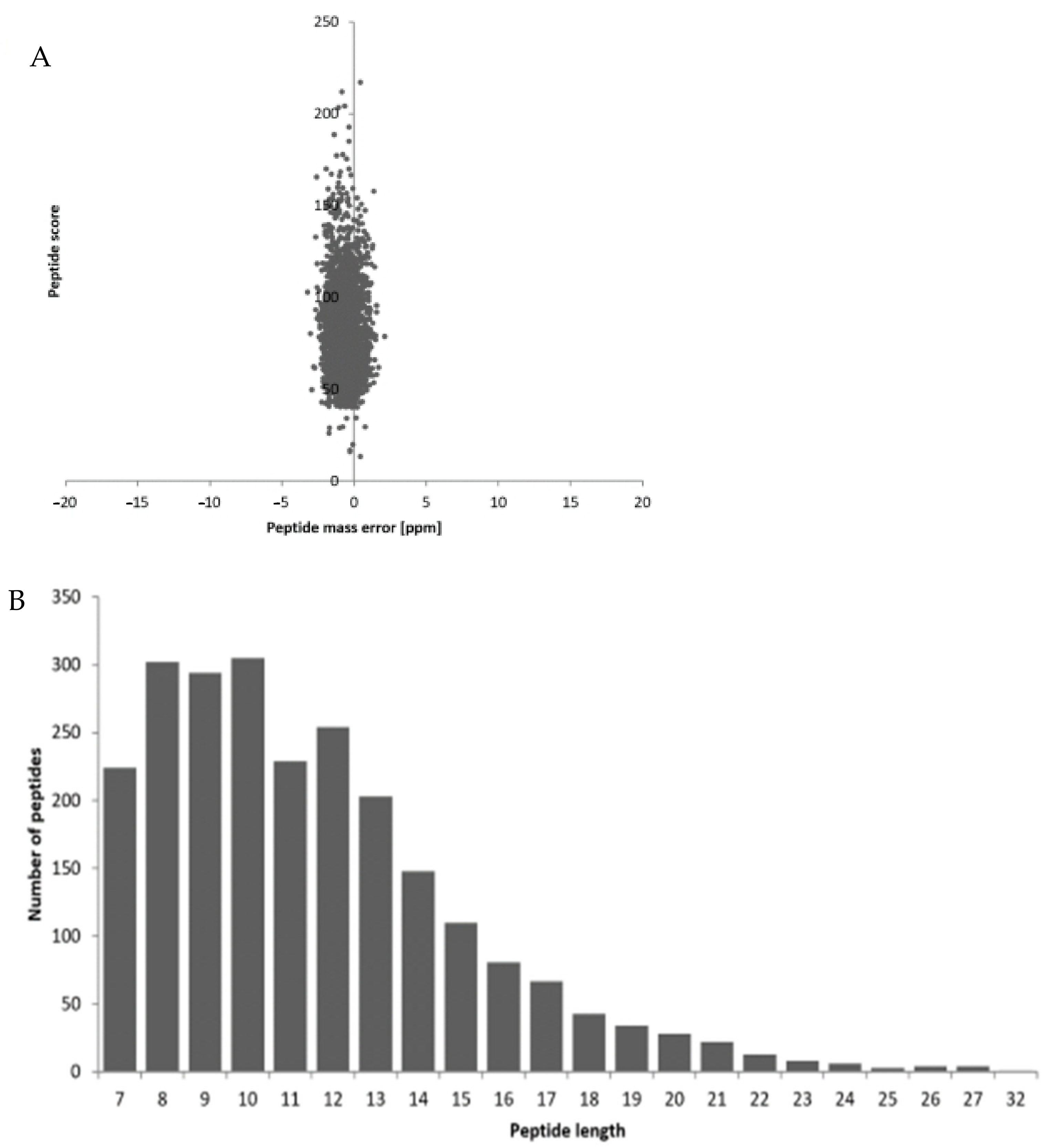
3.1.1. Quality Control Testing by Mass Spectrometry
3.1.2. Proteome-Wide Analysis of Kac Sites in Rapeseed
3.1.3. GO Analysis of Acetylated Proteins
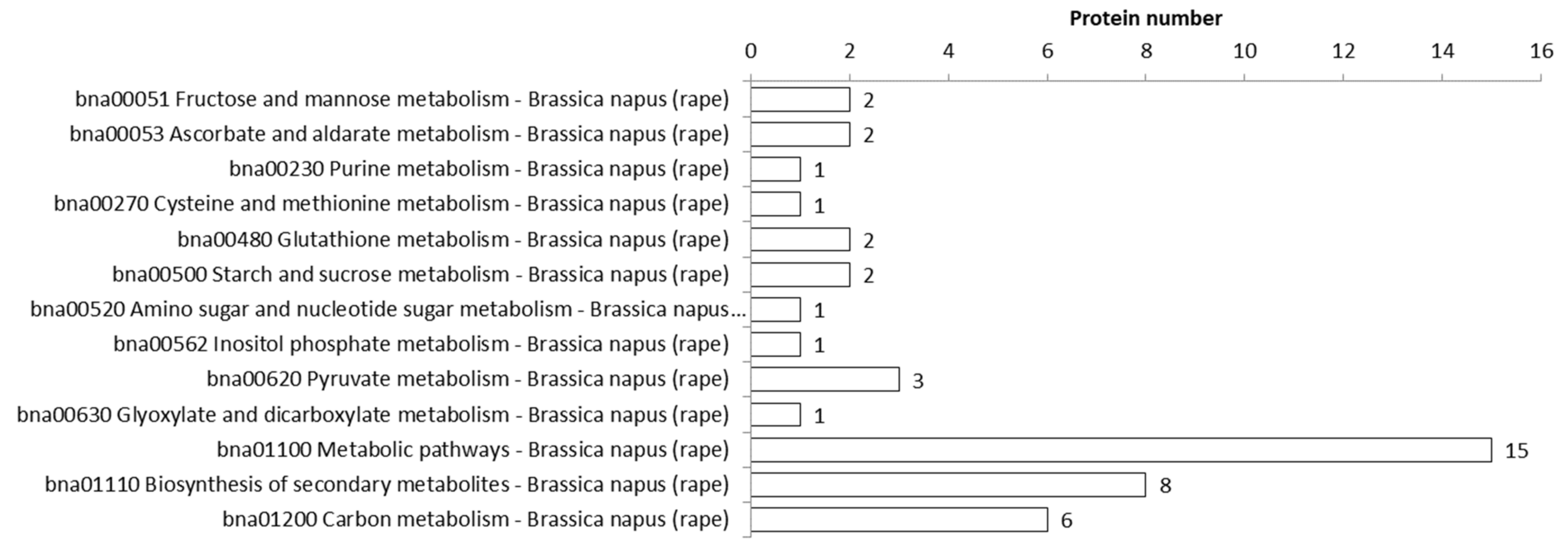
3.1.4. Protein Acetylation Regulates Diverse Metabolic Pathways in Rapeseed
3.2. Verification Analysis of Sequencing Result
3.2.1. Validation of Protein Expression by Western Blot
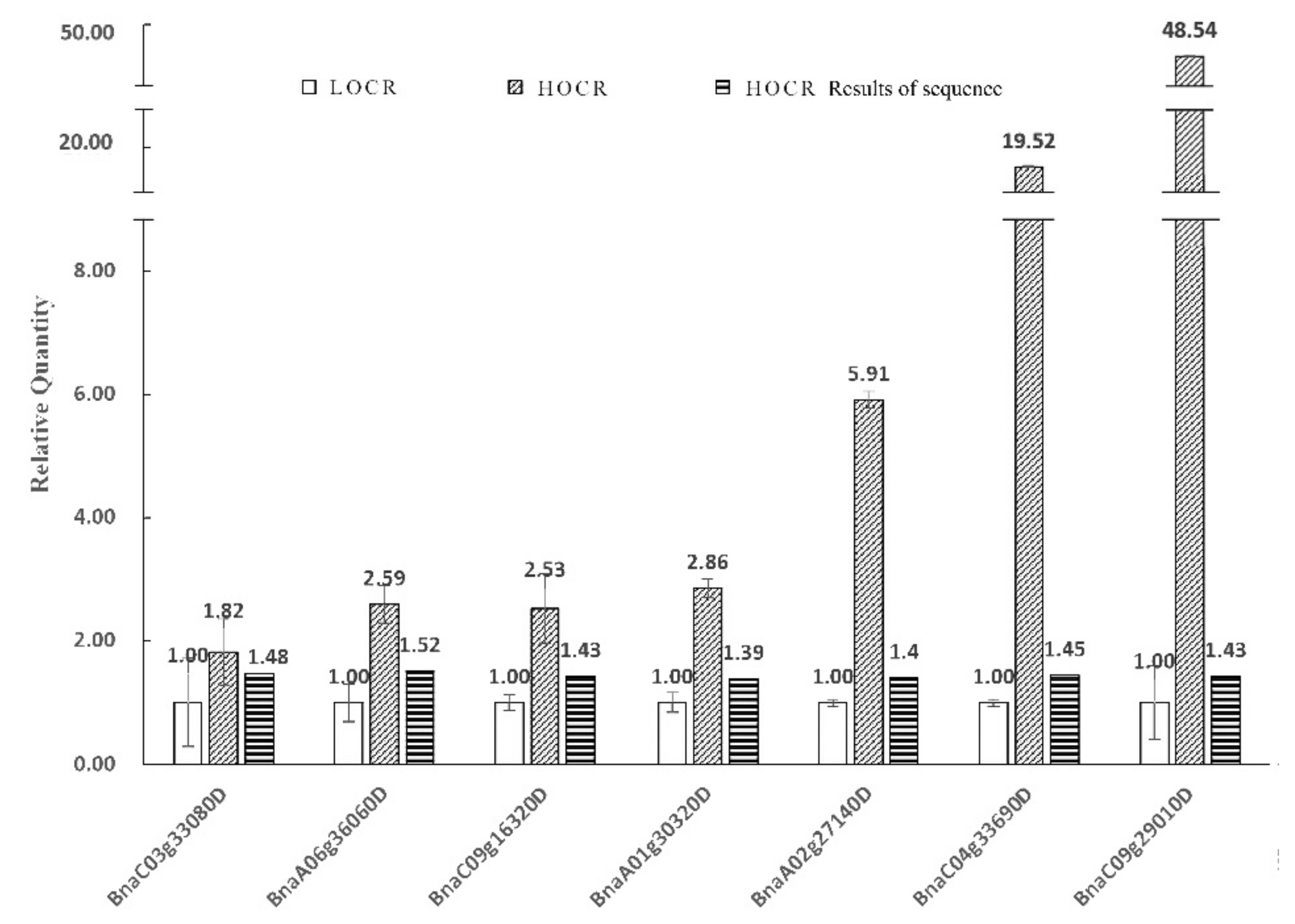
3.2.2. Gene Expression Validation by qPCR
3.3. Gene Function Verification
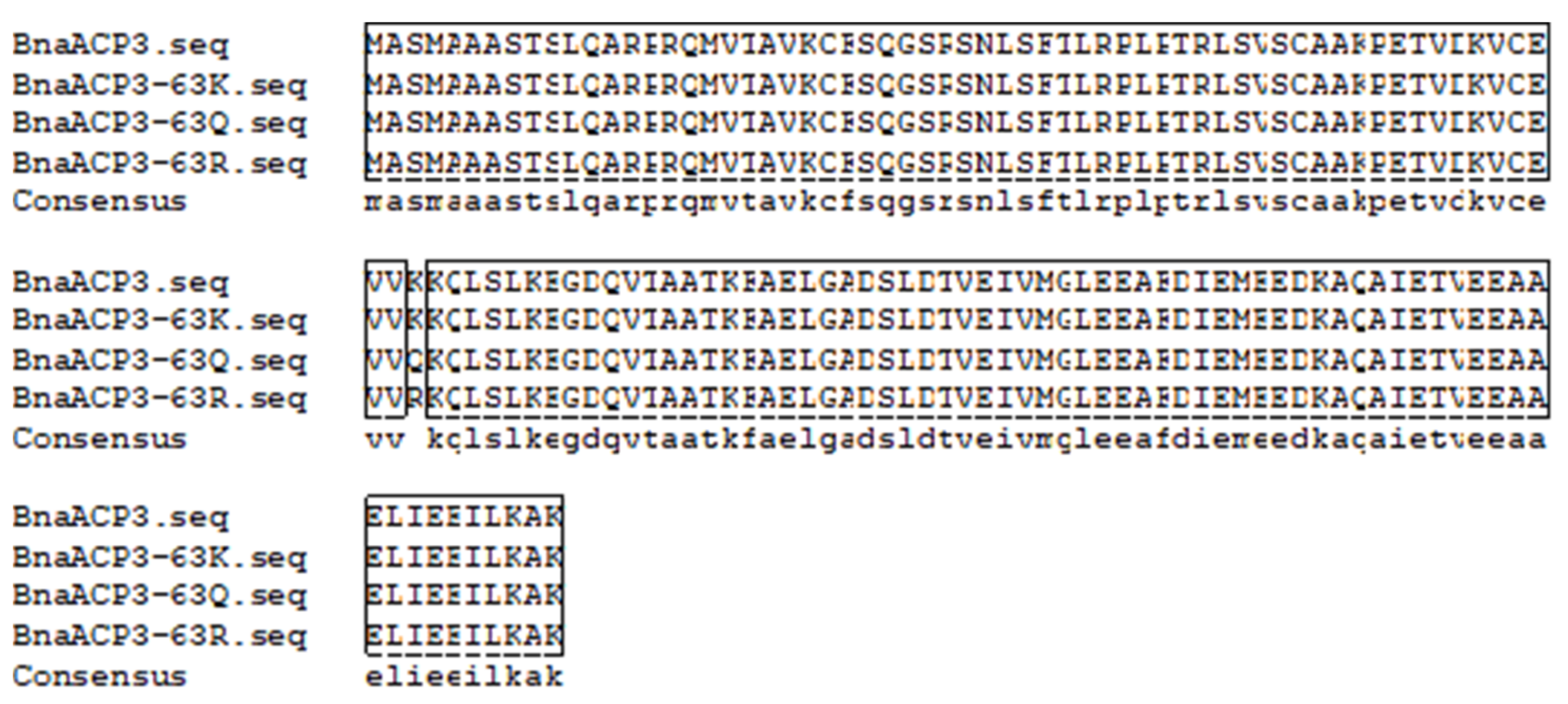
3.3.1. Cloning of Fatty Acid Metabolism-Related Genes and Introduction of Base-Directed Mutations
3.3.2. Overexpression Vector Construction and Arabidopsis Transformation
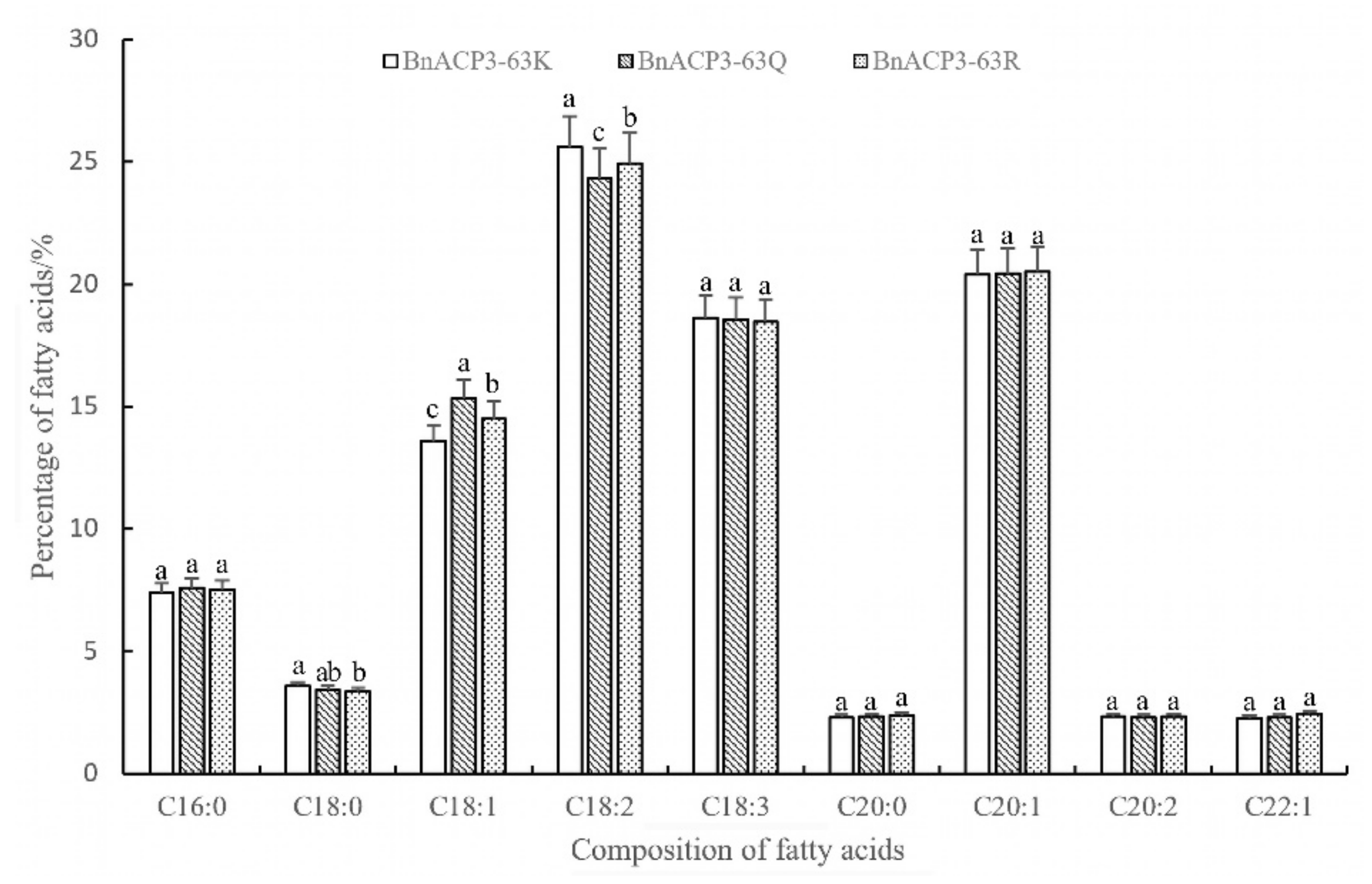
3.3.3. Fatty Acid Composition Analysis in Transgenic Arabidopsis
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Appendix A
References
- Sharma, M.M.M.; Ramekar, R.V.; Park, N.-I.; Choi, I.-Y.; Choi, S.-K.; Park, K.-C. Characterization of transcription factor genes related to cold tolerance in Brassica napus. Genom. Inform. 2021, 19, e45. [Google Scholar] [CrossRef] [PubMed]
- Xiong, Q.F.; Zhang, X.M.; Wen, J. Comparation of nutritional values between rapeseed oil and several other edible vegetable oils-discussion of rapeseed quality genetic improvement. J. Chin. Cereals Oils Assoc. 2014, 29, 122–128. [Google Scholar]
- Kapoor, B.; Kapoor, D.; Gautam, S.; Singh, R.; Bhardwaj, S. Dietary Polyunsaturated Fatty Acids (PUFAs): Uses and Potential Health Benefits. Curr. Nutr. Rep. 2021, 10, 232–242. [Google Scholar] [CrossRef] [PubMed]
- Rome, F. Fats and fatty acids in human nutrition. Report of an expert consultation. FAO Food Nutr. Pap. 2010, 91, 1. [Google Scholar]
- Los, D.A.; Murata, N. Structure and expression of fatty acid desaturases. Biochim. Biophys. Acta Lipids Lipid Metab. 1998, 1394, 3–15. [Google Scholar] [CrossRef]
- De Azevedo, S.C.; Kim, S.S.; Koch, S.; Kienow, L.; Schneider, K.; McKim, M.S.; Haughn, G.W.; Kombrink, E.; Douglas, C.J. A novel fatty Acyl-CoA synthetase is required for pollen development and sporopollenin biosynthesis in Arabidopsis. Plant Cell 2009, 21, 507–525. [Google Scholar] [CrossRef] [Green Version]
- Liu, R.-Y.; Liu, F.; Guan, C.-Y. Cloning and Expression Analyses for BnFAD2 Genes in Brassica napus. Acta Agron. Sin. 2016, 42, 1000. [Google Scholar] [CrossRef]
- Xiao, G.; Zhang, H.J.; Peng, Q.; Guan, C.Y. Screening and analysis of multiple copy of oleate desaturase gene (fad2) in Brassica napus. Acta Agron. Sin. 2008, 34, 1563–1568. [Google Scholar] [CrossRef]
- Long, W.; Hu, M.; Gao, J.; Chen, S.; Zhang, J.; Cheng, L.; Pu, H. Identification and Functional Analysis of Two New Mutant BnFAD2 Alleles That Confer Elevated Oleic Acid Content in Rapeseed. Front. Genet. 2018, 9, 399. [Google Scholar] [CrossRef]
- Peng, Q.; Hu, Y.; Wei, R.; Zhang, Y.; Guan, C.; Ruan, Y.; Liu, C. Simultaneous silencing of FAD2 and FAE1 genes affects both oleic acid and erucic acid contents in Brassica napus seeds. Plant Cell Rep. 2010, 29, 317–325. [Google Scholar] [CrossRef]
- Sivaraman, I.; Arumugam, N.; Sodhi, Y.S.; Gupta, V.; Mukhopadhyay, A.; Pradhan, A.K.; Burma, P.K.; Pental, D. Development of high oleic and low linoleic acid transgenics in a zero erucic acid Brassica juncea L. (Indian mustard) line by antisense suppression of the fad2 gene. Mol. Breed. 2004, 13, 365–375. [Google Scholar] [CrossRef]
- Lusser, A.; Kölle, D.; Loidl, P. Histone acetylation: Lessons from the plant kingdom. Trends Plant Sci. 2001, 6, 59–65. [Google Scholar] [CrossRef]
- Martin, C.; Zhang, Y. The diverse functions of histone lysine methylation. Nat. Rev. Mol. Cell Biol. 2005, 6, 838–849. [Google Scholar] [CrossRef] [PubMed]
- Xiong, Y.; Guan, K.-L. Mechanistic insights into the regulation of metabolic enzymes by acetylation. J. Cell Biol. 2012, 198, 155–164. [Google Scholar] [CrossRef] [Green Version]
- Kim, E.Y.; Kim, W.K.; Kang, H.J.; Kim, J.-H.; Chung, S.J.; Seo, Y.S.; Park, S.G.; Lee, S.C.; Bae, K.-H. Acetylation of malate dehydrogenase 1 promotes adipogenic differentiation via activating its enzymatic activity. J. Lipid Res. 2012, 53, 1864–1876. [Google Scholar] [CrossRef] [Green Version]
- Yu, W.; Lin, Y.; Yao, J.; Huang, W.; Lei, Q.; Xiong, Y.; Zhao, S. Lysine 88 acetylation negatively regulates ornithine carbamoyltransferase activity in response to nutrient signals. J. Biol. Chem. 2009, 284, 13669–13675. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhao, D.; Zou, S.W.; Liu, Y.; Zhou, X.; Mo, Y.; Wang, P.; Xu, Y.-H.; Dong, B.; Xiong, Y.; Lei, Q.-Y.; et al. Lysine-5 acetylation negatively regulates lactate dehy-drogenase A and is decreased in pancreatic cancer. Cancer Cell 2013, 23, 464–476. [Google Scholar] [CrossRef] [Green Version]
- Wang, T.; Xing, J.; Liu, X.; Liu, Z.S.; Yao, Y.Y.; Hu, Z.R.; Peng, H.; Xin, M.; Zhou, D.-X.; Zhang, Y.; et al. Histone acetyltransferase general control non-repressed protein 5 (GCN5) affects the fatty acid composition of Arabidopsis thaliana seeds by acetylating fatty acid desaturase3 (FAD3). Plant J. 2016, 88, 794–808. [Google Scholar] [CrossRef]
- Wang, T.; Xing, J.; Liu, X.; Yao, Y.; Hu, Z.; Peng, H.; Xin, M.; Zhou, D.-X.; Zhang, Y.; Ni, Z. GCN5 contributes to stem cuticular wax biosynthesis by histone acetylation of CER3 in Arabidopsis. J. Exp. Bot. 2018, 69, 2911–2922. [Google Scholar] [CrossRef] [Green Version]
- Handa, H. The complete nucleotide sequence and RNA editing content of the mitochondrial genome of rapeseed (Brassica napus L.): Comparative analysis of the mitochondrial genomes of rapeseed and Arabidopsis thaliana. Nucleic Acids Res. 2003, 31, 5907–5916. [Google Scholar] [CrossRef] [Green Version]
- Wang, Y.; Yang, Q.; Xiao, G.; Zhang, Z.Q.; Guan, C.Y.; Liu, Z.S. iTRAQ-based quantitative proteomics analysis of an immature high-oleic acid near-isogenic line of rapeseed. Mol. Breed. 2018, 38, 1–14. [Google Scholar] [CrossRef]
- Zhang, Z.Q.; Tan, M.; Xiao, G.; Wu, X.M.; Guan, C.Y. Comparative study of different oleic acid content of rapeseed. J. Biol. 2015, 32, 20–24. [Google Scholar]
- Liao, G.; Xie, L.; Li, X.; Cheng, Z.; Xie, J. Unexpected extensive lysine acetylation in the trump-card antibiotic producer Streptomyces roseosporus revealed by proteome-wide profiling. J. Proteom. 2014, 106, 260–269. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.-Q.; Xiao, G.; Liu, R.-Y.; Tan, T.-L.; Guan, C.-Y.; Wang, G.-H.; Chen, S.-Y.; Wu, X.-M.; Guan, M.; Li, Q. Proteomic analysis of differentially expressed proteins between Xiangyou 15 variety and the mutant M15. Front. Biol. 2014, 9, 234–243. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Clough, S.J.; Bent, A.F. Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 1998, 16, 735–743. [Google Scholar] [CrossRef] [Green Version]
- Lander, E.S.; Linton, L.M.; Birren, B.; Nusbaum, C.; Zody, M.C.; Baldwin, J.; Devon, K.; Dewar, K.; Doyle, M.; FitzHugh, W.; et al. Initial sequencing and analysis of the human genome. Nature 2001, 409, 860–921. [Google Scholar]
- Cox, J.; Mann, M. Is proteomics the new genomics? Cell 2007, 130, 395–398. [Google Scholar] [CrossRef] [Green Version]
- Fattore, E.; Bosetti, C.; Brighenti, F.; Agostoni, C.; Fattore, G. Palm oil and blood lipid–related markers of cardiovascular disease: A systematic review and meta-analysis of dietary intervention trials. Am. J. Clin. Nutr. 2014, 99, 1331–1350. [Google Scholar] [CrossRef] [Green Version]
- Li, Y.P.; Hruby, A.; Bernstein, A.M.; Ley, S.H.; Wang, D.D.; Chiuve, S.E.; Sampson, L.; Rexrode, K.M.; Rimm, E.B.; Willet, W.C.; et al. Saturated fats compared with un-saturated fats and sources of carbohydrates in relation to risk of coronary heart disease: A prospective cohort study. J. Am. Coll. Cardiol. 2015, 66, 1538–1548. [Google Scholar] [CrossRef] [Green Version]
- Van-Rooijen, M.A.; Mensink, R.P. Palmitic acid versus stearic acid: Effects of interesterification and intakes on car-diometabolic risk markers-a systematic review. Nutrients 2020, 12, 615. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nguyen, C.; Haushalter, R.W.; Lee, D.J.; Markwick, P.R.L.; Bruegger, J.; Caldara-Festin, G.; Finzel, K.; Jackson, D.R.; Ishikawa, F.; O’Dowd, B.; et al. Trapping the dynamic acyl carrier protein in fatty acid biosynthesis. Nature 2013, 505, 427–431. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Branen, J.K.; Chiou, T.J.; Engeseth, N.J. Over expression of acyl carrier protein-1 alters fatty acid composition of leaf tissue in Arabidopsis. Plant Physiol. 2001, 127, 222–229. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Branen, J.K.; Shintani, D.K.; Engeseth, N.J. Expression of Antisense Acyl Carrier Protein-4 Reduces Lipid Content in Arabidopsis Leaf Tissue. Plant Physiol. 2003, 132, 748–756. [Google Scholar] [CrossRef] [Green Version]
- Li, Z.L. In Silico Analysis of GDSL Genes in Arabidopsis and Brassica and the Investigation on Their Function in Seeds; Zhejiang University: Hangzhou, China, 2014. [Google Scholar]
- Zhang, Y.Y. Function Analyses of Several Genes Involved in Biosynthesis and Regulation of Glucosinolate in Brassica Napus and Arabidopsis Thaliana; Huazhong Agricultural University: Wuhan, China, 2015. [Google Scholar]


| Differential Proteins | Codes of Corresponding Proteins | Corresponding Gene in B. napus | Primer Sequences (5′ to 3′) |
|---|---|---|---|
| acyl-(acyl-carrier-protein) desaturase 5 | GSBRNA2T00153661001 | BnaC03g33080D | F:TTCGTGGTGCTTGTTGGT R:GGGTTGTTCTCAGTTTTAGG |
| 3-oxoacyl-(acyl-carrier-protein) synthase I | GSBRNA2T00054708001 | BnaA06g36060D | F:GGACTGGTATGGGTGGTTT R:GGTAGCACAAGCGGTAGAG |
| acyl carrier protein 3 | GSBRNA2T00100854001 | BnaC09g16320D | F:GTTCTTCACCCTCCTCTCTTTG R:GCTTTTTGACCACTTCACACACT |
| phosphoglycerate kinase 1 | GSBRNA2T00076479001 | BnaA01g30320D | F:ACAATCACTGACGATACGAGG R:TGGACAGGATGACTTTAGCAC |
| probable fructose-bisphosphate aldolase | GSBRNA2T00069603001 | BnaA02g27140D | F:CTTTCGTCTGGCGGAGTCTTC R:GCAATCGTTTTGGCGGTTT |
| triosephosphate isomerase | GSBRNA2T00108116001 | BnaC04g33690D | F:TCATCTATCCGTCTCGTTTC R:GAGTCCTTAGTCCCGTTACA |
| plastidial pyruvate kinase | GSBRNA2T00009340001 | BnaC09g29010D | F:ATGGCTCAGGTGGTTGCT R:CCTCTTCGCTTCGTTTCC |
| Primer Name | Primer Sequence (5′→3′) |
|---|---|
| F1 | GAATTCTCCTCTCTTTGCCTTTCTCCGC |
| R(R)1 | AGTTGCTTTCTGACCACTTCACACA |
| F(R)2 | TGTGTGAAGTGGTCAGAAAGCAACT |
| R2 | GTCGACGTGGGTTTGGGTTTAGTGGGGTT |
| R(Q)1 | AGTTGCTTTTGGACCACTTCACACA |
| F(Q)2 | TGTGTGAAGTGGTCCAAAAGCAACT |
| Primer Name | Sequence (5′→3′) | PCR Length (bp) |
|---|---|---|
| BnaACP3-Fw | GAATTCTCCTCTCTTTGCCTTTCTCCGC | 536 |
| BnaACP3-Rw | GTCGACGTGGGTTTGGGTTTAGTGGGGTT | |
| M13-Fw | TGTAAAACGACGGCCAGT | 667 |
| M13-Rv | CAGGAAACAGCTATGACC | |
| Detection1-Fw (35s) | AGTGGGATTGTGCGTCAT | 1153 |
| Detection1-Rv | TCAGGCGGGTAGGAAGA | |
| Detection2-Fw (Hyg) | GCTCCATACAAGCCAACC | 670 |
| Detection2-Rv | AGCGTCTCCGACCTGAT |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jia, X.; Xiong, X.; Chen, H.; Xiao, G.; Cheng, Q.; Zhang, Z. Promising Novel Method of Acetylation Modification for Regulating Fatty Acid Metabolism in Brassica napus L. Biology 2022, 11, 483. https://doi.org/10.3390/biology11040483
Jia X, Xiong X, Chen H, Xiao G, Cheng Q, Zhang Z. Promising Novel Method of Acetylation Modification for Regulating Fatty Acid Metabolism in Brassica napus L. Biology. 2022; 11(4):483. https://doi.org/10.3390/biology11040483
Chicago/Turabian StyleJia, Xiaojiang, Xinghua Xiong, Hao Chen, Gang Xiao, Qian Cheng, and Zhenqian Zhang. 2022. "Promising Novel Method of Acetylation Modification for Regulating Fatty Acid Metabolism in Brassica napus L." Biology 11, no. 4: 483. https://doi.org/10.3390/biology11040483
APA StyleJia, X., Xiong, X., Chen, H., Xiao, G., Cheng, Q., & Zhang, Z. (2022). Promising Novel Method of Acetylation Modification for Regulating Fatty Acid Metabolism in Brassica napus L. Biology, 11(4), 483. https://doi.org/10.3390/biology11040483






