Transcription Regulation of Tceal7 by the Triple Complex of Mef2c, Creb1 and Myod
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Plasmid Construction
2.2. Cell Culture and Cell Transfection
2.3. Transcription Assays
2.4. Western Blot and Coimmunoprecipitation Assays
2.5. ChIP Assays
2.6. Statistics
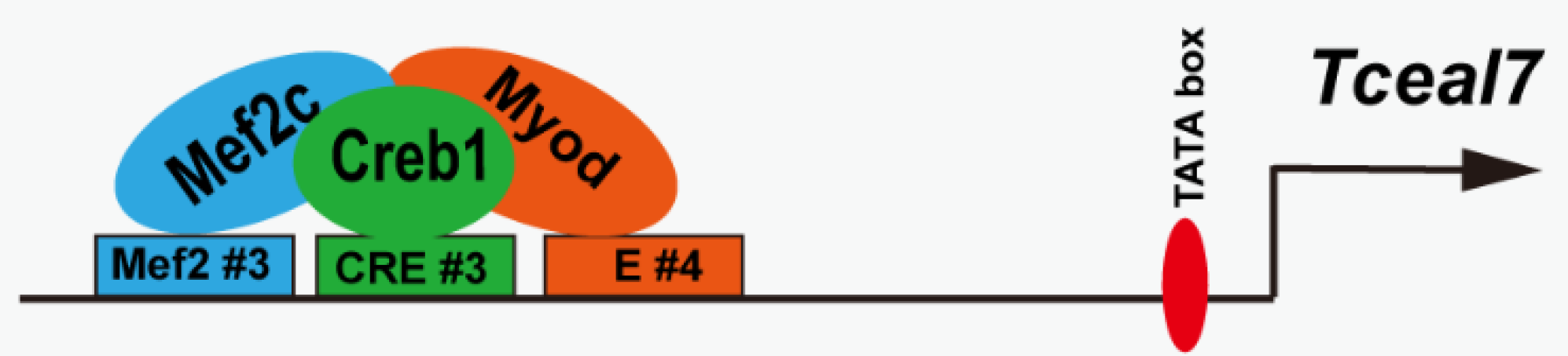
3. Results
3.1. The Functional Role of Mef2 and CRE Motifs in Tceal7 Promoter
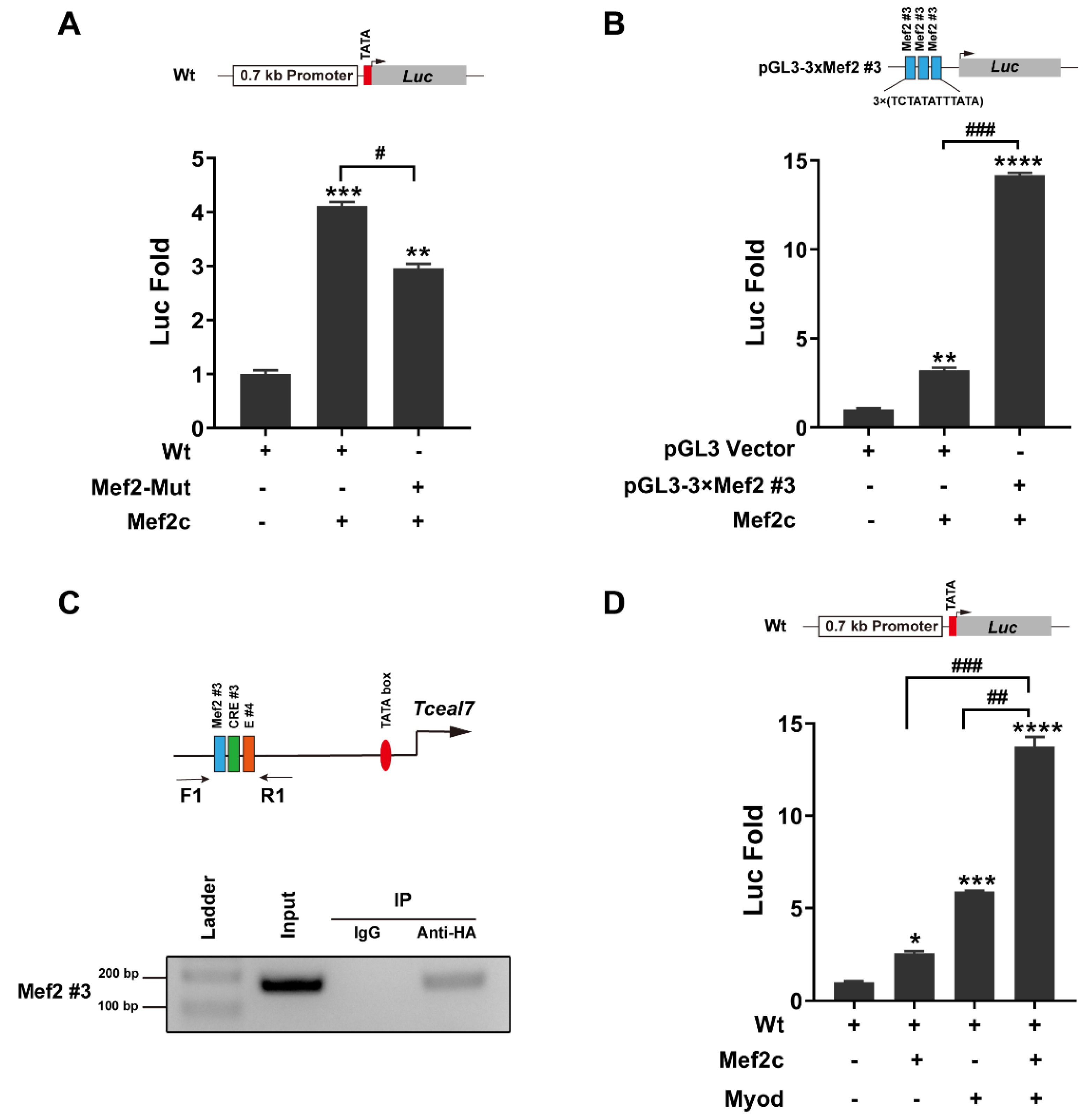
3.2. Transactivation of Tceal7 by Mef2c in the Distal Region
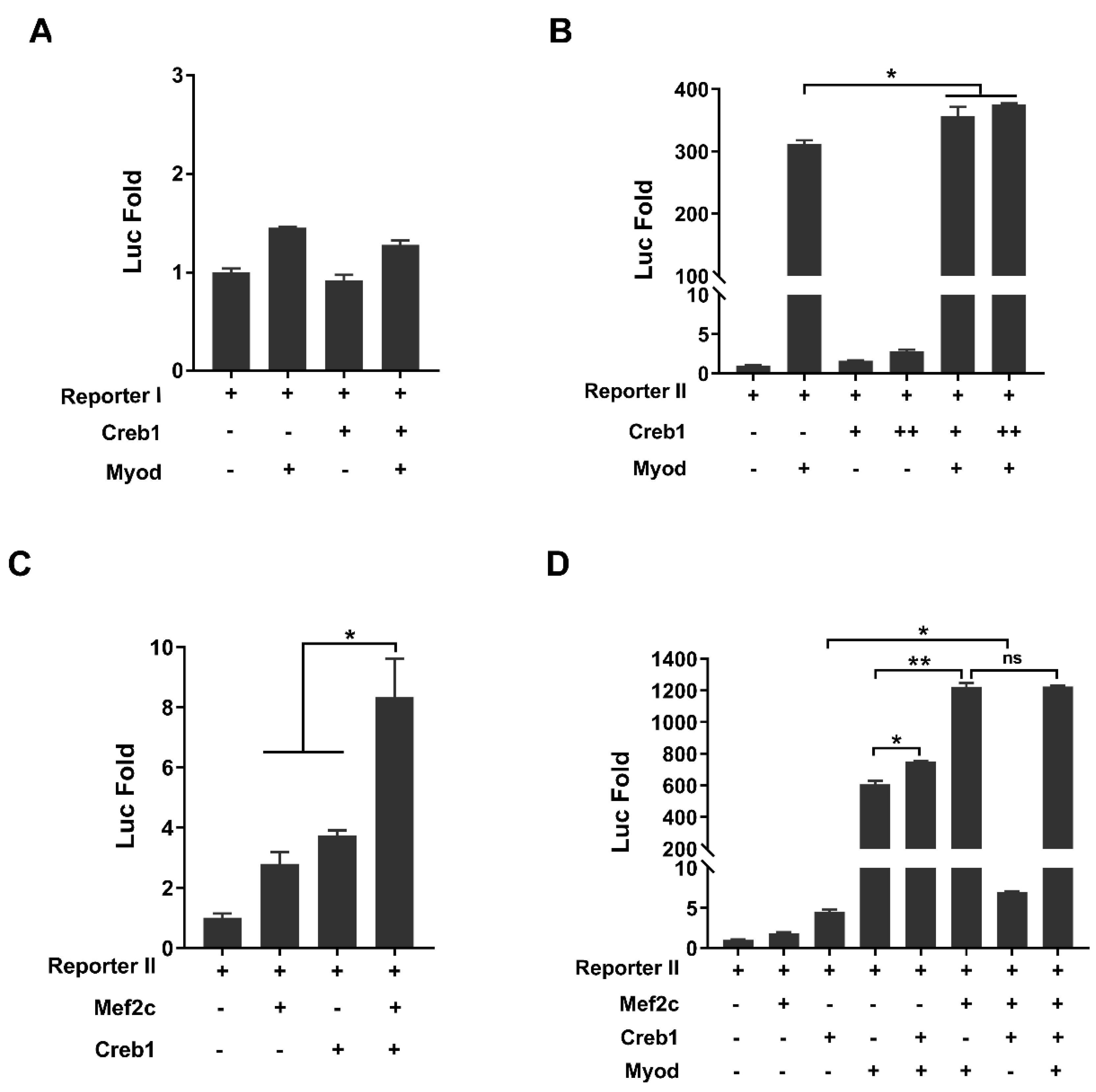
3.3. Myod and Creb1 Transactivate Tceal7 Expression through the Distal Region
3.4. Transactivation of Tceal7 Expression by Myod, Mef2c and Creb1
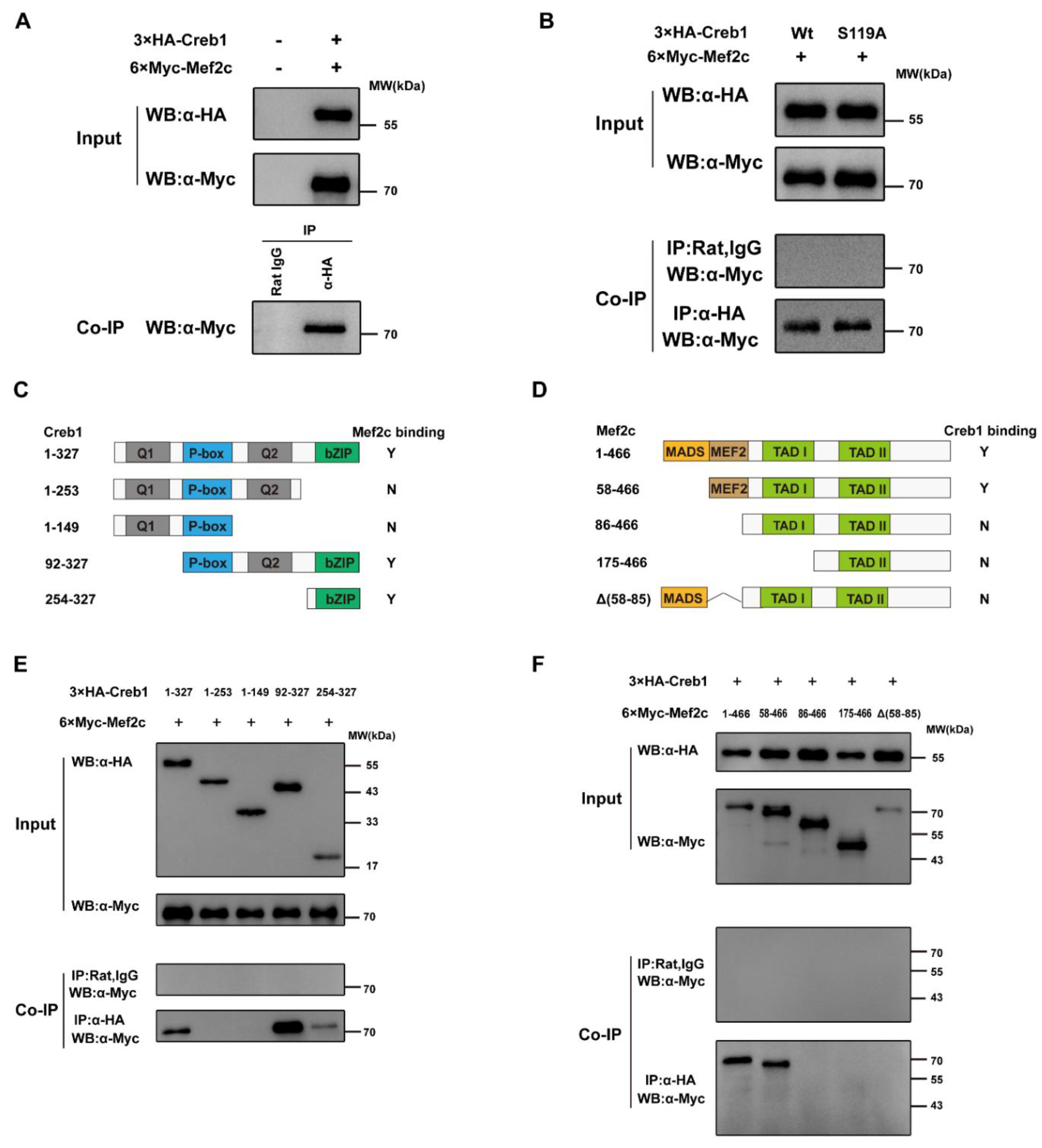
3.5. Mef2c Interacts with Creb1
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Hernández-Hernández, J.M.; García-González, E.G.; Brun, C.E.; Rudnicki, M.A. The myogenic regulatory factors, determinants of muscle development, cell identity and regeneration. Semin. Cell Dev. Biol. 2017, 72, 10–18. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Capetanaki, Y. Regulation of the mouse desmin gene: Transactivated by MyoD, myogenin, MRF4 and Myf5. Nucleic Acids Res. 1993, 21, 335–343. [Google Scholar] [CrossRef] [PubMed]
- Lassar, A.B.; Buskin, J.N.; Lockshon, D.; Davis, R.L.; Apone, S.; Hauschka, S.D.; Weintraub, H. MyoD is a sequence-specific DNA binding protein requiring a region of myc homology to bind to the muscle creatine kinase enhancer. Cell 1989, 5, 823–831. [Google Scholar] [CrossRef]
- Shi, X.; Garry, D.J. Myogenic regulatory factors transactivate the Tceal7 gene and modulate muscle differentiation. Biochem. J. 2010, 428, 213–221. [Google Scholar] [CrossRef]
- Bowlin, K.M.; Embree, L.J.; Garry, M.G.; Garry, D.J.; Shi, X. Kbtbd5 is regulated by MyoD and restricted to the myogenic lineage. Differentiation 2013, 86, 184–191. [Google Scholar] [CrossRef]
- You, S.S.; Xiong, Z.Z.; Chen, X.Y.; Shi, X. The identification of the promoter and enhancer of Rb1 gene. Chin. Sci. Bull. 2021, 66, 4677–4690. [Google Scholar] [CrossRef]
- Rudnicki, M.A.; Braun, T.; Hinuma, S.; Jaenisch, R. Inactivation of MyoD in mice leads to up-regulation of the myogenic HLH gene Myf-5 and results in apparently normal muscle development. Cell 1992, 71, 383–390. [Google Scholar] [CrossRef]
- Wang, C.; Liu, W.; Nie, Y.; Qaher, M.; Horton, H.E.; Yue, F.; Asakura, A.; Kuang, S. Loss of MyoD Promotes Fate Transdifferentiation of Myoblasts Into Brown Adipocytes. EBioMedicine 2017, 16, 212–223. [Google Scholar] [CrossRef]
- Weskamp, K.; Olwin, B.B.; Parker, R. Post-Transcriptional Regulation in Skeletal Muscle Development, Repair, and Disease. Trends Mol. Med. 2021, 27, 469–481. [Google Scholar] [CrossRef]
- Hirai, H.; Verma, M.; Watanabe, S.; Tastad, C.; Asakura, Y.; Asakura, A. MyoD regulates apoptosis of myoblasts through microRNA-mediated down-regulation of Pax3. J. Cell Biol. 2010, 191, 347–365. [Google Scholar] [CrossRef]
- Chen, J.F.; Tao, Y.; Li, J.; Deng, Z.; Yan, Z.; Xiao, X.; Wang, D.Z. microRNA-1 and microRNA-206 regulate skeletal muscle satellite cell proliferation and differentiation by repressing Pax7. J. Cell Biol. 2010, 190, 867–879. [Google Scholar] [CrossRef] [PubMed]
- Peng, X.L.; So, K.K.; He, L.; Zhao, Y.; Zhou, J.; Li, Y.; Yao, M.; Xu, B.; Zhang, S.; Yao, H.; et al. MyoD- and FoxO3-mediated hotspot interaction orchestrates super-enhancer activity during myogenic differentiation. Nucleic Acids Res. 2017, 45, 8785–8805. [Google Scholar] [CrossRef] [PubMed]
- Wang, R.; Chen, F.; Chen, Q.; Wan, X.; Shi, M.; Chen, A.K.; Ma, Z.; Li, G.; Wang, M.; Ying, Y.; et al. MyoD is a 3D genome structure organizer for muscle cell identity. Nat. Commun. 2022, 13, 205. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Zhou, J.; He, L.; Li, Y.; Yuan, J.; Sun, K.; Chen, X.; Bao, X.; Esteban, M.A.; Sun, H.; et al. MyoD induced enhancer RNA interacts with hnRNPL to activate target gene transcription during myogenic differentiation. Nat. Commun. 2019, 10, 5787. [Google Scholar] [CrossRef]
- Black, B.L.; Olson, E.N. Transcriptional control of muscle development by myocyte enhancer factor-2 (MEF2) proteins. Annu. Rev. Cell Dev. Biol. 1998, 14, 167–196. [Google Scholar] [CrossRef]
- Taylor, M.V.; Hughes, S.M. Mef2 and the skeletal muscle differentiation program. Semin. Cell Dev. Biol. 2017, 72, 33–44. [Google Scholar] [CrossRef] [PubMed]
- Cserjesi, P.; Olson, E.N. Myogenin induces the myocyte-specific enhancer binding factor MEF-2 independently of other muscle-specific gene products. Mol. Cell Biol. 1991, 11, 4854–4862. [Google Scholar]
- Edmondson, D.G.; Lyons, G.E.; Martin, J.F.; Olson, E.N. Mef2 gene expression marks the cardiac and skeletal muscle lineages during mouse embryogenesis. Development 1994, 120, 1251–1263. [Google Scholar] [CrossRef]
- Lin, Q.; Lu, J.; Yanagisawa, H.; Webb, R.; Lyons, G.E.; Richardson, J.A.; Olson, E.N. Requirement of the MADS-box transcription factor MEF2C for vascular development. Development 1998, 125, 4565–4574. [Google Scholar] [CrossRef]
- Potthoff, M.J.; Arnold, M.A.; McAnally, J.; Richardson, J.A.; Bassel-Duby, R.; Olson, E.N. Regulation of skeletal muscle sarcomere integrity and postnatal muscle function by Mef2c. Mol. Cell Biol. 2007, 27, 8143–8151. [Google Scholar] [CrossRef]
- Naya, F.J.; Olson, E.N. MEF2: A transcriptional target for signaling pathways controlling skeletal muscle growth and differentiation. Curr. Opin. Cell Biol. 1999, 11, 683–688. [Google Scholar] [CrossRef]
- Wang, D.Z.; Valdez, M.R.; McAnally, J.; Richardson, J.; Olson, E.N. The Mef2c gene is a direct transcriptional target of myogenic bHLH and MEF2 proteins during skeletal muscle development. Development 2001, 128, 4623–4633. [Google Scholar] [CrossRef] [PubMed]
- Steven, A.; Friedrich, M.; Jank, P.; Heimer, N.; Budczies, J.; Denkert, C.; Seliger, B. What turns CREB on? And off? And why does it matter? Cell Mol. Life Sci. 2020, 77, 4049–4067. [Google Scholar] [CrossRef]
- Fujii, Y.; Shimizu, T.; Toda, T.; Yanagida, M.; Hakoshima, T. Structural basis for the diversity of DNA recognition by bZIP transcription factors. Nat. Struct. Biol. 2000, 7, 889–893. [Google Scholar]
- Montminy, M.R.; Bilezikjian, L.M. Binding of a nuclear protein to the cyclic-AMP response element of the somatostatin gene. Nature 1987, 328, 175–178. [Google Scholar] [CrossRef] [PubMed]
- Du, K.; Asahara, H.; Jhala, U.S.; Wagner, B.L.; Montminy, M. Characterization of a CREB gain-of-function mutant with constitutive transcriptional activity in vivo. Mol. Cell Biol. 2000, 20, 4320–4327. [Google Scholar] [CrossRef] [PubMed]
- Shi, X.; Zirbes, K.M.; Rasmussen, T.L.; Ferdous, A.; Garry, M.G.; Koyano-Nakagawa, N.; Garry, D.J. The transcription factor Mesp1 interacts with cAMP-responsive element binding protein 1 (Creb1) and coactivates Ets variant 2 (Etv2) gene expression. J. Biol. Chem. 2015, 290, 9614–9625. [Google Scholar] [CrossRef]
- Altarejos, J.Y.; Montminy, M. CREB and the CRTC co-activators: Sensors for hormonal and metabolic signals. Nat. Rev. Mol. Cell Biol. 2011, 12, 141–151. [Google Scholar] [CrossRef]
- Mayr, B.; Montminy, M. Transcriptional regulation by the phosphorylation-dependent factor CREB. Nat. Rev. Mol. Cell Biol. 2001, 2, 599–609. [Google Scholar] [CrossRef]
- Felinski, E.A.; Kim, J.; Lu, J.; Quinn, P.G. Recruitment of an RNA polymerase II complex is mediated by the constitutive activation domain in CREB, independently of CREB phosphorylation. Mol. Cell Biol. 2001, 21, 1001–1010. [Google Scholar] [CrossRef]
- Rudolph, D.; Tafuri, A.; Gass, P.; Hämmerling, G.J.; Arnold, B.; Schütz, G. Impaired fetal T cell development and perinatal lethality in mice lacking the cAMP response element binding protein. Proc. Natl. Acad. Sci. USA 1998, 95, 4481–4486. [Google Scholar] [CrossRef] [PubMed]
- Berdeaux, R.; Goebel, N.; Banaszynski, L.; Takemori, H.; Wandless, T.; Shelton, G.D.; Montminy, M. SIK1 is a class II HDAC kinase that promotes survival of skeletal myocytes. Nat. Med. 2007, 13, 597–603. [Google Scholar] [CrossRef] [PubMed]
- Stewart, R.; Flechner, L.; Montminy, M.; Berdeaux, R. CREB is activated by muscle injury and promotes muscle regeneration. PLoS ONE 2011, 6, e24714. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Fan, C.M. A CREB-MPP7-AMOT Regulatory Axis Controls Muscle Stem Cell Expansion and Self-Renewal Competence. Cell Rep. 2017, 21, 1253–1266. [Google Scholar] [CrossRef]
- Koyano-Nakagawa, N.; Shi, X.; Rasmussen, T.L.; Das, S.; Walter, C.A.; Garry, D.J. Feedback Mechanisms Regulate Ets Variant 2 (Etv2) Gene Expression and Hematoendothelial Lineages. J. Biol. Chem. 2015, 290, 28107–28119. [Google Scholar] [CrossRef]
- Magenta, A.; Cenciarelli, C.; De Santa, F.; Fuschi, P.; Martelli, F.; Caruso, M.; Felsani, A. MyoD stimulates RB promoter activity via the CREB/p300 nuclear transduction pathway. Mol. Cell Biol. 2003, 23, 2893–2906. [Google Scholar] [CrossRef]
- Matsuo, N.; Tanaka, S.; Gordon, M.K.; Koch, M.; Yoshioka, H.; Ramirez, F. CREB-AP1 protein complexes regulate transcription of the collagen XXIV gene (Col24a1) in osteoblasts. J. Biol. Chem. 2006, 281, 5445–5452. [Google Scholar] [CrossRef]
- Shi, X.; Bowlin, K.M.; Garry, D.J. Fhl2 interacts with Foxk1 and corepresses Foxo4 activity in myogenic progenitors. Stem Cells. 2010, 28, 462–469. [Google Scholar] [CrossRef]
- Cramer, P. Organization and regulation of gene transcription. Nature 2019, 573, 45–54. [Google Scholar] [CrossRef]
- Francois, M.; Donovan, P.; Fontaine, F. Modulating transcription factor activity: Interfering with protein-protein interaction networks. Semin. Cell Dev. Biol. 2020, 99, 12–19. [Google Scholar] [CrossRef]
- Li, Y.; He, J.; Sui, S.; Hu, X.; Zhao, Y.; Li, N. Clenbuterol upregulates histone demethylase JHDM2a via the β2-adrenoceptor/cAMP/PKA/p-CREB signaling pathway. Cell Signal. 2012, 24, 2297–2306. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.Y.; Tse, M.C.L.; Hu, X.; Jia, W.H.; Du, G.H.; Chan, C.B. Interaction of CREB and PGC-1α Induces Fibronectin Type III Domain-Containing Protein 5 Expression in C2C12 Myotubes. Cell Physiol. Biochem. 2018, 50, 1574–1584. [Google Scholar] [CrossRef] [PubMed]
- Sands, W.A.; Palmer, T.M. Regulating gene transcription in response to cyclic AMP elevation. Cell Signal. 2008, 20, 460–466. [Google Scholar] [CrossRef] [PubMed]
- Dittmer, S.; Kovacs, Z.; Yuan, S.H.; Siszler, G.; Kögl, M.; Summer, H.; Geerts, A.; Golz, S.; Shioda, T.; Methner, A. TOX3 is a neuronal survival factor that induces transcription depending on the presence of CITED1 or phosphorylated CREB in the transcriptionally active complex. J. Cell Sci. 2011, 124, 252–260. [Google Scholar] [CrossRef]
- Ernst, P.; Wang, J.; Huang, M.; Goodman, R.H.; Korsmeyer, S.J. MLL and CREB bind cooperatively to the nuclear coactivator CREB-binding protein. Mol. Cell Biol. 2001, 21, 2249–2258. [Google Scholar] [CrossRef]
- Rasmussen, T.L.; Shi, X.; Wallis, A.; Kweon, J.; Zirbes, K.M.; Koyano-Nakagawa, N.; Garry, D.J. VEGF/Flk1 signaling cascade transactivates Etv2 gene expression. PLoS ONE 2012, 7, e50103. [Google Scholar] [CrossRef]
- Chen, A.E.; Ginty, D.D.; Fan, C.M. Protein kinase A signalling via CREB controls myogenesis induced by Wnt proteins. Nature 2005, 433, 317–322. [Google Scholar] [CrossRef]
- Kim, C.H.; Xiong, W.C.; Mei, L. Inhibition of MuSK expression by CREB interacting with a CRE-like element and MyoD. Mol. Cell Biol. 2005, 25, 5329–5338. [Google Scholar] [CrossRef][Green Version]
- Perdiguero, E.; Ruiz-Bonilla, V.; Gresh, L.; Hui, L.; Ballestar, E.; Sousa-Victor, P.; Baeza-Raja, B.; Jardí, M.; Bosch-Comas, A.; Esteller, M.; et al. Genetic analysis of p38 MAP kinases in myogenesis: Fundamental role of p38alpha in abrogating myoblast proliferation. EMBO J. 2007, 26, 1245–1256. [Google Scholar] [CrossRef]
- Sin, T.K.; Zhang, G.; Zhang, Z.; Zhu, J.Z.; Zuo, Y.; Frost, J.A.; Li, M.; Li, Y.P. Cancer-Induced Muscle Wasting Requires p38β MAPK Activation of p300. Cancer Res. 2021, 81, 885–897. [Google Scholar] [CrossRef]
- Zhang, G.; Anderson, L.J.; Gao, S.; Sin, T.K.; Zhang, Z.; Wu, H.; Jafri, S.H.; Graf, S.A.; Wu, P.C.; Dash, A.; et al. Weight Loss in Cancer Patients Correlates With p38β MAPK Activation in Skeletal Muscle. Front. Cell Dev. Biol. 2021, 9, 784424. [Google Scholar] [CrossRef] [PubMed]
- Lluís, F.; Perdiguero, E.; Nebreda, A.R.; Muñoz-Cánoves, P. Regulation of skeletal muscle gene expression by p38 MAP kinases. Trends Cell Biol. 2006, 16, 36–44. [Google Scholar] [CrossRef] [PubMed]
- Yang, S.H.; Galanis, A.; Sharrocks, A.D. Targeting of p38 mitogen-activated protein kinases to MEF2 transcription factors. Mol. Cell Biol. 1999, 9, 4028–4038. [Google Scholar] [CrossRef]
- Dilworth, F.J.; Seaver, K.J.; Fishburn, A.L.; Htet, S.L.; Tapscott, S.J. In vitro transcription system delineates the distinct roles of the coactivators pCAF and p300 during MyoD/E47-dependent transactivation. Proc. Natl. Acad. Sci. USA 2004, 101, 11593–11598. [Google Scholar] [CrossRef] [PubMed]
- Felinski, E.A.; Quinn, P.G. The CREB constitutive activation domain interacts with TATA-binding protein-associated factor 110 (TAF110) through specific hydrophobic residues in one of the three subdomains required for both activation and TAF110 binding. J. Biol. Chem. 1999, 274, 11672–11678. [Google Scholar] [CrossRef]
- Malecova, B.; Dall’Agnese, A.; Madaro, L.; Gatto, S.; Coutinho Toto, P.; Albini, S.; Ryan, T.; Tora, L.; Puri, P.L. TBP/TFIID-dependent activation of MyoD target genes in skeletal muscle cells. Elife 2016, 5, e12534. [Google Scholar] [CrossRef]
- Guo, Y.; Liao, Y.; Jia, C.; Ren, J.; Wang, J.; Li, T. MicroRNA-182 promotes tumor cell growth by targeting transcription elongation factor A-like 7 in endometrial carcinoma. Cell Physiol. Biochem. 2013, 32, 581–590. [Google Scholar] [CrossRef]
- Yue, X.; Lan, F.; Xia, T. Hypoxic Glioma Cell-Secreted Exosomal miR-301a Activates Wnt/β-catenin Signaling and Promotes Radiation Resistance by Targeting TCEAL7. Mol. Ther. 2019, 27, 1939–1949. [Google Scholar] [CrossRef]
- Yu, L.; Luan, W.; Feng, Z.; Jia, J.; Wu, Z.; Wang, M.; Li, F.; Li, Z. Long non-coding RNA HAND2-AS1 inhibits gastric cancer progression by suppressing TCEAL7 expression via targeting miR-769-5p. Dig. Liver Dis. 2021, 53, 238–244. [Google Scholar] [CrossRef]
- Yan, Z.; Sheng, Z.; Zheng, Y.; Feng, R.; Xiao, Q.; Shi, L.; Li, H.; Yin, C.; Luo, H.; Hao, C.; et al. Cancer-associated fibroblast-derived exosomal miR-18b promotes breast cancer invasion and metastasis by regulating TCEAL7. Cell Death Dis. 2021, 12, 1120. [Google Scholar] [CrossRef]
- Liu, X.; Song, X.; Li, H. Transcription elongation factor A-like 7, regulated by miR-758-3p inhibits the progression of melanoma through decreasing the expression levels of c-Myc and AKT1. Cancer Cell Int. 2021, 21, 43. [Google Scholar] [CrossRef] [PubMed]
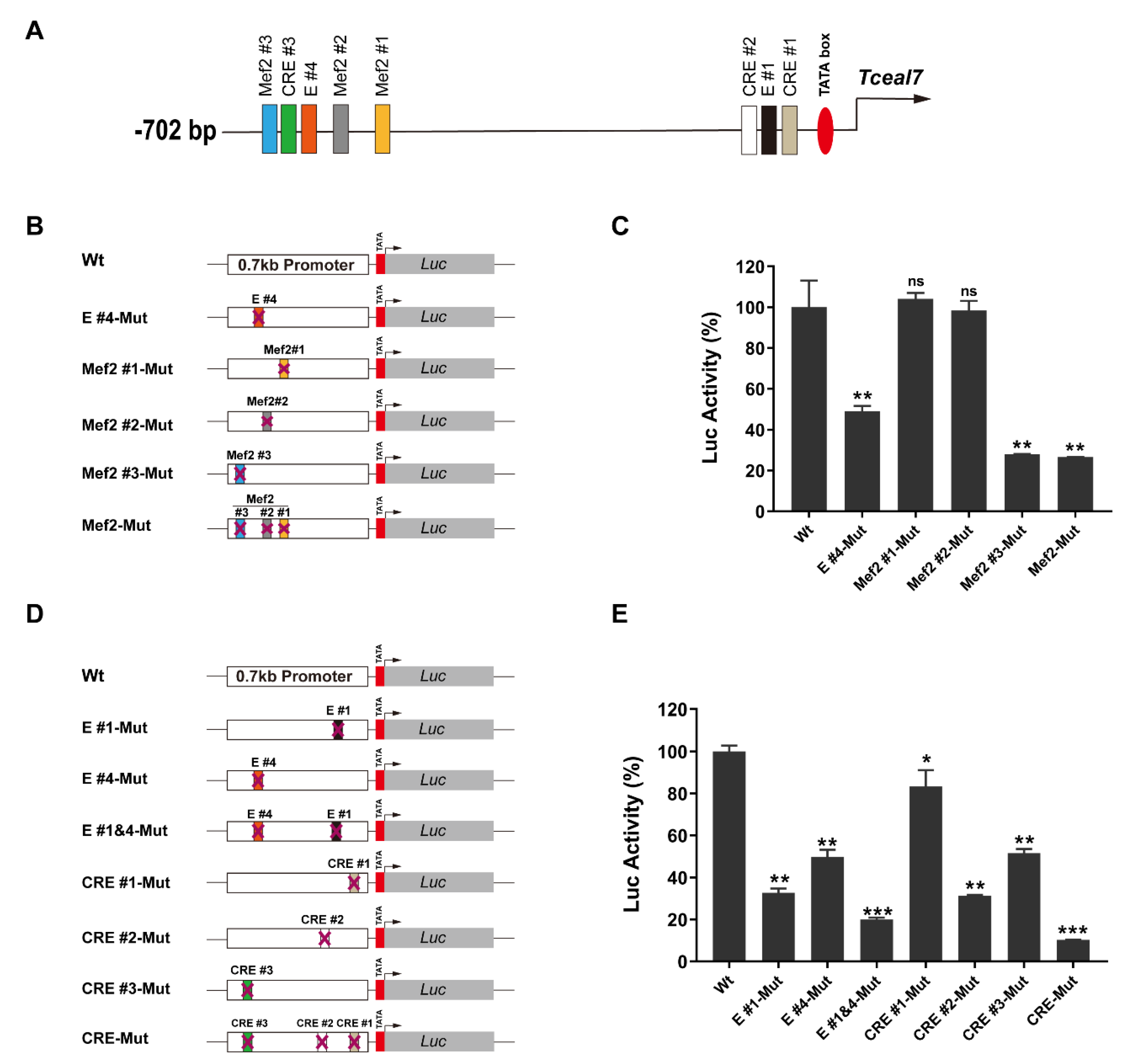

| Motifs | Species | Sequence |
|---|---|---|
| Mef2#1 | MMU | GTAAAAATAA |
| HSA | CTATATTTAT | |
| Mef2#2 | MMU | CTATTTTTAA |
| HSA | CTATATTTAT | |
| Mef2#3 | MMU | CTATATTTAT |
| HSA | CTATATTTAT | |
| CRE#1 | MMU | TGACGTGA |
| HSA | TGACGTGA | |
| CRE#2 | MMU | TGACATAA |
| HSA | TGACATAA | |
| CRE#3 | MMU | TGACTTCA |
| HSA | TGACTTCA | |
| E#1 | MMU | CACGTG |
| HSA | CACGTG | |
| E#4 | MMU | CAGGTG |
| HSA | CAGGTG |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Xiong, Z.; Wang, M.; You, S.; Chen, X.; Lin, J.; Wu, J.; Shi, X. Transcription Regulation of Tceal7 by the Triple Complex of Mef2c, Creb1 and Myod. Biology 2022, 11, 446. https://doi.org/10.3390/biology11030446
Xiong Z, Wang M, You S, Chen X, Lin J, Wu J, Shi X. Transcription Regulation of Tceal7 by the Triple Complex of Mef2c, Creb1 and Myod. Biology. 2022; 11(3):446. https://doi.org/10.3390/biology11030446
Chicago/Turabian StyleXiong, Zhenzhen, Mengni Wang, Shanshan You, Xiaoyan Chen, Jiangguo Lin, Jianhua Wu, and Xiaozhong Shi. 2022. "Transcription Regulation of Tceal7 by the Triple Complex of Mef2c, Creb1 and Myod" Biology 11, no. 3: 446. https://doi.org/10.3390/biology11030446
APA StyleXiong, Z., Wang, M., You, S., Chen, X., Lin, J., Wu, J., & Shi, X. (2022). Transcription Regulation of Tceal7 by the Triple Complex of Mef2c, Creb1 and Myod. Biology, 11(3), 446. https://doi.org/10.3390/biology11030446






