1. Introduction
Healthcare has become a crucial part of the human lifestyle now. Following that, the change and development of healthcare systems have become very dominant in terms of technologies. Identifying diseases has also become very dependent on biomedical technologies, such as ultrasound, X-rays, particle beams, and MRI, etc. With more use of technologies, the excessive growth of biomedical data is a problem for healthcare professionals. Nevertheless, high-computing tools have increased the speed of analyzing biomedical data and reduced work for healthcare professionals. In addition to that, these advancements allowed the researchers to have more audacity to work with more complex clinical patterns. Healthcare further points out disorders of human abnormality, inhibiting or altering the vital functions of several human-body areas. Cardiovascular, genetic, psychiatric, brain, skin, trauma, infectious, tissue, and digestive problems are only a few of the many types of human disorders [
1].
Neurological diseases (NDs) are a fragment of human disorders that identify complications of the brain. Neurological illnesses, often known as brain, behavioral, or cognitive disorders, affect people’s abilities to walk, speak, learn, and move [
2]. As the brain is the control center of the human nerves, affecting the brain can threaten one’s life. Awareness of these diseases has lessened the mortality rate; however, some chronic NDs can cause permanent and partial disability or suffering. The global prevalence of these disorders accounted for 10.2% of the cases. Furthermore, these illnesses have a high causality rate of 16.8 % per year, respectively. These percentages indicate that neurological and neuropsychiatric disorders have higher disability rates than other human disorders [
3]. In addition, the diagnosis of neurological illnesses is a developing problem and one of the most complex challenges. For the identification, monitoring, and treatment of neurological diseases, current diagnosis technologies, reviewed in
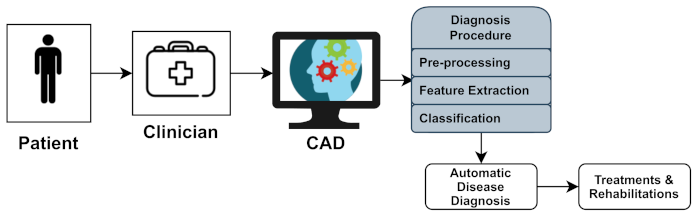
Section 4, produce massive amounts of data. Experts generally perform a manual analysis of big medical data to find and comprehend problems. Recently, an advanced notion of an automated computer-aided diagnosis (CAD) [
4] system for experts or neurologists to detect neurological disorders from big medical data has been proposed. The algorithms of significant CAD systems are built using pattern recognition techniques and theories, and consequently, CAD is considered one of the pattern recognition domains [
5]. The techniques used by CAD systems are illustrated in
Figure 1, and include data pre-processing, feature extraction, and classification. The CAD solutions assist specialists in effectively evaluating big medical data, improving diagnosis accuracy and consistency while reducing analysis time. The CAD system is cost-effective and efficient, and it may be utilized by professionals in the diagnosis and treatment of neurological illnesses as a decision support system. The acquired medical data (e.g., medical image data or medical signal data) were processed during the pre-processing period to remove noise and reduce the complexity and computation time of CAD algorithms. One of the essential elements of the CAD system is the feature extraction section, which extracts disease bio-markers from the source data. The extracted feature vectors are utilized as input in the classifier model for allocating the candidate to one of the available categories (e.g., healthy or normal) based on the output of a classifier in the classification process for CAD systems [
6].
However, a few automated computerized categorization approaches for diagnosing neurological illnesses have recently been proposed. They are sufficiently tough to handle data points from various scanners in various applications. Additionally, many developed CAD techniques have been reviewed in a single article. As a result, this study presents a quick overview of some of the essential and recent research on neurological diseases and diagnosing neurological illnesses. Following that, there are some studies, where Nadeem et al. [
7] presented an article that aimed to create a significant deep learning concept relevant to brain tumor analysis, reflecting the large variety of deep learning applications. This study looked at brain tumors segmentation, classification, prediction, and evaluation using deep learning. The significant characteristics of this developing subject were reviewed and studied, and a comprehensive taxonomy of the study landscape was based on the existing literature. In addition, Muhammad et al. [
8] addressed the fundamental concepts of deep learning-based brain tumor classification (BTC), such as pre-processing, feature extraction, and classification, as well as its accomplishments and deficiencies. This overview outlines the bench-marking datasets that have been used to evaluate BTC. Fundamental problems, such as a lack of public data and end-to-end deep learning techniques, have also been emphasized, and comprehensive suggestions for future research in the BTC field have been made. Shoeibi et al. [
9] investigated a wide range of studies centered on automated epilepsy and seizure detection by applying DL approaches and neuroimaging modalities. Several strategies for autonomously diagnosing epileptic seizures utilizing EEG and MRI modalities are outlined. The significant challenges of integrating DL with EEG and MRI modalities to detect automated epileptic seizures accurately were explored. In addition, the most promising DL models were proposed, along with probable future developments. With suitable signposting, Noor et al. [
10] showed an overview of different DL designs and pre-processing strategies for detecting anomalies in MRI data, namely a comprehensive review of existing studies based on detection using MRI scans and classification using neural network methods for NDs. In addition, he also provided a comprehensive analyses of accessible datasets, including their origins and extensive data for the subjects (e.g., patients, age, gender, and MRI scan modalities). Yolcu et al. [
11] proposed a DL method for automatic facial expression recognition. This paper is the initial step to develop a non-invasive computational system for neurological disease diagnosis, with the primary goal of increasing the quality of service. The proposed framework integrates part-based and holistic information for effective face expression identification. A new framework based on deep learning techniques was suggested (ENDs) by Attallah et al. [
12]. The methodology relies on transfer learning and deep feature fusion to recognize ENDs. It utilized raw embryo brain images to develop three deep convolutional neural networks (DCNNs) with distinct architectures. Gautam et al. [
3] provided a thorough examination of various deep learning algorithms for diagnosing severe neurological and neuropsychiatric illnesses. This study discovered that EEG- and MRI-based data could be more beneficial for diagnosing epilepsy, stroke, Parkinson’s disease, and Alzheimer’s disease. A summarized information of these related studies are tabulated in
Table 1.
Here, a thorough study on the prevalence and diagnosis of major human neurological and neuropsychiatric illnesses was conducted using a systematic review of methodologies.
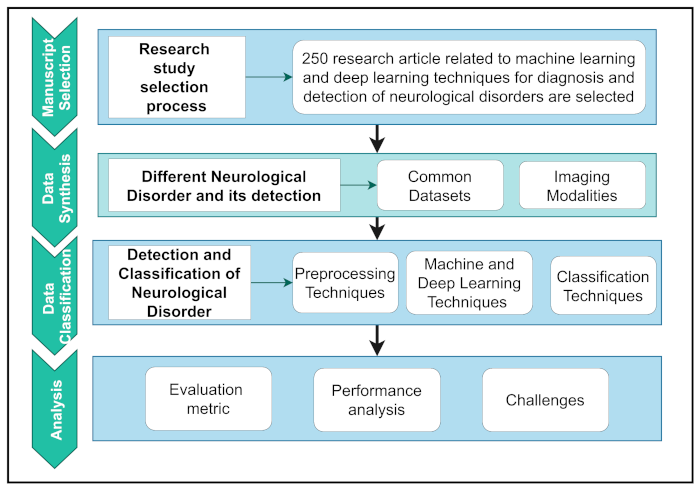
Figure 2 depicts the overall workflow of this study.
To organize the workflow, it was first summarized into four separate parts. First, a logical selection methodology is used to extract relevant articles based on research motives. Second, the data synthesis section explored NDs’ datasets and details and the detection of NDs using modalities. Data classification includes a basic introduction and a critical assessment of pre-processing techniques and various ML and DL techniques. Finally, the analysis section shows the evaluation and interpretation of performance analysis and the challenges related to major human neurological and neuropsychiatric disorders. The overall contributions of this study are as follows:
A concise introduction with the appropriate workflow of the different neurological disease detections of other DL and ML architectures and the pre-processing techniques used in detecting abnormalities from different neuroimaging modalities. It specified the background for a new entrant to the field and was performed as a future reference;
Thorough interpretation of the existing studies, we reported the purposes and limitations for detecting and classifying neurological diseases. To the best of our knowledge, this is the first attempt to review the ML- and DL-based classification approaches of different neurological disorders from other imaging modalities;
A comprehensive study on the most popular open-access datasets and their sources, and extensive information on participants in various modalities. We will use open-access datasets to verify and compare the implementation of the proposed technique;
A robust discussion on recent research issues and future directions to assist entrants in making an impact.
The rest of the paper is organized as follows:
Section 2 provides an overview of datasets. A detailed overview of the diseases and their symptoms is reported in
Section 3.
Section 4 describes the commonly used imaging modalities and their categories. Pre-processing and methods are covered in
Section 5 and
Section 6, respectively. In
Section 7, common categories of machine learning and deep learning techniques are presented. A total of the performance metrics for the analysis of the results of previous studies are presented in
Section 8. Finally, an overview of the challenges related to this study is presented in
Section 9, and
Section 10 concludes the article.
2. Dataset
The literature or review on the diagnosis and detection of neurological disorders mainly focuses on techniques, technologies, and results. Therefore, various datasets on neurological disorders are considered vital for a better analysis of these techniques and technologies. However, these datasets also contain specific categories or types. For example, MRI images for detecting neurological disorders and archiving them are vast. Magnetic resonance imaging (MRI) is a non-invasive medical imaging technology for the brain that is utilized to measure and visualize the brain’s anatomical structure, assess brain abnormalities, identify diseased regions, and perform surgical planning and image-guided procedures. MRI pictures are subjected to various image-processing techniques to identify, detect, and classify illnesses and anomalies in the brain. Another popular category is the EEG datasets of brain signals. The electrical activities of brain behaviors were reflected in the electroencephalogram (EEG) data. EEG signals reflect the electrical impulses or disorders of neurons in the human brain. EEG signal investigation is a signal-processing strategy critical for monitoring and diagnosing neurological brain disorders, such as autism spectrum disorder (ASD) and epilepsy. Such actions in the human brain define brain illnesses, such as ASD and epileptic conditions. Currently, brain disorder diagnosis is mainly performed manually by neurologists or competent clinicians by looking at EEG patterns. Parkinson’s disease applications based on speech pattern analysis for developing predictive telediagnosis and telemonitoring models are catching attention. A collection of voice samples was compiled from a set of speaking exercises for people with Parkinson’s disease, comprising sustained vowels, words, and sentences. Two key challenges are learning from a dataset with many speech recordings per participant. First, the accuracy of voice samples of various forms, such as sustained vowels versus words, in diagnosing cases of Parkinson’s disease. Second, the accuracy of the central tendency and dispersion of metrics represents all of a subject’s sample recordings. In addition, the handwriting and facial images of patients with disorders were used to detect diseases. This study has presented various summary tables,
Table 2,
Table 3,
Table 4,
Table 5,
Table 6 and
Table 7 pointing out the number of patients, modality, and available links of datasets of Alzheimer’s disease, Parkinson’s disease, Cerebral palsy, Brain tumor, Epilepsy, respectively.
This study focused on some of the most commonly used datasets in neurological disease detection. These are ADNI, OASIS for Alzheimer’s disease; for Parkinson’s disease, the most frequently utilized is PPMI with a very high number of subjects. Br35H, BraTS (MRI), Temple University EEG corpus dataset has the highest number of subject/patients data suffering from epilepsy. In addition, COBRE [
18] dataset of schizophrenia patients is the most common dataset and reliably excellent resource.
3. Neurological Diseases
The illnesses of the peripheral and central nervous systems are known as neurological disorders. Muscle weakness, paralysis, convulsions, discomfort, poor coordination, and loss of consciousness are common symptoms. There are more than 600 illnesses that affect the neurological system, including brain tumors, Parkinson’s disease (PD), Alzheimer’s disease (AD), multiple sclerosis (MS), epilepsy, dementia, headache disorders, neuro infections, stroke, or traumatic brain injury. Neuropathological examinations of patients are widely used to identify aberrant or atypical neurological diseases. However, most people have abnormal neurological abnormalities that are not usually linked to a neurological illness [
19]. Therefore, a brief review of the conditions and the related parts of the growing severity of the diseases is given in
Table 7.
3.1. Parkinson’s Disease (PD)
Parkinson’s disease (PD) is one of the most common neurological disorders worldwide, involving one to two individuals per 1000 and with a prevalence rate of 1% in the population over 60 years old [
20]. Between 1990 and 2016, the anticipated global population affected by PD more than quadrupled (from 2.5 million to 6.1 million), increasing the number of older persons and age-standardized prevalence rates [
21]. PD is a degenerative neurological condition that affects several elements of movement, particularly planning, initiation, and execution [
22,
23]. Before cognitive and behavioral abnormalities, including dementia, movement-related symptoms such as tremors, rigidity, and initiating problems, can be noted [
24]. PD has a significant impact on patient quality of life, social functions, family relationships, and it imposes high financial costs for individuals and societies [
25,
26,
27]. Khojasteh et al. [
14] investigated the effectiveness of a deep convolutional neural network (DCNN) in discriminating between PD and healthy voices using spectral data. In addition, the influence of various DCNN architecture designs and characteristics, such as frame size and the number of convolutional layers and feature maps, were examined on raw pathological and healthy voices of differing lengths. Zhang et al. [
28] used a PD screening challenge with multiview data, which attempts to employ an MRI data diagnosis to prevent and delay the progression of Parkinson’s disease. They presented a new DL architecture dubbed DNN with broad views to accomplish this goal, based on Wasserstein generative adversarial networks (WGAN), and ResNeXt can influence multiview data simultaneously. Finally, Yuvaraj et al. [
29] obtained high-order features using higher-order spectra (HOS) to develop the PD diagnosis index (PDDI), which is a single value that can discriminate between two classes. They also used various classifier techniques to aid clinicians in their diagnosis and help test the efficacy of drugs. The efficiency of supervised classification techniques, such as deep neural networks, in reliably diagnosing people with the condition is investigated in this research(reference [
30]). Wodzinski et al. [
31] showed how to diagnose Parkinson’s illness using vowels with prolonged phonation, and a ResNet architecture was designed for picture classification. They estimated the audio recording’s spectra and used them as an image input to the ResNet architecture, previously trained with the ImageNet and SVD databases. Tagaris et al. [
32] created a novel system that can make predictions and judgments based on a dataset. Their core approach is to use deep learning approaches, which are the state of the art in image analysis and computer vision-based CNN and RNN. The study by Sivaranjini et al. [
33] aimed to use a deep learning neural network to categorize the MR images of healthy control and Parkinson’s disease participants. AlexNet, a convolutional neural network design, was utilized to refine Parkinson’s disease diagnosis. The transfer learning network trained and tested the MR images to determine accuracy measures. The research by Shivangi et al. [
34] aims to create a deep learning model with two modules: a VGFR spectrogram detector and a voice impairment classifier. These modules use convolutional neural networks (CNN) and artificial neural networks (ANN) to provide a cheaper and more accurate objective diagnosis of PD early.
3.2. Dementia
Dementia is linked to the impairment of the elderly all over the world. Dementia affects almost 50 million individuals worldwide, with an estimated 10 million new cases diagnosed each year. Dementia is a syndrome in which cognitive performance, such as thinking, remembering, and reasoning, deteriorates to the point where it conflicts with daily life and tasks. Many dementia patients lose emotional control, and even personality shifts occur. Memory loss, task difficulty, disorientation, language problems, behavioral abnormalities, and lost opportunities for initiative are the most common indications and symptoms of dementia. Dementia symptoms and signs were divided into three stages: early, middle, and late. Due to the steady progression of 226 diseases, the early stage is reasonably vague. It involves losing track of time, amnesia, and becoming lost in familiar surroundings. The middle stage is more evident in events and identities. Additional signs include communication difficulties and an increased need for personal care. With persistent inquiry and roaming, behaviors are altered. The later stage is characterized by atypical symptoms, such as near-total reliance and inactivity due to significant memory problems. Difficulties walking, drastic behavioral changes, failures to recognize time and location, and failures to identify relatives and friends are all detailed symptoms and indicators. The five most common forms of dementia are as follows.
3.2.1. Alzheimer’s Disease
Alzheimer’s disease (AD) is a prevalent type of dementia, and a concern of healthcare in the 21st century. It is a degenerative brain condition characterized by the loss of cognitive function, and is without a proper cure [
35]. As a result, much work has been in developing early detection tools, particularly in the pre-symptomatic phases, to reduce or prevent disease progression [
36,
37]. Advanced neuroimaging technologies, such as magnetic resonance imaging (MRI) and positron emission tomography (PET), have been established and used to detect structural and molecular bio-markers associated with AD [
38]. However, integrating large-scale, high-dimensional multimodal neuroimaging data has become difficult because of the rapid advancements in neuroimaging techniques. Lodha et al. [
39] created a machine learning model that can reliably forecast a person’s risk of AD based on a set of characteristics that include both cognitive and medical aspects. Ebrahimighahnavieh et al. [
40] developed a systematic literature review on DL to detect AD from neuroimaging studies. A review of a new ML method for identifying Alzheimer’s disease is also shown by Liu et al. [
41].
3.2.2. Frontotemporal Dementia
Frontotemporal dementia (FTD) is a rare type of dementia that affects behavior and communication and is detected in individuals aged <60. FTD is associated with aberrant levels or types of tau and TDP-43 proteins. The most typical symptoms are extreme personality changes, such as swearing, theft, or worsening personal cleanliness standards. Behavior patterns that are socially improper, impulsive, or recurrent with impaired judgment, lack of empathy, and lack of self-awareness are symptoms of this disease [
42].
3.2.3. Lewy Body Dementia (LBD)
Lewy body dementia is a type of dementia characterized by Lewy bodies and aggregates of alpha-synuclein. Following Alzheimer’s disease, it is the second most frequent type of progressive dementia. It grows in the nerve cells in the parts of the brain that control thinking, memory, and movement (motor control). It differs from Alzheimer’s disease, which has less serious memory problems and more significant impairments in visuospatial, attentional, and frontal-executive skills [
43]. Only a post-mortem brain autopsy could validate the probable diagnosis of LBD. However, researchers are looking for techniques to detect LBD earlier in life and more accurately.
3.2.4. Vascular Dementia (VD)
Vascular dementia (VD) is a broad term that refers to reasoning, planning, judgment, memory, and other thought processes [
44]. This is generally caused by brain damage resulting from reduced blood flow to the brain. It is a chronic condition encompassing a wide range of cognitive dysfunctions produced by brain tissue damage induced by vascular diseases [
45]. VD is also a serious concern because of its significant incidence and absence of effective treatments [
46]. Though cognitive impairment caused by stroke usually improves with time, vascular dementia caused by SVD is often progressive. Therefore, brain scans such as computerized tomography (CT) or magnetic resonance imaging (MRI) are usually performed on someone thought to be carrying VD to detect any alterations in the brain.
3.2.5. Mixed Dementia
Mixed dementia is a disorder in which the brain shows signs of more than one types of dementias. The most prevalent forms are plaques and tangles associated with Alzheimer’s disease and blood vessel alterations due to vascular dementia. When FTD is combined with motor neuron disease, dementia progresses significantly more quickly with a mobility problem. The typical life expectancy for persons with both illnesses was 2–3 years after identification.
3.3. Multiple Sclerosis
“Scar tissue in various places” is the definition of multiple sclerosis. A scar or sclerosis forms whenever the myelin sheath vanishes or is damaged in many locations. These regions are also known as plaques or lesions. The brain stem, cerebellum (which regulates movement), balance, spinal cord, optic nerves, and white matter in specific brain areas, were affected. It is a potentially fatal brain and spinal cord condition (central nervous system). In MS, the immune system attacks the protective sheath (myelin) that surrounds nerve fibers, causing communication issues between the brain and the rest of the body. Four types of MS are generally seen. The first is a clinically isolated syndrome (CIS), with symptoms persisting for at least 24 h. Relapse–remitting MS (RRMS) was the most frequent one. It appears with episodes of new or worsening symptoms, followed by the symptoms being subsided partially or entirely during periods of remission. Thirdly, primary progressive MS (PPMS) cases are characterized by the persistent worsening of symptoms with no early relapses or remissions. Fifteen percent of patients with MS had PPMS. Lastly, the secondary progressive MS (SPMS) initially shows relapses and remissions in patients, regardless of whether the disease proceeds slowly. Shoeibi et al. [
47] reviewed DL techniques and the applications of automated MS detection using MRI. Ye et al. [
48] also developed a study for the classification of multiple sclerosis lesions on deep learning using diffusion-based spectrum imaging. In addition, an imaging-based machine learning approach to predict conversion from a clinically isolated syndrome to multiple sclerosis was proposed by Zhang et al. [
49].
3.4. Cerebral Palsy (CP)
Cerebral palsy is a collection of neurological illnesses that begin in infancy or early childhood and impact physical movement and muscle coordination for the rest of one’s life. Damage or abnormalities in the developing brain create CP, which weakens the brain’s capacity to control movements, maintain posture, and balance. Palsy is related to the impairment of motor function, and cerebral refers to the brain. It affects the brain’s motor region’s outer layer (also known as the cerebral cortex), which controls muscular action. Zhang et al. [
50] described the application of supervised machine learning algorithms in the classification of the sagittal gait patterns of cerebral palsy in children with spastic diplegia in their study. Bertoncelli et al. [
51] identified factors associated with the autism spectrum disorder in adolescents with cerebral palsy using artificial intelligence (AI). The medical diagnosis of cerebral palsy rehabilitation using eye images in ML techniques was proposed by Illavarason et al. [
52].
3.5. Brain Tumor
A brain tumor is a collection of abnormal cells called neurons that form a mass. There are many distinct types of brain tumors. Certain brain tumors were benign (noncancerous), whereas the others were cancerous (malignant). The indications for a brain tumor vary based mainly on tumor size and location. Many tumors infiltrate the brain tissue and inflict direct injury, while others damage the surrounding brain. Missing borders, noise, and low-contrast factors affected brain tumor segmentation in medical image processing. MRI segmentation utilizing learning algorithms and patterns recognition technologies for analyzing brain data is particularly effective. The technique is a parametric model that considers functions chosen based on the density function [
53]. With modern clinical imaging modalities, the early detection of these brain tumors is critical for accessible therapy and healthy living. Particle emission tomography (PET), MRI, and computed tomography (CT) are the most popular modalities used to examine brain tumors [
54]. Anil et al. [
55] proposed a brain tumor detection method from a brain MRI using deep learning that classified into two classes: with tumor and without tumor. Wu et al. [
56] used an artificial intelligence algorithm to diagnose pregnancies complicated by brain tumors using ultrasonic diagnostics. The role of AI in the study of pediatric brain tumor imaging was also investigated in a comprehensive review by Huang et al. [
57].
3.6. Epilepsy and Seizures
Epilepsy is a neurological condition characterized by recurrent seizures. It is a prevalent long-term mental illness. A seizure is an abrupt shift in behavior caused by a moment of alteration in the brain’s electrical activity. Typically, the brain sends out small electric signals regularly. This produces epileptic seizures, which are electrical bursts in the brain. Epilepsy can be classified into four types: focal, generalized, combination-focal, and undetermined. The kind of seizure a person experiences depends upon the type of epilepsy they experience. When a seizure occurs while an EEG is recorded, the usual pattern of brain activity is disrupted, and unusual bradycardia patterns emerge. When a seizure occurs, EEG is recorded, the regular pattern of brain activity is disrupted, and unusual brain activity can be observed. The electrodes on the brain area where the seizure is occurring can show brain changes during focal seizures. Kaur et al. [
58] provided a synopsis of studies on the application of AI systems for real-time pattern detection in EEG for the clinical diagnosis of epileptic seizures. By replicating brain network dynamics, An et al. [
59] evaluated artificial intelligence and computational methodologies for the automatic diagnosis and optimal treatment for each epilepsy patient.
With the detailed analysis of NDs and their symptoms, the main focus is detecting NDs with AI. For detection, images of the brain or other parts of the nervous system are required, which is briefly described in the following section.
4. Neuroimaging Modalities
Neuroimaging modalities are screening procedures used to diagnose neurological diseases. For clinicians and neurologists, functional neuroimaging is critical information regarding brain function during the development of any condition [
60,
61,
62]. Specialists can learn a lot about persons who may have neurological issues using structural neuroimaging techniques [
63]. The most used neuroimaging modalities include magnetic resonance imaging (MRI) [
64], electroencephalography (EEG) [
65], magnetoencephalography (MEG) [
66,
67], positron emission tomography (PET) [
68,
69], single-photon emission computed tomography (SPECT) [
70,
71,
72], functional MRI (fMRI) [
73,
74,
75], computed tomography (CT) [
76], and functional near-infrared spectroscopy (fNIRS) [
77,
78,
79,
80], etc. In the following subsection, we discuss the 370 most commonly used neuroimaging modalities.
4.1. Magnetic Resonance Imaging (MRI)
MRI is the best clinical procedure for diagnosing and analyzing various diseases, including brain tumors and epilepsy [
81,
82]. Typically, a system controlled by hardware or computers aids in automating a procedure to produce precise and timely results. It is a painless and secure test that employs a magnetic field and radio waves to create high-resolution two-dimensional or three-dimensional images of the brain stem. The brain, spinal cord, and vascular anatomy have been described. Some advantages include witnessing anatomy in all three planes: axial, sagittal, and coronal. MRI outperforms CT in detecting circulating blood and cryptic vasculature abnormalities. It can also detect demyelinating disease and does not have the beam-hardening artifacts of the CT images. The posterior fossa was more prominent. As a result, MRI allows for a better visualization of the posterior fossa than CT. Ionizing radiation was not used during the imaging process.
4.2. Electroencephalography (EEG)
Electrical activity on the skull is recorded using EEG, and brain neurons play an influential part in stimulation. EEG is the most widely used method for studying the brain’s functional anatomy throughout neurological disorders and collecting brain activity. This is prevalent because of its superior temporal resolution, safety, and affordability [
83,
84]. These non-Gaussian and non-stationary signals are used to determine the type of brain disease by covering the brain’s electrical activity. Implementing either inbuilt amplifiers or external amplifiers has the primary goal of reducing the influence of ambient noise. The readings can distinguish normal and pathological brain processes. Experienced Neurologists investigate epilepsy by analyzing continuously recorded EEG signals. One of the problems with EEG is that it requires gels or saline liquids to reduce the skin–electrode resistance. In addition, it requires a significant amount of human effort and time over days, weeks, or even months.
4.3. Magnetoencephalography (MEG)
MEG scanning, or magnetoencephalography, is a brain imaging technology that detects and analyzes small magnetic fields created in the brain [
85,
86,
87]. The scanning produces a magnetic source image (MSI), which was used to identify the beginning of the seizures. MEG also monitors current flow in the brain to estimate the magnetic areas. Electric fields go through the skull more frequently than magnetic fields, and they have a better spatial resolution than EEG. The brain’s magnetic field was measured and evaluated using a neuroimaging approach. It works on the outside of the head and is now routinely used in clinical treatment. MEG has grown increasingly important, particularly for individuals suffering from epilepsy and brain malignancies. It could help discover brain regions with normal functions in epilepsy, tumors, or other mass lesions. MEG captures motions with an extremely high temporal and spatial resolution as well. As a result, scanners must be placed near the brain’s surface to detect the cerebral activity that produces small magnetic fields.
4.4. Positron Emission Tomography (PET)
Positron emission tomography (PET) is a functional imaging modality that visualizes with radioactive chemicals called radio tracers [
88,
89,
90]. It is a high-tech imaging technology that examines brain activity in real-time and accomplishes the non-invasive monitoring of cerebral blood flow, metabolism, and receptor binding. A PET-CT scan combined 3D-generated images for a more precise diagnosis. Initially, PET was utilized only in research due to the comparatively high costs and complexity of the associated equipment, including cyclotrons, PET scanners, and radio-chemistry laboratories. Owing to technological advancements and the ubiquity of PET scanners, PET scanning has been increasingly used in clinical neurology in recent decades to enhance our knowledge of illness etiology and facilitate diagnosis.
4.5. Functional Magnetic Resonance Imaging (fMRI)
Functional magnetic resonance imaging, or functional MRI, defines brain activity by distinguishing the variations in blood flow. The concept that cerebral blood flow and neuronal activity are connected is the foundation of this method. When a part of the brain is used, blood flow to that part of the brain increases. Because fMRI has a high spatial resolution, it is useful for detecting active brain regions [
91]. The fMRI method has a low time resolution of one to two [
92]. It also has a low head-movement resolution, which might cause distortions. Scans from fMRI are based on the same atomic physics principles as MRI scans. On the other hand, MRI scans depict anatomical structures, whereas fMRI scans measure metabolic function. As a result, the MRI scan results resemble three-dimensional depictions of anatomic structures. It is used to track the progression of brain cancers, assess how well the brain functions after a stroke or Alzheimer’s diagnosis, and detect where seizures originated in the brain.
4.6. Functional Near-Infrared Spectroscopy (fNIRS)
Similar to fMRI, functional near-infrared spectroscopy (fNIRS) is a non-invasive brain imaging technology that monitors variations in blood oxygenation [
93]. The approach detects variations in the absorption of light emitted by sources onto the surface of the head and is monitored by the detectors. Any brain surgery requires extra oxygen. Capillary red blood cells provide this extra oxygen to the neurons and increase blood flow in the active brain regions.
4.7. Computed Tomography (CT)
One of the most often-utilized diagnostics in neurology is computed tomography (CT) [
76]. In the 1970s, it revolutionized neurology by allowing the high-resolution viewing of cerebral structures. MRI has primarily replaced CT in the examination of several neurological conditions. However, it still plays a role in the crucial evaluation of stroke and head trauma patients. It assesses head trauma, severe headaches, dizziness, and other symptoms of an aneurysm, hemorrhage, stroke, and brain tumors using specialized X-ray equipment.
4.8. Single-Photon Emission Computed Tomography (SPECT)
A single-photon emission computed tomography (SPECT) scan is an imaging examination. This illustrates how blood flows through the tissues and organs [
70,
94,
95]. Seizures, strokes, stress fractures, infections, and spinal malignancies may all be diagnosed using the test. This scanning technique combines computed tomography (CT) and a radioactive tracer to produce a nuclear imaging scan. Experts can see how blood travels to tissues and organs using the tracers. It is primarily used to examine blood flow through the brain’s arteries and veins. It may detect diminished blood flow in wounded regions. It has been demonstrated to be more responsive to brain damage than both MRI or CT scanning in tests. This test differs from a PET scan. The tracer remains in the bloodstream of humans instead of being absorbed by surrounding tissues, limiting the images to areas where blood flows. SPECT scans are cheaper and more readily available than higher resolution PET scans.
The images or brain signals extracted from these modalities contain so much noise that they must be removed to classify better. The following section shows a standard number of pre-processing techniques.
9. Challenges and Opportunities
As previously noted, AI has played a crucial role in identifying neurological illnesses. It has transformed the massive amounts of data collected into clinically relevant data [
286]. However, there were significant restrictions and uncertain legal repercussions despite these advantages. Even with the most powerful algorithms, it is impossible to account for all the potentially beneficial or adverse side effects [
287]. DL algorithms prefer to minimize adverse reactions and confusing factors, such as test results, to reach their aim. This can impair patient safety and outcomes [
287].
Table 21 presents several advantages and the limitations of the DL and ML methods. Additionally, different reasoning methods, or a combination of approaches, such as case-based, rule-based, model-based, fuzzy logic, genetic algorithms, natural language processing, and neural networks, have been used by various computers in a literature review. Each method has its capabilities and limitations [
288]. The efficacy of each technique differed, limiting their application in detecting rare and complex disorders, such as multiple sclerosis [
288]. Nonetheless, they could assist patients and clinicians in making a rapid clinical diagnosis [
288].
9.1. Mostly Faced Challenges
DL-based frameworks for NLD prediction have become attractive with the significant growth in computational capacity and the remarkable development of DL tools. However, more studies should be performed to fine-tune DL algorithms to improve inferences. The following are some of the concerns, along with potential prospects.
9.1.1. Lack of Standard Data
In machine learning, data standards and open data repositories are lacking. For instance, the non-integration of motor and non-motor features and the lack of open data storage and available programming have hampered the integration of existing commercial medical instruments, such as Parkinson’s Kinetigraph TM, which have moderate healthcare coverage [
289].
9.1.2. Small Sample Size
The ML view implies that sample sizes ought to be a multiple of the number of input and output variables [
290]. However, studies have been conducted using small sample sizes of patients [
291,
292,
293]. Creating an extensive dataset of patients with these mental disorders can be highly beneficial to physicians for accurately diagnosing diseases. Further attention and more practical research in this field can fulfill the necessity for longitudinal studies or a follow-up study on a patient’s transition.
9.1.3. DL Algorithms Need a Large Trained Dataset
For massive datasets, DL algorithms provide impactful and accurate solutions. However, high-dimensional CNNs such as 2D-CNN and 3D-CNN for big and multimodal neuroimages will yield high accuracy. A generative adversarial network (GAN) can create a synthetic neuroimage utilized with a CNN. The availability of training datasets was one of the most significant impediments to the application procedure of DL in neuroimaging, which also comes in as a consequence of maintaining patient privacy. Simultaneously, annotating these data is a significant challenge that necessitates expert assistance. As a result, the dataset for uncommon diseases discovered was mainly unbalanced. Medical practitioners and data analysts must collaborate to solve dataset development and annotation challenges. Simultaneously, data augmentation techniques can be used to solve the problem of unbalanced data by altering the volume and quality of the data
9.1.4. Bias-Free Neuroimaging Dataset
It is challenging to construct a bias-free neuroimaging dataset because it is a legacy of a learning system that could result in a computational artifact. However, the risk can be addressed by incorporating an extensive dataset into the model, examining the link between the extracted features and fine-tuning the model’s parameters.
9.1.5. Limitation of ML Clinical Presentation
The clinical significance of ML in confounding groups with similar neurological, psychological, or pathological manifestations are limited: for example, ML’s ability to differentiate PNES not only from epileptic patients but also from patients based on psychopathological displays, including major depression, or ML’s ability to discern epilepsy from healthy controls versus application in patients with a panic disorder [
294], schizophrenia [
295], and drug-induced memory deficits with similar alterations in microstate C.
9.1.6. Non-Standardized Acquisition of Images
This resulted in discrepancies in the photos from different databases. It is a significant challenge when using DL to evaluate neuroimages. To solve this difficulty, it is suggested that transfer learning be used. These extensive training datasets are considered the foundation for attaining better results with ML and DL techniques. The lack of these datasets is one of the most significant impediments to the application process.
9.1.7. DL Models Are Black Box
A deep learning model is a black box that learns from data and models the data collection process. Instead of being explainable, these models can be interpreted. However, when the model is used to forecast with data that do not belong to the database, the black box fails miserably. When the mechanism is employed to foresee an explainable process, according to Rudin, DL is overly sophisticated, highly recursive, and challenging to comprehend [
296]. Consequently, the explanation frequently fails to provide sufficient information to understand the DL mechanism. As a result, there is a frequent transition between explainable and interpretable DL models.
9.1.8. Ethical and Legal Ramifications
While the ethical and legal implications are beyond the focus of this work, maintaining patient trust would be critical in supporting collaboration and AI application [
297]. AI-enabled computer-aided diagnostic (CAD) solutions [
298] are unfeasible in black-box situations. The legal implications are unclear, especially whether the manufacturer or the practitioner is to blame [
299]. Therefore, standards for evaluating AI technologies must be developed. It is debatable whether AI can replace doctors. Nonetheless, AI will play a more significant role in integrating healthcare.
9.1.9. Limitation of Supervised Architecture
Considering the time and labor necessary to manufacture label data, low scalability, and the selection of optimal bias levels, the supervised architecture was excluded. For image analyses, unsupervised learning was not a standard option. On the other hand, unsupervised architecture learns features from a dataset. It creates a data-driven decision support system from it. Consequently, an unsupervised deep architecture can overcome medical imaging-related issues.
9.1.10. Adversarial Noise
It can enhance neuroimages while also lowering classification accuracy. As a result, canceling adversarial errors is difficult.
9.1.11. Lack of Sufficient Hardware Resources
The most significant aspect of CAD is the identification of differentiating traits that can lead to the identification of the valuable bio-markers of NDs. In addition, owing to a lack of access to hardware resources and data availability, developing DL architectures to diagnose NDs is challenging. Although tools such as Google Colab and others now provide researchers with powerful computing processors, implementing and using these methodologies in the real world still presents numerous challenges [
300].
9.2. Future Research Directions
To overcome the existing challenges, approaches based on graph theory and machine learning have quickly emerged as a significant trend to understand better paths to appropriately diagnose and adequately handle disorders given the availability of high-dimensional data and increased computing capacities. Under the umbrella of machine learning-based solutions to neurological illnesses, deep learning has recently gained an ever-increasing position in the era of health and medical studies. Our best hope for treating neurological diseases in children is to combine applied deep learning with graph theory on a tailored scale. However, doctors or clinicians must practically diagnose mental disorders by acknowledging symptoms. These symptoms can be very similar to other diseases, which can be confusing. For example, symptoms such as difficulties in movements and memory or awareness loss were identical in several NDs. This is why doctors must be particular in distinguishing diseases. DL in neuroimaging is also derived from the desire to protect patient privacy. Simultaneously, annotating these data is a significant issue that necessitates expert assistance. As a result, the datasets discovered for uncommon diseases are typically unbalanced. These data primarily depend on brain signals that contain noise and artifacts. Therefore, the health industry, medical practitioners, and data scientists must solve dataset development and annotation challenges. Simultaneously, data augmentation techniques can be used to address the issue of unbalanced data by altering data volume and quality [
10].
Some other aspects need to be handled in the case of the neurological disorder detection directions. These are indicated in the following section.
9.2.1. Deep Brain Stimulation (DBS)
It is a safe and effective surgical therapy option for patients with tremor manifestations, including Parkinson’s disease (PD) [
301,
302] and essential tremor (ET) [
303,
304]. A neurologist currently determines DBS leads, and as a result, interpersonal variability is a factor. AI could assist in making an objective analysis in this aspect, provided that regulatory standards are met and medical clearance is obtained [
291].
9.2.2. Open Data Portals
Open data portals may be in people’s best interests [
86]. Study models, frameworks, algorithms, and anonymous data samples have all been made open source by some researchers, making it easier to replicate in future investigations [
291]. The use of low-cost smartphones with widely available customized apps boosts their usefulness [
291].
9.2.3. Testing Multiple Hypotheses
Compared with human skills, newer algorithms can test several hypotheses in an acceptable amount of time and make prior assumptions [
287]. It can be used to treat various diseases and specialties [
287]. ML can evaluate massive datasets at considerably faster rates with an improved accuracy. ML methods such as the self-organizing map (SOM) can be extended to include non-vestibular factors, including previous concussions, neuropsychological results, and various other variables [
305]. It also aids in the improvement of diagnostic criteria by identifying characteristics across a wide range of patient populations [
306].
9.2.4. Utilizing Methodology in Brain Signal Analysis
Researchers still could not figure out the ultimate solution for brain signal analysis. There is the requirement for making brain signals readable by using a method that can lead us to the solution, which is identifying the early stage diseases that can occur in the brain or neuron. There is massive scope for improving the classification methods in terms of the neurological disease detection process.
We must establish the best practices for evaluating AI tools [
307]. It is uncertain whether AI will replace physicians, but it will undoubtedly play a more prominent role in health care integration [
194].