Simple Summary
Fetuses with hypotrophy (FGR, fetal growth restriction) are too small for their gestational age and may be prone to various diseases and loss of life. This study aimed to determine the role of single nucleotide polymorphisms (SNPs), located in two homeotic and two angiogenesis-related genes, in the occurrence of FGR, by analyzing blood samples from 380 women in singleton pregnancies. We found that the AT heterozygotes in HLX rs868058 were significantly associated with an approximately two-fold increased risk of FGR, diagnosed before 32 weeks of gestation (early-onset FGR). AT heterozygotes were significantly more frequent in women with early-onset FGR than in those with late-onset FGR (diagnosed from 32 weeks of gestation) and compared with healthy subjects. In conclusion, the AT genotype in HLX rs868058 may be a significant risk factor for the development of early-onset FGR. So far, the only therapeutic strategy for the management of early-onset FGR is to monitor and terminate pregnancy when the risk of fetal immaturity is lower than the risk of intrauterine death. Therefore, the disclosure of the mechanisms of action of the heterozygous AT state in HLX rs868058 would be important to identify plausible targets for new therapeutic approaches to treat the condition.
Abstract
Fetal growth restriction (FGR) is a condition that characterizes fetuses as too small for their gestational age, with an estimated fetal weight (EFW) below the 10th percentile and abnormal Doppler parameters and/or with EFW below the 3rd percentile. We designed our study to demonstrate the contribution of single nucleotide polymorphisms (SNPs) from DLX3 (rs11656951, rs2278163, and rs10459948), HLX (rs2184658, and 868058), ANGPT2 (−35 G > C), and ITGAV (rs3911238, and rs3768777) genes in maternal blood in FGR. A cohort of 380 women with singleton pregnancies consisted of 190 pregnancies with FGR and 190 healthy full-term controls. A comparison of the pregnancies with an early-onset FGR and healthy subjects showed that the AT heterozygotes in HLX rs868058 were significantly associated with an approximately two-fold increase in disease risk (p ≤ 0.050). The AT heterozygotes in rs868058 were significantly more frequent in the cases with early-onset FGR than in late-onset FGR in the overdominant model (OR 2.08 95% CI 1.11–3.89, p = 0.022), and after being adjusted by anemia, in the codominant model (OR 2.45 95% CI 1.23–4.90, p = 0.034). In conclusion, the heterozygous AT genotype in HLX rs868058 can be considered a significant risk factor for the development of early-onset FGR, regardless of adverse pregnancy outcomes in women.
1. Introduction
Fetal growth restriction (FGR, fetal hypotrophy) is a condition that characterizes fetuses as too small for their gestational age that reveal an estimated fetal weight (EFW) below the 10th percentile and abnormal Doppler parameters and/or with EFW below the 3rd percentile, diagnosed in approximately 3–10% of all pregnancies [1,2]. FGR results from impaired genetic growth potential due to a pathological process of various etiologies, which leads to hypoxia and malnutrition of the fetus, imposing a serious threat to its health and life [3]. An FGR-affected newborn may be unable to maintain normal body temperature and may present respiratory distress, hypo- or hyperglycemia, susceptibility to infections, as well as cognitive delays plus neurological and psychiatric disorders in childhood [4,5,6].
Homeotic (homeobox) genes are the most important transcription factors that play a fundamental role in body structure pattern formation [7]. Several homeotic genes have been reported to be involved in placenta and embryo development [2,7]. In mouse models, the disrupted functions of distal-less homeobox 3 (Dlx3), extraembryonic, spermatogenesis, homeobox 1 (Esx1) and TGFB-induced factor homeobox 1 (Tgif1) genes resulted in an abnormal placenta development and embryonic hypotrophy [8,9]. In women with idiopathic FGR, significant differences were noted in the expression of the DLX3 and H2.0-like homeobox (HLX) genes, as well as in HLX downstream targets, compared to healthy controls without fetal hypotrophy [10,11,12]. Reduced expressions of HLX and ESX homeobox 1 (ESX1L) genes were found in a placenta with idiopathic FGR compared to those from control pregnancies [11]. On the other hand, in case of the DLX3, DLX4 and TGIF1 genes, an increased expression was shown in the FGR-affected placenta [7,13]. It is worth mentioning that TGIF1 has been shown to participate in the regulation of the expression of several genes, including angiopoietin 2 (ANGPT2) and the integrin subunit alpha V (ITGAV) involved in angiogenesis [14]. In the case of the DLX3 gene, its contribution has also been confirmed in the regulation of the expression of the peroxisome proliferator activated receptor gamma (PPARγ) transcription factor, involved in placenta development, trophoblast differentiation, and the occurrence of FGR [3,15].
Considering the genetic changes localized in homeotic genes previously related to FGR, today’s literature indicates single nucleotide polymorphisms (SNPs), both from DLX3 and HLX genes [16,17,18]. For the DLX3 gene, a certain association was demonstrated between the rs2278163 polymorphism and the occurrence of dental caries in children with higher loads of Streptococcus mutans and Streptococcus sobrinus [18]. Additionally, a weak correlation was found between the incidence of alleles in DLX3 rs10459948, localized near rs2278163, and a susceptibility to dental caries in children with higher loads of Streptococcus mutans [18]. In the case of rs2278163, some involvement was also demonstrated in the susceptibility to molar–incisor hypomineralization [19]. With regard to the HLX gene, several polymorphisms were associated with the clinical course of Graves’ disease (GD), the expression and secretion of type 1/2 T helper (Th1/Th2) cell line cytokines in neonates after birth, the onset of asthma in children, and the development of treatment-dependent acute myeloid leukemia [16,17,20,21]. In the case of HLX rs3806325 and rs2184648 polymorphisms, the presence of allelic variants −1407 T and 2742 G, respectively, was significantly associated with a reduced HLX promoter transactivation, which was accompanied by an almost complete decrease in the binding of the specificity protein-transcription factors to that region [21]. Regarding the angiogenesis-related genes, the ANGPT2 −35 G > C polymorphism was recently shown to be significantly correlated with high C-reactive protein levels and severity scores in patients with sepsis [22]. Among the ITGAV polymorphisms, both rs3911238 and rs3768777 were associated with rheumatoid arthritis, while rs3768777 was also correlated with a severe progression of primary biliary cirrhosis [23,24,25].
No studies have previously been performed to determine the possible role of SNPs from the DLX3 and HLX homeotic genes, as well as from the angiogenesis-related ANGPT2 and ITGAV genes, in the pathogenesis of FGR. Therefore, we designed a case–control genetic association study to demonstrate the contribution of DLX3 (rs11656951, rs2278163, rs10459948), HLX (rs2184658 and 868058), ANGPT2 (−35 G > C), and ITGAV (rs3911238 and rs3768777) polymorphisms in the occurrence of FGR.
2. Materials and Methods
2.1. Characteristics of Pregnant Women
A cohort of 380 women with singleton pregnancies included in our study consisted of 190 individuals with FGR and 190 healthy full-term (from 37 to 42 weeks) controls (see Table 1), being inpatients of the Department of Perinatology, Obstetrics and Gynecology, as well as of the Department of Obstetrics and Gynecology of the Polish Mother’s Memorial Hospital-Research Institute (PMMH-RI), in Lodz, Poland. Maternal blood samples were prospectively collected from all the pregnant women on admission during the period from August 2016 to March 2021. Among the diagnosed FGR cases, 58 (30.5%) women were characterized as early-onset FGRs, with the diagnosis obtained between the 18th and the 32nd week of gestation, and 129 (67.9%) were late-onset FGRs, where the diagnosis was obtained from the 32nd week to the 40th week of pregnancy (see Table S1). The women with FGR and early-onset FGR were 15 to 43 years old, while the control group was 19 to 43 years old. The patients with late-onset FGR ranged from 16 to 43 years of age. Early-onset preeclampsia (PE) was diagnosed in 7 (12.1%) women with early-onset FGR. The control group included women after 37 weeks of pregnancy and without FGR, admitted to the department for delivery. The exclusion criteria from the study included: multiple pregnancy, congenital anomalies, genetic syndrome, structural uterine defects, endometriosis, two-vessel umbilical cord, and fetal abnormalities. FGR was diagnosed by ultrasound when EFW was below the 10th percentile in relation to the gestational age and Doppler abnormalities were found, and/or EFW was below the 3rd percentile. The Fetal Medicine Barcelona (FMB) calculator [26] was used to assess percentiles based on ultrasound EFW, determined from biparietal diameter (BPD), head circumference (HC), abdominal circumference (AC), femur length (FL), and humerus length (HL), as well as umbilical artery (UA) and middle cerebral artery (MCA) pulsation indices, UA diastolic flow, and uterine artery (Ut.A) flows, estimated by Doppler ultrasound.

Table 1.
Characteristics of women with fetal growth restriction and healthy controls.
See Table 1 and Table S1 for detailed data on the number of pregnancies and the occurrence of certain pregnancy disorders among the studied pregnant women, including anemia, asthma and respiratory infections, bleeding, diabetes mellitus (DM), hypothyroidism, threatened miscarriage, thrombocytopenia and urogenital infections. The activated partial thromboplastin time (APTT) and platelet (PLT) parameters, including PLT count, platelet distribution width (PDW), mean platelet volume (MPV) and plateletcrit (PCT), are also presented for the women enrolled into the study. EFW was below the 10th percentile in FGR cases, while among the controls, it was between the 11th and the 100th percentiles, estimated by the FMB calculator. The study was approved by the Research Ethics Committee at the PMMH-RI (approval numbers 31/2018 and 13/2019). Informed consent forms were signed by all the invited pregnant women, as recommended by the Research Ethics Committee.
2.2. Blood Sample Collection and Analysis
Peripheral venous blood samples were collected on admission from each pregnant woman enrolled into the study, both for diagnostic and research purposes, being anonymized in the latter instance. Nine NC/1.4 mL coagulation tubes were used to evaluate APTT using the HemosIL APTT-SP reagent on an ACL TOP 550 CTS automated system (Instrumentation Laboratory, Werfen Company, Bedford, MA, USA). The APTT reference range was 23 to 36.9 s, according to the manufacturer. EDTA KE/1.2 mL tubes were used for complete blood count (CBC) and DNA extraction. PLT parameters were estimated as a part of the CBC complex using the Fluorocell PLT reagent on a Sysmex XN-2000 Automated Hematology System (Sysmex, Kobe, Japan). The PLT count was referenced between 150 × 109/L and 400 × 109/L, and the MPV was normal from 8.0 to 10.0 fL, as reported by the manufacturer (Sysmex, Kobe, Japan).
Total DNA was purified from 200 μL of whole-blood samples using a Syngen Blood/Cell DNA Mini Kit (Syngen Biotech, Wroclaw, Poland). The obtained DNA was eluted from a mini spin column in 100 μL of DE buffer and stored at −20 °C until further analysis.
2.3. PCR-RFLP Assays for ANGPT2, HLX, and ITGAV Polymorphisms
ANGPT2 −35 G > C, HLX rs2184658 and rs868058, as well as ITGAV rs3911238 and rs3768777 polymorphisms, were assayed by PCR-RFLP, as previously described [17,24,27,28,29]. The European minor allele frequencies (MAFs) for SNPs, localized on the HLX and ITGAV genes, were >10.0%, according to the NCBI Allele Frequency Aggregator (ALFA) project. The primer sequences and PCR-RFLP assay parameters are presented in Table S2. Briefly, PCR mixtures contained up to 0.5 μg of purified DNA, 0.2 mM dNTPs mix, 0.4 μM of each SNP-specific primer, 1 × polymerase B buffer, and 0.5 U of Perpetual Taq DNA Polymerase (EURx, Gdańsk, Poland). The PCR program included initial denaturation at 95 °C for 3 min, 40 cycles of denaturation at 95 °C for 30 s, annealing at 52.7–61.5 °C, depending on polymorphism, for 40 s, an extension at 72 °C for 1 min and a final extension at 72 °C for 7 min. The PCR products were digested with 10 U of the appropriate endonuclease at defined temperatures for 16 h. PCR and restriction digestions were performed on a T100 Thermal Cycler (Bio-Rad, Singapore). The PCR and RFLP products were separated in 1.0–3.4% agarose gels (see Figure 1), prepared in 1 × TAE buffer, depending on the length of analyzed DNA fragments, and visualized in a ChemiDoc XRS+ imaging system (Bio-Rad, Hercules, CA, USA).
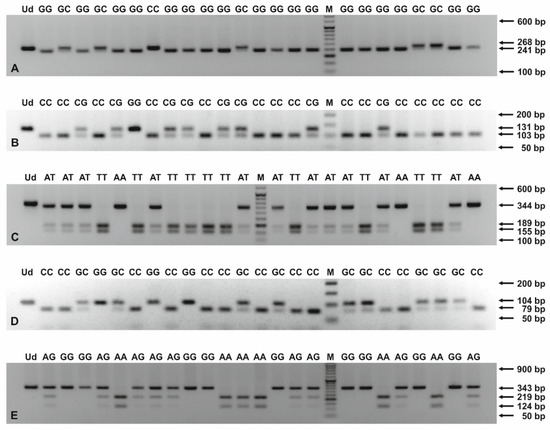
Figure 1.
PCR-RFLP products for genotyping of ANGPT2 −35 G > C (A), HLX rs2184658 (B), rs868058 (C), ITGAV rs3911238 (D), and rs3768777 (E) polymorphisms. The PCR products were digested with endonucleases: HindIII (A), MnlI (B), VspI (C), MvaI (D), and NlaIII (E), and then separated in 2.5–3.4% agarose gels, stained with ethidium bromide. The numbers to the right of the electropherograms show the lengths of separated DNA fragments. M: 50 bp DNA marker; Ud: undigested PCR product; AA, AG, AT, CC, CG, GC, GG, TT: genotypes in studied SNPs.
2.4. Sanger Sequencing for the SNPs Localized on DLX3 Gene
The primer sequences used for genotyping of the three SNPs, localized on DLX3 gene, i.e., rs11656951, rs2278163 and rs10459948, were self-designed using the PerlPrimer v1.1.21 software, and their specificity was checked by the Primer-BLAST tool [30]. The European MAFs for the tested DLX3 SNPs were >5.0%, according to the NCBI ALFA project. For rs11656951, PCR products of 331 bp length were obtained, using the following primers For: 5′-CCCACCTTAGACCATCTCTTTCC-3′ and Rev: 5′-CTCTCGCTCCTATGCTCTCC-3′ (primer concentration: 0.4 μM, annealing at 58 °C, 40 cycles). For rs2278163 and rs10459948, 337 bp PCR products were obtained with the primers: For: 5′-CATTTGATTGTGGCTTGGGAC-3′ and Rev: 5′-GTGACAGAAGACTCGGGCAG-3′ (primer concentration: 0.3 μM, annealing at 64 °C, 36 cycles). PCR was performed using Perpetual Taq DNA Polymerase (EURx, Gdańsk, Poland), similarly to the PCR-RFLP assays described in this study. PCR products were verified in 1.0% agarose gels and purified using the ExoSAP-IT™ PCR Product Cleanup Reagent (Thermo Fisher Scientific, Waltham, MA, USA). Sanger sequencing was performed with forward primers, using the BigDye™ Terminator v3.1 Cycle Sequencing Kit (Thermo Fisher Scientific), on a 3500 Genetic Analyzer (Thermo Fisher Scientific). Chromatograms (see Figure 2 and Figure 3) were analyzed using the Sequence Scanner 1.0 software (Applied Biosystems, Waltham, MA, USA).
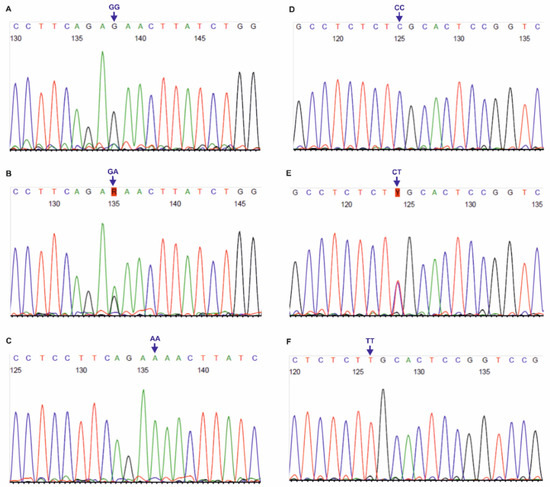
Figure 2.
Chromatograms of DNA forward strands for DLX3 rs11656951 (A–C) and rs2278163 (D–F) SNPs. AA, CC, CT, GA, GG, TT: genotypes in the tested polymorphisms.
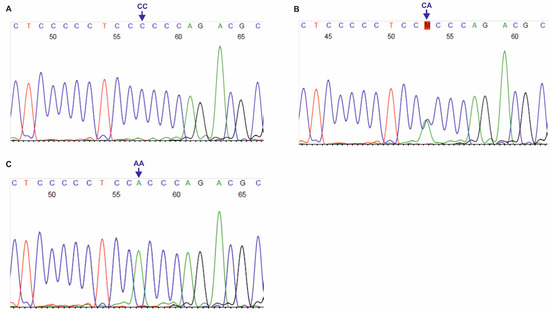
Figure 3.
Chromatograms of DNA forward strands for DLX3 rs10459948 polymorphism (A–C). AA, CA, CC: genotypes in the tested SNP.
2.5. Statistical Analysis
Descriptive statistics of the pregnant women were performed using NCSS 2004 software. Pearson’s chi-square test was used to determine differences between the studied groups of pregnant women in the number of pregnancies, the occurrence of pregnancy disorders, the methods of delivery, and the fetal sex. In order to compare the women in terms of age, APTT, PLT parameters, gestational age at delivery, as well as birth weight and Apgar scores of their offspring, Student’s t-test or Mann–Whitney U test were performed, depending on the normality level of the examined variables. The Hardy–Weinberg equilibrium and the frequencies of alleles and genotypes in the ANGPT2, DLX3, HLX, and ITGAV gene polymorphisms were determined using SNPStats software [31]. Relationships between the genotypes in the tested SNPs and the occurrence of FGR, as well as the models of inheritance, were estimated using logistic regression analyses. The distribution of alleles from the studied polymorphisms between the pregnant groups was determined using Pearson’s chi-square test. The results were considered statistically significant at the significance level of p ≤ 0.050.
3. Results
3.1. Females and Offspring with FGR and Healthy Controls
The pregnant women in the study groups were of a similar age (see Table 1 and Table S1). Among FGR cases, multiparity occurred much more frequently compared to healthy controls (p = 0.021). Considering pregnancy disorders, anemia, threatened miscarriage, and thrombocytopenia were significantly more frequent in women with FGR than in healthy subjects (p ≤ 0.050). Similarly, anemia was more common in the women with early-onset FGR compared to those with late-onset disease (p = 0.001). The APTT and PLT parameters reached similar values in the studied groups. A lower gestational age at delivery was observed among the women with FGR compared to healthy ones, and in the women with early-onset FGR than in those with late-onset FGR (p ≤ 0.001). Among the FGR cases, caesarean section was significantly more frequent than vaginal delivery vs. the healthy controls (p ≤ 0.050). The fetal sex of the offspring had a similar distribution between the studied groups of women. Neonatal birth weight and Apgar scores at 1 and 5 min were significantly lower in the FGRs than in healthy controls and in the early-onset FGR compared to the late-onset disease (p ≤ 0.001).
3.2. Hardy–Weinberg Equilibrium
The Hardy–Weinberg (H-W) equilibrium was maintained for the genotypes from DLX3, HLX and ITGAV polymorphisms in all the groups of pregnant women (p > 0.050). In ANGPT2 −35 G > C SNP, H-W was observed in FGR cases (p > 0.050), while it was not obtained in healthy controls (p = 0.027).
3.3. Genetic Alterations from ANGPT2, DLX3, HLX, and ITGAV Polymorphisms
The genotypes, defined in ANGPT2 −35 G > C, DLX3 rs11656951, rs2278163, rs10459948, HLX rs2184658 and rs868058, as well as in ITGAV rs3911238 and rs3768777, were similarly distributed among the women with FGR and the healthy controls (see Table S3). A comparison of the women with early-onset FGR and the healthy subjects showed that the AT heterozygotes in HLX rs868058 were significantly associated with an approximately two-fold increase in disease risk in the codominant (OR 2.18 95% CI 1.16–4.09, p = 0.045, see Table 2) and overdominant models (OR 2.11 95% CI 1.16–3.83, p = 0.014). The relationship was significant in the overdominant model (OR 1.99 95% CI 1.07–3.70, p = 0.030) and also after the cases of being large for their gestational age (54/190 (28.4%)) were excluded from the control group. That association was significant in both the codominant (OR 2.21 95% CI 1.08–4.53, p = 0.028) and overdominant models (OR 2.42 95% CI 1.21–4.82, p = 0.011) after the adjustment imposed by anemia. Similarly, the AT heterozygotes in rs868058 were significantly more frequent in the women with early-onset FGR than in those with the late-onset disease in the overdominant model (OR 2.08 95% CI 1.11–3.89, p = 0.022, see Table 3). The results from the study also remained significant in the codominant and/or overdominant models when corrected for pregnancy disorders including anemia, asthma and respiratory infections, bleeding, DM, hypothyroidism, miscarriage risk, thrombocytopenia, and urogenital infections (see Table 4 and Table S4). After the adjustment by anemia, the AT heterozygotes in HLX rs868058 were found significantly more often in the women with early-onset FGR, compared to those with late-onset disease, also in the codominant model (OR 2.45 95% CI 1.23–4.90, p = 0.034). The alleles, localized in all the analyzed SNPs, had a similar distribution pattern among the studied groups of pregnant women (see Tables S5 and S6).

Table 2.
Relationships among the polymorphisms in ANGPT2, DLX3, HLX, and ITGAV genes, and the early-onset fetal growth restriction.

Table 3.
Distribution of genotypes from ANGPT2, DLX3, HLX, and ITGAV polymorphisms between the women with early and late-onset FGR.

Table 4.
Association of HLX rs868058 genotypes with early-onset fetal growth restriction, corrected for adverse pregnancy outcomes.
3.4. Study Size Calculation
Based on the allele prevalence rates determined for all the polymorphisms analyzed in our populations of women with FGR and healthy controls, a minimum necessary sample size should have been 184 subjects, with a 95% confidence level and a 5% margin of error. Considering the analyses performed for the women with early-onset FGR and healthy controls, the number of enrolled individuals should have been at least 147, while for comparisons of early-onset FGR and late-onset disease, the minimum sample size should have been 123 women. All those results were obtained with respect to the allele frequencies, found for ITGAV rs3768777.
Taking into account the European MAFs, provided for SNPs from the DLX3, HLX, and ITGAV genes, as part of the NCBI ALFA project, we calculated that at least 338 pregnant women should have been included in our study. That minimum sample size was obtained for both ITGAV rs3768777 and HLX rs868058.
4. Discussion
The reported study showed that the AT heterozygotes for HLX rs868058 had contributed to an approximately two-fold increase in the risk of early-onset FGR in Caucasian pregnant women. It was previously determined that the homeobox gene HLX was mainly expressed in proliferating cytotrophoblastic cells during early placenta development [32]. It was then suggested that decreased HLX levels were necessary for cytotrophoblast differentiation, while altered gene expression could be associated with placental pathologies [32]. In term placental explants and the BeWo trophoblast cell line, lowered HLX expression was correlated with a knockdown of the insulin-like growth factor 2 receptor (IGF2R) involved in the regulation of villous trophoblast survival and apoptosis [33]. Noteworthy was the significantly reduced expression of both HLX and ESX1L, noted from the 27th week of pregnancy, which could be associated with a declined growth rate of the fetus, observed in the third trimester [34,35]. A decreased expression of HLX mRNA and protein was found in the placentas from the pregnancies with idiopathic FGR [11,12]. Similarly, a significantly lowered expression of the HLX gene at both mRNA and protein levels was determined in the placenta of FGR twins compared to the normal control cases [36]. HLX was suggested to have been involved in abnormal placenta development, found in discordant twin pregnancies [36].
Regarding the rs868058 polymorphism of the HLX gene, it was only investigated in relation to the development of asthma in children, but no relationship was found [21]. Our results, as reported here, were the first outcomes to suggest a possible clinical relevance of the presented SNP. Rs868058 is located in the third intron of the gene and is included in the non-coding transcript exon variant HLX-203 (transcript ID: ENST0000054919.2), according to the Ensembl genome browser. Moreover, rs868058 is located in the regulatory region ENSR00000020421, typed as a promoter, which—among others—contributes to trophoblast suppression and stable placenta. Regarding the ENSR00000020421 regulatory function, the T allele in rs868058 is indicated as a regulatory region variant, in line with the Ensembl browser. Based on the results of our study, AT heterozygotes appear to be involved in a deregulation process of the trophoblast function, resulting in placental imbalance and followed by early-onset FGR. For other SNPs localized in HLX, the association of some polymorphisms with GD, childhood asthma, and therapy-related acute myeloid leukemia (t-AML) was previously confirmed [17,20,21,37]. The G allele in rs2184658, associated with decreased HLX expression was more frequently identified in patients with intractable GD, compared to those with GD in remission [17]. We found no relationship in our study between this SNP and FGR. An association with two tagging SNPs, rs3806325 and rs12141189, representing seven HLX polymorphisms, was reported in childhood asthma [21]. In turn, an over three-fold increased risk of t-AML was found in carriers of the CT genotype or at least one polymorphic T allele in the rs2738756 polymorphism [20].
The genetic results of our study remained also significant when corrected by pregnancy disorders, including anemia, asthma and respiratory infections, bleeding, DM, hypothyroidism, threatened miscarriage, thrombocytopenia, and urogenital infections. Previously, maternal anemia was, among other things, correlated with preterm birth (PTB), low birth weight, perinatal mortality, and maternal death [38]. The incidence of gestational iron deficiency anemia was significantly much higher in the women with FGR, as well as in PTB [39]. Similarly, maternal anemia was in our study significantly more common in the FGRs, and especially in those with early-onset disease, compared to the healthy controls. Considering pregnant women with moderate or severe asthma, a higher incidence of spontaneous abortion, fetal structural anomalies, PTB, PE, FGR, oligohydramnios, gestational diabetes, and intrauterine fetal death was recently confirmed [40]. In pregnant women with symptomatic asthma, maternal hypoxia was suggested as a possible mechanism of FGR [41]. In our study, asthma and respiratory infections had a similar distribution pattern among the study groups, although the disorder was more frequent in cases of early-onset FGR compared to healthy controls. Conversely, decidual hemorrhage observed in the first trimester was previously associated with later adverse pregnancy outcomes, including fetal loss, PE, abruption, FGR, and PTB [42]. During the second half of pregnancy, independent risk factors for bleeding were reported, including oligo- and polyhydramnios, FGR, previous abortions, and advanced maternal age [43]. Bleeding was in our study more frequent among FGR cases, when compared to healthy controls, but the difference was statistically insignificant. It was also suggested from the first trimester that adverse pregnancy conditions, possibly involved in fetal nutrient restriction, such as smoking, cocaine use, chronic hypertension, anemia, and chronic DM, lead to symmetrical FGR [44]. However, in our study, DM was similarly distributed between the women with FGR and healthy controls.
Regarding subclinical hypothyroidism (SCH), it has recently been linked to FGR, although no effects on the risk of the disease were found for SCH, thyroid peroxidase antibody and isolated hypothyroxinemia [45]. In Bangladesh, overt hypothyroidism predisposed women to pregnancy-induced hypertension, FGR, and gestational diabetes, compared to the subclinical disease [46]. In line with our outcomes, hypothyroidism had a similar distribution among the groups of enrolled pregnant women, although a slightly higher prevalence was observed for FGR, as compared to control cases. Previously, no differences were observed in the incidence of FGR between the groups of women without bleeding and threatened abortion [47]. In our study, threatened miscarriage was found only in the women with FGR, as cases with that diagnosis were excluded from the healthy control group. Considering the mean platelet count, it was observed that the values decreased during gestation in women with pregnancy-related complications, as well as in healthy subjects, starting from the first trimester [48]. However, gestational thrombocytopenia at delivery was more common in the women with adverse pregnancy outcomes compared to healthy controls [48]. In the current study, gestational thrombocytopenia was also more prevalent among the women with FGR, particularly those with the early-onset disease, compared to the healthy controls. Regarding urogenital infections, an increased risk of FGR was previously determined in cases of vaginal and cervical infections with Bacteroides, Prevotella, Porphyromonas spp., Mycoplasma hominis, Ureaplasma urealyticum, and Trichomonas vaginalis [49]. In another study, a colonization of the maternal genital tract with Chlamydia trachomatis and Candida albicans was also associated with FGR [50]. Currently, we observed a similar distribution of urogenital infections in the studied groups of pregnant women.
In conclusion, the heterozygous AT genotype in HLX rs868058 can be considered a significant risk factor for the development of early-onset FGR, regardless of adverse pregnancy outcomes in women. Although early-onset FGR is now diagnosed without difficulty, it is still an incurable disorder of pregnancy. The only available therapeutic strategy for the management of early-onset FGR is to monitor and terminate pregnancy when the risk associated with fetal immaturity is lower than the risk of intrauterine death. It would be extremely important to understand the signaling pathways involved in the heterozygous AT state in HLX rs868058 in order to identify targets for new therapeutic approaches that are still needed to treat early-onset FGR.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/biology11030447/s1. Table S1: Characteristics of women with early and late-onset fetal growth restriction. Table S2: Selected PCR-RFLP parameters for genotyping of single nucleotide polymorphisms, localized in HLX, ITGAV, and ANGPT2 genes. Table S3: Distribution of the genotypes from ANGPT2, DLX3, HLX, and ITGAV polymorphisms between women with FGR and healthy controls. Table S4: Distribution of HLX rs868058 genotypes between women with early-onset and late-onset FGR, adjusted by adverse pregnancy outcomes. Table S5: Distribution of the alleles from the ANGPT2, DLX3, HLX, and ITGAV SNPs between women with fetal growth restriction and healthy controls. Table S6: Distribution of the alleles from the ANGPT2, DLX3, HLX, and ITGAV polymorphisms between women with early and late-onset FGR.
Author Contributions
Conceptualization, W.I.W. and M.G.; Data curation, W.I.W., M.K. (Michał Krekora), P.K. and M.G.; Formal analysis, W.I.W., M.K. (Marian Kacerovsky), P.K. and M.G.; Funding acquisition, W.I.W. and M.G.; Investigation, W.I.W., M.K. (Michał Krekora), P.K. and M.G.; Methodology, W.I.W., M.K. (Michał Krekora), P.K. and M.G.; Project administration, W.I.W. and M.G.; Resources, W.I.W., M.K. (Michał Krekora), P.K. and M.G.; Supervision, W.I.W. and M.G.; Visualization, W.I.W.; Writing—Original draft, W.I.W.; Writing—Review and editing, W.I.W., M.K. (Marian Kacerovsky), M.K. (Michał Krekora), P.K., B.L. and M.G. All authors have read and agreed to the published version of the manuscript.
Funding
The study was funded by the Polish Ministry of Science and Higher Education, Polish Mother’s Memorial Hospital-Research Institute (Statutory Research Task No. 2018/I/22-SZB).
Institutional Review Board Statement
The study was conducted according to the guidelines of the Declaration of Helsinki and approved by the Research Ethics Committee of the Polish Mother’s Memorial Hospital–Research Institute (approval numbers 31/2018 and 13/2019).
Informed Consent Statement
Informed consent was obtained from all subjects involved in the study.
Data Availability Statement
All data and materials as well as software application support the published claims and comply with field standards.
Acknowledgments
We thank: Agnieszka Gach, Department of Genetics (DG), PMMH-RI, for providing the 3500 Genetic Analyzer; Anna Nykel, DG, PMMH-RI, for providing the Sanger sequencing method; Anna Nykel and Kinga Sałacińska, DG, PMMH-RI, for their invaluable assistance in the selection of primers, the preliminary assessment of sample chromatograms and the overall technical support in the use of the 3500 Genetic Analyzer.
Conflicts of Interest
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
References
- Murthi, P.; Said, J.M.; Doherty, V.L.; Donath, S.; Nowell, C.J.; Brennecke, S.P.; Kalionis, B. Homeobox gene DLX4 expression is increased in idiopathic human fetal growth restriction. Mol. Hum. Reprod. 2006, 12, 763–769. [Google Scholar] [CrossRef] [PubMed]
- Murthi, P.; Rajaraman, G.; Brennecke, S.P.; Kalionis, B. The role of placental homeobox genes in human fetal growth restriction. J. Pregnancy 2011, 2011, 548171. [Google Scholar] [CrossRef] [PubMed]
- Chui, A.; Kalionis, B.; Abumaree, M.; Cocquebert, M.; Fournier, T.; Evain-Brion, D.; Brennecke, S.P.; Murthi, P. Downstream targets of the homeobox gene DLX3 are differentially expressed in the placentae of pregnancies affected by human idiopathic fetal growth restriction. Mol. Cell Endocrinol. 2013, 377, 75–83. [Google Scholar] [CrossRef] [PubMed]
- Karowicz-Bilińska, A. Intrauterine growth restriction. Gin Perinat Prakt. 2018, 3, 93–102. [Google Scholar]
- Majewska, M.; Lipka, A.; Paukszto, L.; Jastrzebski, J.P.; Szeszko, K.; Gowkielewicz, M.; Lepiarczyk, E.; Jozwik, M.; Majewski, M.K. Placenta Transcriptome Profiling in Intrauterine Growth Restriction (IUGR). Int. J. Mol. Sci. 2019, 20, 1510. [Google Scholar] [CrossRef]
- Wixey, J.A.; Chand, K.K.; Colditz, P.B.; Bjorkman, S.T. Review: Neuroinflammation in intrauterine growth restriction. Placenta 2017, 54, 117–124. [Google Scholar] [CrossRef]
- Murthi, P.; Kalionis, B.; Rajaraman, G.; Keogh, R.J.; Da Silva Costa, F. The role of homeobox genes in the development of placental insufficiency. Fetal Diagn. Ther. 2012, 32, 225–230. [Google Scholar] [CrossRef]
- Bartholin, L.; Melhuish, T.A.; Powers, S.E.; Goddard-Leon, S.; Treilleux, I.; Sutherland, A.E.; Wotton, D. Maternal Tgif is required for vascularization of the embryonic placenta. Dev. Biol. 2008, 319, 285–297. [Google Scholar] [CrossRef]
- Murthi, P. Review: Placental homeobox genes and their role in regulating human fetal growth. Placenta 2014, 35, S46–S50. [Google Scholar] [CrossRef]
- Chui, A.; Tay, C.; Cocquebert, M.; Sheehan, P.; Pathirage, N.A.; Donath, S.; Fournier, T.; Badet, J.; Evain-Brion, D.; Brennecke, S.P.; et al. Homeobox gene Distal-less 3 is a regulator of villous cytotrophoblast differentiation and its expression is increased in human idiopathic foetal growth restriction. J. Mol. Med. 2012, 90, 273–284. [Google Scholar] [CrossRef]
- Murthi, P.; Doherty, V.; Said, J.; Donath, S.; Brennecke, S.P.; Kalionis, B. Homeobox gene HLX1 expression is decreased in idiopathic human fetal growth restriction. Am. J. Pathol. 2006, 168, 511–518. [Google Scholar] [CrossRef] [PubMed]
- Rajaraman, G.; Murthi, P.; Pathirage, N.; Brennecke, S.P.; Kalionis, B. Downstream targets of homeobox gene HLX show altered expression in human idiopathic fetal growth restriction. Am. J. Pathol. 2010, 176, 278–287. [Google Scholar] [CrossRef]
- Pathirage, N.A.; Cocquebert, M.; Sadovsky, Y.; Abumaree, M.; Manuelpillai, U.; Borg, A.; Keogh, R.J.; Brennecke, S.P.; Evain-Brion, D.; Fournier, T.; et al. Homeobox gene transforming growth factor beta-induced factor-1 (TGIF-1) is a regulator of villous trophoblast differentiation and its expression is increased in human idiopathic fetal growth restriction. Mol. Hum. Reprod. 2013, 19, 665–675. [Google Scholar] [CrossRef] [PubMed]
- Gunatillake, T.; Yong, H.E.; Dunk, C.E.; Keogh, R.J.; Borg, A.J.; Cartwright, J.E.; Whitley, G.S.; Murthi, P. Homeobox gene TGIF-1 is increased in placental endothelial cells of human fetal growth restriction. Reproduction 2016, 152, 457–465. [Google Scholar] [CrossRef] [PubMed]
- Murthi, P.; Kalionis, B.; Cocquebert, M.; Rajaraman, G.; Chui, A.; Keogh, R.J.; Evain-Brion, D.; Fournier, T. Homeobox genes and down-stream transcription factor PPARgamma in normal and pathological human placental development. Placenta 2013, 34, 299–309. [Google Scholar] [CrossRef] [PubMed]
- Casaca, V.I.; Illi, S.; Suttner, K.; Schleich, I.; Ballenberger, N.; Klucker, E.; Turan, E.; Von Mutius, E.; Kabesch, M.; Schaub, B. TBX21 and HLX1 polymorphisms influence cytokine secretion at birth. PLoS ONE 2012, 7, e31069. [Google Scholar] [CrossRef]
- Morita, M.; Watanabe, M.; Inoue, N.; Inaoka, C.; Akamizu, T.; Tatsumi, K.I.; Hidaka, Y.; Iwatani, Y. Functional polymorphisms in TBX21 and HLX are associated with development and prognosis of Graves’ disease. Autoimmunity 2012, 45, 129–136. [Google Scholar] [CrossRef]
- Ohta, M.; Nishimura, H.; Asada, Y. Association of DLX3 gene polymorphism and dental caries susceptibility in Japanese children. Arch. Oral Biol. 2015, 60, 55–61. [Google Scholar] [CrossRef]
- Jeremias, F.; Pierri, R.A.; Souza, J.F.; Fragelli, C.M.; Restrepo, M.; Finoti, L.S.; Bussaneli, D.G.; Cordeiro, R.C.; Secolin, R.; Maurer-Morelli, C.; et al. Family-Based Genetic Association for Molar-Incisor Hypomineralization. Caries Res. 2016, 50, 310–318. [Google Scholar] [CrossRef]
- Jawad, M.; Seedhouse, C.H.; Russell, N.; Plumb, M. Polymorphisms in human homeobox HLX1 and DNA repair RAD51 genes increase the risk of therapy-related acute myeloid leukemia. Blood 2006, 108, 3916–3918. [Google Scholar] [CrossRef][Green Version]
- Suttner, K.; Ruoss, I.; Rosenstiel, P.; Depner, M.; Pinto, L.A.; Schedel, M.; Adamski, J.; Illig, T.; Schreiber, S.; von Mutius, E.; et al. HLX1 gene variants influence the development of childhood asthma. J. Allergy Clin. Immunol. 2009, 123, 82–88. [Google Scholar] [CrossRef] [PubMed]
- Szederjesi, J.; Lazar, A.; Petrisor, M.; Hutanu, A.; Tripon, F.; Georgescu, A.M.; Azamfirei, L. Genetic variability of ANG2 -35G>C gene as a predictor factor in sepsis. Rev. Romana Medicina Lab. 2020, 28, 175–184. [Google Scholar] [CrossRef]
- Huang, J.M.; Pang, Z.Y.; Qi, G.B.; Wang, Z.; Lv, Z.T. Association of ITGAV polymorphisms and risk of rheumatoid arthritis: Evidence from a meta-analysis. Expert. Rev. Clin. Immunol. 2020, 16, 631–640. [Google Scholar] [CrossRef] [PubMed]
- Inamine, T.; Nakamura, M.; Kawauchi, A.; Shirakawa, Y.; Hashiguchi, H.; Aiba, Y.; Taketomi, A.; Shirabe, K.; Nakamuta, M.; Hayashi, S.; et al. A polymorphism in the integrin αV subunit gene affects the progression of primary biliary cirrhosis in Japanese patients. J. Gastroenterol. 2011, 46, 676–686. [Google Scholar] [CrossRef] [PubMed]
- Shakiba, E.; Tavilani, H.; Goodarzi, M.T.; Kiani, A.; Pourmotabbed, T.; Vaisi-Raygani, A. The ITGAV-rs3911238 polymorphism is associated with disease activity in rheumatoid arthritis. Iran. J. Allergy Asthma Immunol. 2014, 13, 356–363. [Google Scholar]
- Calculadoras|Medicina Fetal Barcelona. 2021. Available online: http://medicinafetalbarcelona.org/calc/ (accessed on 25 January 2022).
- Bányász, I.; Bokodi, G.; Vannay, A.; Szebeni, B.; Treszl, A.; Vásárhelyi, B.; Tulassay, T.; Szabó, A. Genetic polymorphisms of vascular endothelial growth factor and angiopoietin 2 in retinopathy of prematurity. Curr. Eye Res. 2006, 31, 685–690. [Google Scholar] [CrossRef]
- Jacq, L.; Garnier, S.; Dieudé, P.; Michou, L.; Pierlot, C.; Migliorini, P.; Balsa, A.; Westhovens, R.; Barrera, P.; Alves, H.; et al. The ITGAV rs3738919-C allele is associated with rheumatoid arthritis in the European Caucasian population: A family-based study. Arthritis Res. Ther. 2007, 9, R63. [Google Scholar] [CrossRef]
- Suttner, K.L. Charakterisierung von Genetischen Varianten in den Immunregulatorischen Transkriptionsfaktoren TBX21, HLX1 und GATA3 und Deren Funktionelle Rolle Bei der Entstehung von Asthma Bronchiale; Technische Universität München: München, Germany, 2009. [Google Scholar]
- Primer-BLAST Tool. 2021. Available online: https://www.ncbi.nlm.nih.gov/tools/primer-blast/index.cgi (accessed on 25 January 2022).
- SNPStats Software. 2021. Available online: https://www.snpstats.net/start.htm (accessed on 25 January 2022).
- Rajaraman, G.; Murthi, P.; Quinn, L.; Brennecke, S.P.; Kalionis, B. Homeodomain protein HLX is expressed primarily in cytotrophoblast cell types in the early pregnancy human placenta. Reprod Fertil Dev. 2008, 20, 357–367. [Google Scholar] [CrossRef]
- Harris, L.K.; Pantham, P.; Yong, H.E.J.; Pratt, A.; Borg, A.J.; Crocker, I.; Westwood, M.; Aplin, J.; Kalionis, B.; Murthi, P. The role of insulin-like growth factor 2 receptor-mediated homeobox gene expression in human placental apoptosis, and its implications in idiopathic fetal growth restriction. Mol. Hum. Reprod. 2019, 25, 572–585. [Google Scholar] [CrossRef]
- Boudreau, N.J.; Varner, J.A. The homeobox transcription factor Hox D3 promotes integrin alpha5beta1 expression and function during angiogenesis. J. Biol. Chem. 2004, 279, 4862–4868. [Google Scholar] [CrossRef]
- Gorski, D.H.; Leal, A.J. Inhibition of endothelial cell activation by the homeobox gene Gax. J. Surg. Res. 2003, 111, 91–99. [Google Scholar] [CrossRef]
- Yuen, N.; Brennecke, S.P.; Umstad, M.P.; Yong, H.E.J.; Borg, A.J.; Rajaraman, G.; Kalionis, B.; Murthi, P. Expression of Homeobox Gene HLX and its Downstream Target Genes are Altered in Placentae from Discordant Twin Pregnancies. Twin Res. Hum. Genet. 2018, 21, 42–50. [Google Scholar] [CrossRef] [PubMed]
- Suttner, K.; Rosenstiel, P.; Depner, M.; Schedel, M.; Pinto, L.A.; Ruether, A.; Adamski, J.; Klopp, N.; Illig, T.; Vogelberg, C.; et al. TBX21 gene variants increase childhood asthma risk in combination with HLX1 variants. J. Allergy Clin. Immunol. 2009, 123, 1062–1068. [Google Scholar] [CrossRef] [PubMed]
- Jung, J.; Rahman, M.M.; Rahman, M.S.; Swe, K.T.; Islam, M.R.; Rahman, M.O.; Akter, S. Effects of hemoglobin levels during pregnancy on adverse maternal and infant outcomes: A systematic review and meta-analysis. Ann. N. Y. Acad. Sci. 2019, 1450, 69–82. [Google Scholar] [CrossRef]
- Kemppinen, L.; Mattila, M.; Ekholm, E.; Pallasmaa, N.; Törmä, A.; Varakas, L.; Mäkikallio, K. Gestational iron deficiency anemia is associated with preterm birth, fetal growth restriction, and postpartum infections. J. Perinat Med. 2020, 49, 431–438. [Google Scholar] [CrossRef] [PubMed]
- Tanacan, A.; Fadiloglu, E.; Celebioglu, E.D.; Orhan, N.; Unal, C.; Celik, T.; Kalyoncu, A.F.; Beksac, M.S. The Effect of Asthma Severity on Perinatal Outcomes: A Tertiary Hospital Experience. Z Geburtshilfe Neonatol. 2021, 225, 333–340. [Google Scholar] [CrossRef]
- Sims, C.D.; Chamberlain, G.V.; de Swiet, M. Lung function tests in bronchial asthma during and after pregnancy. Br. J. Obstet. Gynaecol. 1976, 83, 434–437. [Google Scholar] [CrossRef]
- Schatz, F.; Guzeloglu-Kayisli, O.; Arlier, S.; Kayisli, U.A.; Lockwood, C.J. The role of decidual cells in uterine hemostasis, menstruation, inflammation, adverse pregnancy outcomes and abnormal uterine bleeding. Hum. Reprod. Update 2016, 22, 497–515. [Google Scholar] [CrossRef]
- Koifman, A.; Levy, A.; Zaulan, Y.; Harlev, A.; Mazor, M.; Wiznitzer, A.; Sheiner, E. The clinical significance of bleeding during the second trimester of pregnancy. Arch. Gynecol. Obstet. 2008, 278, 47–51. [Google Scholar] [CrossRef]
- Chew, L.C.; Verma, R.P. Fetal Growth Restriction. StatPearls Publishing LLC. 2021. Available online: https://www.ncbi.nlm.nih.gov/books/NBK562268/ (accessed on 27 December 2021).
- Tong, Z.; Xiaowen, Z.; Baomin, C.; Aihua, L.; Yingying, Z.; Weiping, T.; Zhongyan, S. The Effect of Subclinical Maternal Thyroid Dysfunction and Autoimmunity on Intrauterine Growth Restriction: A Systematic Review and Meta-Analysis. Medicine 2016, 95, e3677. [Google Scholar] [CrossRef]
- Sharmeen, M.; Shamsunnahar, P.A.; Laita, T.R.; Chowdhury, S.B. Overt and subclinical hypothyroidism among Bangladeshi pregnant women and its effect on fetomaternal outcome. Bangladesh Med. Res. Counc. Bull. 2014, 40, 52–57. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Petriglia, G.; Palaia, I.; Musella, A.; Marchetti, C.; Antonilli, M.; Brunelli, R.; Ostuni, R.; Panici, P.B. Threatened abortion and late-pregnancy complications: A case-control study and review of literature. Minerva Ginecol. 2015, 67, 491–497. [Google Scholar] [PubMed]
- Reese, J.A.; Peck, J.D.; Deschamps, D.R.; McIntosh, J.J.; Knudtson, E.J.; Terrell, D.R.; Vesely, S.K.; George, J.N. Platelet Counts during Pregnancy. N. Engl. J. Med. 2018, 379, 32–43. [Google Scholar] [CrossRef] [PubMed]
- Germain, M.; Krohn, M.A.; Hillier, S.L.; Eschenbach, D.A. Genital flora in pregnancy and its association with intrauterine growth retardation. J. Clin. Microbiol. 1994, 32, 2162–2168. [Google Scholar] [CrossRef]
- Association of Chlamydia trachomatis and Mycoplasma hominis with intrauterine growth retardation and preterm delivery. The John Hopkins Study of Cervicitis and Adverse Pregnancy Outcome. Am. J. Epidemiol. 1989, 129, 1247–1257.
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).