Simple Summary
JASMONATE ZIM-DOMAIN (JAZ) proteins are key components of the jasmonate (JA) signaling pathway in response to biotic and abiotic stresses in plants. Information about the JAZ gene family in pineapple (AcJAZ) is limited. In this study, 14 AcJAZ genes were identified in the pineapple genome. A complete overview of AcJAZ genes is presented, including the chromosome locations, phylogenetic relationships, gene structures, conserved motifs and cis-regulatory elements, and expression patterns at different developmental stages and under various stress conditions, and their possible involvement in diverse functions is suggested. Furthermore, the BiFC analysis revealed direct binary interactions between AcJAZs and crucial JA-signaling regulators in vivo. These results suggest that AcJAZs and other vital players function in the JA-signaling pathway in response to abiotic stresses in pineapple.
Abstract
JASMONATE ZIM-DOMAIN (JAZ) proteins are negative regulators of the jasmonate (JA)-signaling pathway and play pivotal roles in plant resistance to biotic and abiotic stresses. Genome-wide identification of JAZ genes has been performed in many plant species. However, systematic information about pineapple (Ananas comosus L. Merr.) JAZ genes (AcJAZs) is still not available. In this study, we identified 14 AcJAZ genes and classified them into five groups along with the Arabidopsis and rice orthologs. The AcJAZ genes have 3–10 exons, and the putative AcJAZ proteins have between two and eight conserved regions, including the TIFY motif and Jas domain. The cis-acting element analysis revealed that the putative promoter regions of AcJAZs contain between three and eight abiotic stress-responsive cis-acting elements. The gene-expression analysis suggested that AcJAZs were expressed differentially during plant development and subjected to regulation by the cold, heat, salt, and osmotic stresses as well as by phytohormones. Moreover, the BiFC analysis of protein interactions among the central JA-signaling regulators showed that AcJAZ4, AcMYC2, AcNINJA, and AcJAM1 could interact with AcJAZ5 and AcJAZ13 in vivo, indicating a canonical JA-signaling pathway in pineapple. These results increase our understanding of the functions of AcJAZs and the responses of the core players in the JA-signaling pathway to abiotic stresses.
1. Introduction
Jasmonates (JAs), the endogenous plant metabolites that exist ubiquitously in plants, primarily include Jasmonic acid (JA), Methyl cis-jasmonate (MeJA), (+)-7-iso-jasmonoyl-L-isoleucine (JA-Ile), and its precursors such as OPDA (12-oxo Phytodienoic acid), dinor-OPDA (dinor-12-oxo-Phytodienoic acid), and oxygenated fatty acids (oxylipins). JAs play an important role in regulating plant growth, development, reproduction, and response to biotic and abiotic stresses along with other plant hormones [1,2,3,4]. It has been shown that in tobacco Bright Yellow-2 cells, JAs play a vital role in maintaining cells at the G1 stage by preventing DNA replication [5]. In Arabidopsis, wound-induced jasmonates inhibit mitosis and reduce the cell number, leading to stunted plant growth [2]. Consistently, JAs adversely affect leaf growth by repressing cell proliferation and the onset of endoreduplication [6]. JAs also regulate the biosynthesis of ethylene and control the timing of leaf senescence through 1-aminocyclopropane-1-carboxylic acid (ACC) conjugation [7,8]. They are well known as transducers of elicitor signals for the accumulation of plant secondary metabolites [9]. Further, JAs play critical regulatory roles in plant biotic and abiotic stresses, as shown by many studies [1,3,10,11,12,13,14,15,16,17,18].
Previous studies have suggested that JA-Ile is a bioactive jasmonate of the JA-signaling pathway. It has a similar structure and function as coronatine (a bacterial toxin), activated in JA signaling as a substrate [19,20,21]. The basic helix–loop–helix (bHLH) transcription factors, MYC2, MYC3, MYC4, and MYC5, are core transcription factors in the JA-signaling pathway which redundantly promote the expression of JA-responsive genes [22,23,24]. COI1, an F-box protein subunit of the SKP1-CUL1-F-box (SCF) complex with E3 ubiquitin ligase activity, is the receptor of JA-Ile [25,26]. On the other hand, JASMONATE ZIM-DOMAIN (JAZ) proteins are the repressors of JA signaling, inhibiting the expression of JA-responsive genes by repressing the transcriptional activity of MYC2, MYC3, MYC4, and MYC5 [27]. JAZ proteins belong to the TIFY gene family, harboring a TIFY motif (TIF[F/Y]XG), previously known as Zim domain, in the N-terminal region and a Jas or CCT_2 motif (SLX2FX2KRX2RX5PY) in the C-terminal region [28]. In the absence of active JAs, JAZ proteins, through their Jas domains, bind to MYC transcription factors and recruit NINJA (NOVEL INTERACTOR OF JAZ) proteins through their TIFY motif. NINJA proteins further interact with TPL (TOPLESS) proteins, transcription repressors with an EAR domain. The JAZ–NINJA–TPL complex represses the expression of the MYC2/3/4/5-target genes [29]. In the presence of the external or endogenous stimulus, JA-Ile accumulates in plants and is subsequently received by the SCFCOI1 preceptor [30]. The JA-Ile then mediates the interaction of SCFCOI1 with JAZ proteins, which are then ubiquitinated by SCFCOI1. The ubiquitinated JAZ proteins are targeted to 26S proteasomes for degradation, releasing MYC2/3/4/5 and resulting in the expression of JA-responsive genes.
JAZ proteins are the core regulators of the JA-signaling pathway that are also involved in phytohormonal crosstalk [31]. In Arabidopsis and rice (Oryza sativa), 13 and 15 JAZ genes have been identified, respectively. In recent years, genome-wide identification of the JAZ gene family has been made in several plants, including Petunia axillaris (12), Petunia inflate (16), Triticum aestivum (14), Solanum lycopersicum (13), Vitis vinifera (11), Hevea brasiliensis (18), and sugarcane (7), and the functions of JAZ genes were extensively investigated, especially in Arabidopsis [32,33,34,35,36,37]. For example, the decuple jaz mutant exhibits enhanced resistance to biotic stresses, attenuated vegetative growth, and reduced the seed setting, suggesting the important functions of Arabidopsis JAZs in plant development and stress response [38]. It has also been reported that the jaz10 knockout mutant has enhanced JA sensitivity and increased susceptibility to DC3000 in Arabidopsis [39].
Pineapple (Ananas comosus L. Merr.), a perennial monocot belonging to the family Bromeliaceae, is one of the most important tropical fruit crops around the world with edible and decorative values [40]. For edible usage, pineapples are rich in vitamins, organic acids, saccharides, and trace elements such as calcium and iron. For decoration, pineapples have a perfect flower-shaped structure, bright and gorgeous color, and long flowering time.
Moreover, pineapple plants have excellent stress tolerance to adverse conditions, including drought and salt stress. Since JA signaling plays a pivotal role in plant response to stresses and JAZ proteins are a core component of JA signaling, it is important to explore the JA-signaling pathway based on the JAZ gene family in pineapple (AcJAZ). In this study, the genes involved in the JA-signaling pathway were identified in pineapple, and an extensive investigation of the AcJAZs was conducted. These results provide a comprehensive understanding of the JA-signaling pathway in pineapple, which should facilitate further research related to pineapple stress response and tolerance.
2. Materials and Methods
2.1. Identification of JAZ Gene Family in Pineapple
The protein sequences of pineapple (variety F153) were downloaded from the Pineapple Genomics database (PGD, http://pineapple.angiosperms.org/pineapple/html/index.html (accessed on 20 December 2020)). The proteins harboring TIFY (PF06200) and Jas (PF09425) domains were identified using the HMMER3.0 (http://hmmer.org (accessed on 20 December 2021)) program with default parameters. Furthermore, 13 Arabidopsis JAZ (AtJAZ) protein sequences were downloaded from TAIR (http://www.arabidopsis.org/ (accessed on 20 December 2021)), and 15 rice (Oryza sativa) JAZ (OsJAZ) proteins were downloaded from TIGR (release 5; http://rice.plantbiology.msu.edu (accessed on Dec. 20, 2021)) [41]. Those sequences were used as queries to search for JAZ proteins against the pineapple protein database using the blastp algorithm with an e-value of 1 × 10−5. Then, Pfam (http://pfam.xfam.org/search#tabview=tab1 (accessed on 20 December 2021)), SMART (http://smart.embl.de/smart/batch.pl (accessed on 20 December 2021)) [42], and NCBI CDD (https://www.ncbi.nlm.nih.gov/cdd/ (accessed on 20 December 2021)) were used to determine the correct JAZ proteins in pineapple. The detailed information, including gene location on chromosomes, intron number, and CDS length of AcJAZ genes, and the amino acid number, molecular weight, and isoelectric point (pI) of AcJAZ proteins, was acquired from Phytozome v12.1 (http://phytozome.jgj.doe.gov/pz/portal.html (accessed on 20 December 2021)) and ExPasy (http://web.expasy.org/protparam/ (accessed on 20 December 2021)). The information of related chromosome length and AcJAZ location was obtained from the PGD, and the location map was constructed by MapChart [43].
2.2. Phylogenetic Analysis
The protein sequences of 14 AcJAZs, 13 AtJAZs, and 15 OsJAZs were aligned by MEGA7 using the Clustalw method. Then, the alignment file was used to construct a Neighbor-Joining-based phylogenetic tree with the bootstrap value of 1000. The phylogenetic tree was annotated using the online tool Evolview (https://evolgenius.info//evolview-v2/#login (accessed on 20 December 2021)). All protein sequences used for phylogenetic analysis are listed in Supplementary File S1.
2.3. Analysis of Conserved Motif and Gene Structure
To show sequence similarity between JAZ proteins of pineapple, 14 AcJAZ sequences were used to create a phylogenetic tree by MEGA7 and present conserved motifs and gene structures. The MEME software (Multiple EM for Motif Elicitation, http://meme-suite.org/tools/meme (accessed on 20 December 2021)) was used to identify conserved motifs with the maximum searching number set to 15. All motif information of pineapple JAZ proteins is listed in Supplementary File S2. Moreover, the exon–intron organization data were extracted from the Pineapple Genomics database (PGD). The visualization of the phylogenetic tree, conserved motifs, and gene structures was conducted with Tbtools [44].
2.4. Analysis of Cis-Acting Elements
The 2000 bp upstream sequences from the translation start codon (ATG) were obtained from the Phytozome for all the AcJAZs and used to analyze the cis-acting elements using PlantCARE (http://bioinformatics.psb.ugent.be/webtools/plantcare/html/ (accessed on 20 December 2021)). The following abiotic responsive elements: abscisic acid-responsive element (ABRE, ABRE3a, ABRE4, AT-ABRE), MeJA-responsive elements (CGTCA-motif, TGACG-motif), wound-responsive elements (W-box, WRE3, WUN-motif), ethylene-responsive elements [29], drought-responsive elements (DRE core), salicylic acid-responsive elements (TCA), low-temperature-responsive elements (LTRE), gibberellin-responsive elements (GARE-motif, TATC-box, P-box), auxin-responsive elements (AuRR-core, TGA-element, TGA-box), and defense and stress elements (TC-rich repeats) were searched.
2.5. Plant Materials and Stress Treatments
Regenerated plantlets of pineapple (variety MD2) with approximately 10 cm length were transferred to the greenhouse at 25 °C and under 16 h light/8 h dark photoperiod [45]. After 30 days of the transfer, plants/seedlings were treated with cold (4 °C), heat (45 °C), osmotic stress (15% PEG), salt (150 mM NaCl), IAA (100 μM), MeJA (100 μM), ABA (100 μM), SA (100 μM), and 6-BA (100 μM) followed by sample harvesting at 0, 2, 4, 12, and 36 h for RNA extraction [45,46].
2.6. Total RNA Extractions and RT-qPCR
E.Z.N.A Total RNA Kit II (OMEGA, Guangzhou, China) was used for total RNA extraction, followed by cDNA synthesis using AMV reverse transcriptase (Takara, Beijing, China). The cDNAs were then used for RT-qPCR analysis using 2 × TranStart Top Green qPCR SuperMix (TransGen Biotech, Beijing, China). A 20 μL reaction which included 8 μL nuclease-free water, 0.5 μL (10 mM) forward primer, 0.5 μL (10 mM) reverse primer, 1 μL cDNA, and 10 μL 2 × TranStart Top Green qPCR SuperMix was used for qPCR in a CFX96 Touch real-time PCR machine with the following parameters: 95 °C for 30 s, 40 repeats of 95 °C for 5 s, and 60 °C for 40 s. The relative expression levels of targets were calculated using the delta–delta Ct method using pineapple housekeeping gene PP2A as reference. The online tool IDT (https://sg.idtdna.com/pages/products/qpcr-and-pcr/gene-expression/primetime-primer-only-assays-and-primers (accessed on 20 December 2021)) was used to design real-time PCR primers (Table S1). For expression profiling of the JAZ gene family in pineapple, the RNA-seq data for Calyx (C1–C4), Gynoecium (G1–G7), Ovule (O1–O7), Petal (P1–P3), Stamen (S1–S6), Fruit (S1–S7), Flower, Leaf, and Root were used (Accession Number: PRJEB38680) [47].
2.7. BiFC Assay
The YFP-N end-tagged pSPYNE and YFP-C end-tagged pSPYCE vectors were used to transform Agrobacterium tumefaciens strain GV3101 and then infiltrate tobacco (Nicotiana benthamiana) leaves for BiFC (Bimolecular fluorescence complementation) assay. The tobacco infiltration and inflorescence observations were conducted following the procedure reported by Yuan and Xu [48]. Infusion strategies were adopted for vector preparation using CloneExpress II One Step Cloning Kit (Vazyme, Nanjing, China). The primers used in this assay are listed in Table S1.
2.8. The Cellular Localization of AcJAZ Proteins
The protein sequences of 14 AcJAZs and 15 OsJAZs were analyzed to predict possible subcellular localization using DeepLoc (https://services.healthtech.dtu.dk/service.php?DeepLoc-1.0 (accessed on 20 December 2021)) and MULocDeep (http://mu-loc.org/ (accessed on 20 December 2021)) [49]. All protein sequences used for the prediction of subcellular localization are listed in Supplementary File S1.
3. Results
3.1. Fourteen JAZs Were Identified in the Pineapple Genome
The genome of pineapple variety F153 was used as a reference in this study [50]. In total, 14 JAZ proteins were identified based on BLASTP and HMM searching, and the corresponding genes were designated as AcJAZ1–AcJAZ14 based on their location on pineapple chromosomes from top to bottom, in which AcJAZ1–13 were located on ten pineapple chromosomes (chromosomes 01, 02, 05, 06, 07, 09, 12, 14, 19, 20), and AcJAZ14 was located on an unanchored scaffold_1707 (Figure 1A). Based on the DNA and amino acid sequences, the characteristics of pineapple JAZ genes and the putative proteins, including gene ID, chromosome locations, complete coding sequence (CDS) length, exon numbers, protein length, molecular weight, and isoelectric point (pI) are shown in Table 1. The AcJAZ genes possessed a minimum of three exons, and AcJAZ1 had the maximum (10) exons. The CDSs of AcJAZs ranged from 486 to 1608 bp in length, with the corresponding protein length ranging from 161 to 535 aa, the molecular weight from 16,867.08 Da to 56,897.76 Da, and the isoelectric point between 5.32 and 9.8 (Table 1).
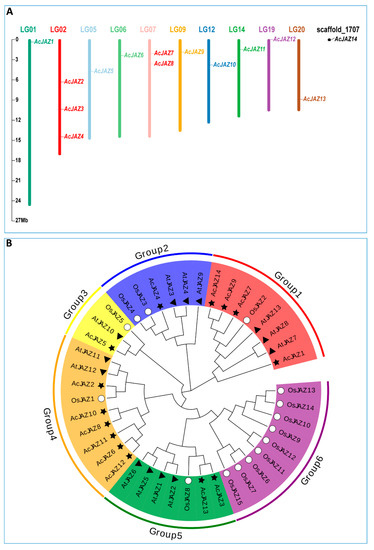
Figure 1.
The JAZ gene family in pineapple. (A) Chromosomal locations of JAZ genes in pineapple. The respective chromosome number is indicated by different colors at the top of each chromosome. The scale on the left is for chromosomes in megabases (B) Phylogenetic relationship of JAZ proteins from Arabidopsis thaliana, Oryza sativa, and pineapple. The phylogenetic tree was created using the neighbor-joining method with 1000 bootstrap replicates by MEGA 7. The diverse groups of JAZ proteins are marked with different colors. The JAZ proteins of A. thaliana, O. sativa, and pineapple are represented by black triangles, white circles, and black stars, respectively.

Table 1.
Information of Pineapple JAZs, including gene ID, chromosome, locations, isoelectric point (pI), molecular weight (MW) protein length, CDS length, and exon number.
3.2. Protein Sequence Analysis Showed the Phylogenetic Relationship of AcJAZs
The evolutionary relationship among the pineapple JAZ proteins was studied by constructing a phylogenetic tree along with 13 Arabidopsis and 15 rice JAZ protein sequences using MEGA X, following the neighbor-joining (NJ) method (Figure 1B). The JAZ proteins were divided into six groups according to the topology of the phylogenetic tree, and the AcJAZs were represented in five groups. In the first group, one OsJAZ (OsJAZ2), three AtJAZs (AtJAZ7, AtJAZ8, and AtJAZ13), and four AcJAZs (AcJAZ1, AcJAZ7, AcJAZ9, and AcJAZ14) were grouped. OsJAZ3, OsJAZ4, AtJAZ3, AtJAZ4, AtJAZ9, and AcJAZ4 belonged to the second group. The third group contained only three genes, i.e., OsJAZ5, AtJAZ10, and AcJAZ5. OsJAZ1, AtJAZ11–AtJAZ12, and six AcJAZs (AcJAZ2, AcJAZ6, AcJAZ8, AcJAZ10, AcJAZ11, and AcJAZ12) belonged to the fourth group. The fifth group was composed of AcJAZs (AcJAZ3 and AcJAZ13), AtJAZs (AtJAZ1, AtJAZ2, AtJAZ5, and AtJAZ6), and OsJAZ8. The sixth group only had OsJAZs (OsJAZ6, OsJAZ7, OsJAZ9–OsJAZ15) (Figure 1B). The phylogenetic relationship of AcJAZs along with AtJAZs and OsJAZs indicated that those JAZ proteins shared a similarity of protein sequences and a common ancestor.
3.3. AcJAZs Are Conserved in Gene Structure and Motif Organization
To understand the gene structure and protein motif in a phylogenetic view, the JAZ proteins of pineapple were used to construct a phylogenetic tree, gene structure, and motif analysis in the same order as in the phylogenetic tree. In the absence of Arabidopsis and rice, the phylogenetic tree showed a similar relationship with the result described above (Figure 2A). According to the pineapple genome release (V3.0), the introns in AcJAZs ranged from two to nine, and exons ranged from 3 to 10. AcJAZ2 had the most extended genomic DNA sequence (12,295 bp) with the longest intron (fourth intron, 9049 bp), while AcJAZ4 had the shortest nucleotide sequence with the shortest intron (third intron, 53 bp). Most of the AcJAZ genes have the 3′ UTR and 5′ UTR annotated, with the exceptions of AcJAZ10 without 5′ UTR, AcJAZ1 and AcJAZ2 without 3′ UTR, and AcJAZ12 lacking both UTRs. AcJAZ7, AcJAZ9, and AcJAZ14 had similar structure and sequence lengths with two introns and three exons.
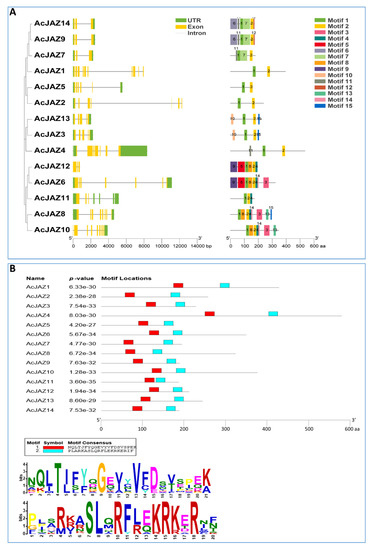
Figure 2.
The JAZ proteins in pineapple. (A) Phylogenetic relationships, gene structures, and motifs of JAZ genes in pineapple. The first lane shows the phylogenetic tree of AcJAZ proteins generated using the neighbor-joining method with 1000 bootstrap replicates by MEGA 7. The second lane shows the exon–intron organization of AcJAZs. 5′ and 3′ UTRs, exons, and introns are indicated by green boxes, yellow boxes, and black lines, respectively. The scale at the bottom represents the number of nucleotides. The third lane shows the conserved motifs of Pineapple JAZ proteins detected by MEME software. Different motifs for AcJAZ proteins are indicated by different colored boxes and numbered 1–15. The scale at the bottom represents the number of amino acids. (B) Conserved motif 1 and motif 2 in AcJAZ proteins. The upper diagram shows the positions of conserved motif 1 and motif 2 in AcJAZs, and the scale below represents the number of amino acids. The lower diagram shows the consensus amino acids for motif 1 (the upper panel) and motif 2 (the lower panel). The height of an amino acid character indicates proportionality to the sequence conservation.
Protein motifs might be assigned putative functions ofproteins. The motifs of pineapple JAZ proteins were identified using Multiple-Expectation maximization for Motif Elicitation (MEME) software. All AcJAZs harbored between two and eight motifs, while AcJAZ1, 2, 4, and 5 had two motifs (Figure 2A). Motif 1 and motif 2 were present in all AcJAZs, whereas motif 3 and motif 4 were present in five AcJAZs (Figure 2A). Two protein pairs, AcJAZ9/AcJAZ14 and AcJAZ3/AcJAZ13, had the same motif features (Figure 2A and Figure S2). Further, AcJAZ protein sequences were subjected to Pfam analysis to identify the conserved motifs, and we found nine Pfam motifs (tify, Jas_motif, CCT, F-box_4, FXR_C1, GATA, zf-Dof, Fz, and TFIIE-A_C) (Figure S3). Motif 1 and motif 2 of the MEME analysis represented the tify and Jas domains, respectively (Figure 2B), and motif 3 represented the GATA zinc finger domain, which uses four cysteine residues to coordinate a zinc ion. However, no Pfam information was detected for motifs 4 (PKKIRYTVRKEVALR) and 7 (VQAVLLLLGG), and they could be new motifs with unknown functions.
To investigate whether those JAZ proteins are localized in the nucleus, we analyzed the possible subcellular localization of AcJAZs and OsJAZ proteins using DeepLoc (https://services.healthtech.dtu.dk/service.php?DeepLoc-1.0 (accessed on 20 December 2021)) and MULocDeep (http://mu-loc.org/ (accessed on 20 December 2021)). Both methods were consistent with each other in predicting that all 14 AcJAZs were localized in the nucleus (Table S2). As a comparison, we also analyzed 15 rice JAZ proteins. The DeepLoc analysis predicted that all 15 OsJAZs were localized in the nucleus, while MULocDeep predicted that 12 OsJAZs were localized in the nucleus and three OsJAZ were mainly localized elsewhere (Table S3). The prediction score of the nucleus localization of AcJAZ1 and AcJAZ3 is relatively low, while the those of the nucleus localization of AcJAZ2, AcJAZ7, AcJAZ8, AcJAZ9, AcJAZ10, AcJAZ11, AcJAZ12, and AcJAZ14 are rather high, indicating the strong nucleus localization of JAZ proteins of pineapple.
3.4. AcJAZs Are Differentially Expressed in Pineapple Organs
FPKM values calculated from RNA-seq generated from nine different tissues were used to create a heatmap and study the expression pattern of AcJAZ genes (Figure 3A) [47]. In the samples analyzed, all the AcJAZ genes except AcJAZ12 were expressed. The clustering based on the expression patterns resulted in three groups. The first group consisted of five AcJAZs (AcJAZ1, AcJAZ2, AcJAZ4, AcJAZ6, and AcJAZ11) with relatively high expression levels. The second group had AcJAZ12, AcJAZ7, AcJAZ5, AcJAZ9, and AcJAZ14, with infrequent representation in the investigated tissues. The third group had four genes (AcJAZ3, AcJAZ8, AcJAZ10, and AcJAZ13) with moderate and varied expression levels. AcJAZ4 had the highest expression level among AcJAZ genes, and its expression level was higher in the calyx, petal, and fruit developmental stages 1–3, whereas AcJAZ12 was rarely expressed. AcJAZ7 showed in petal stage 3 (P3)-specific expression, and AcJAZ5 had a calyx (C1–C4)-specific expression. The expression patterns of AcJAZ8, AcJAZ9, AcJAZ10, and AcJAZ14 were constantly low in all tissues. Two genes, AcJAZ3 and AcJAZ13, had a similar expression pattern and were highly expressed in the calyx (C1–C3), petals (P2–P3), flowers, and leaves (Figure 3A). The RNA-seq results were further verified for AcJAZ2, AcJAZ4, AcJAZ5, AcJAZ11, and AcJAZ13 expression using qPCR (Figure 3B). Consistent with RNA-seq, the qPCR results also showed constitutively high expression of AcJAZ4 and low expression of AcJAZ5 in all the tissues. The expression patterns of these genes in petals, stamen, and fruit also showed a similar expression pattern as observed in the RNA-seq.
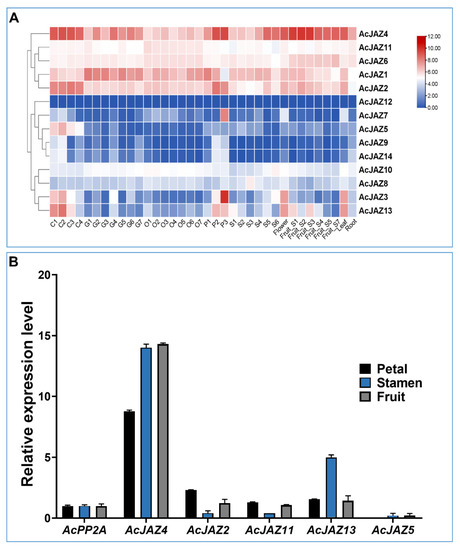
Figure 3.
Expression of AcJAZ genes in various tissues. (A) Heat-map shows the expression profiles of AcJAZ genes in nine different tissues. C1–C4 (Calyx stage 1–4), G1–G7 (Gynoecium stage 1–7), O1–O7 (Ovule stage 1–7), P1–P3 (Petal stage 1–3), S1–S6 (Stamen stage 1–6), Flower, Fruit _S1–S7, leaf, root. The scale indicates log10 transformed gene expression by colors ranging from red (high) to blue (low). (B) The expression of AcJAZ4, AcJAZ2, AcJAZ11, AcJAZ13, and AcJAZ5 in petal, stamen, and fruit determined by RT-qPCR. AcPP2A was used as the reference.
3.5. AcJAZs Are Involved in Responses to Phytohormone and Abiotic Stress
To predict the abiotic stress-responsive cis-elements, the PlantCARE online tool was used to analyze the upstream (2000 bp) sequences of 14 pineapple AcJAZ genes and nine putative abiotic stress-responsive cis-elements were detected (Figure 4A). Each of the 14 AcJAZ genes had between three and eight putative responsive elements except for AcJAZ11, and all other AcJAZs had one or more ABREs (Figure 4A, Table S4). Not surprisingly, all AcJAZs harbor at least one MeJA-responsive element in the 2kb upstream sequences, suggesting their functions in the JA-signaling pathway. The abscisic acid-responsive element (ABRE) and wound-responsive element (WRE3) were the first and third most abundant elements, present in 13 AcJAZ genes, suggesting the involvement of AcJAZ in the ABA and wounding response.
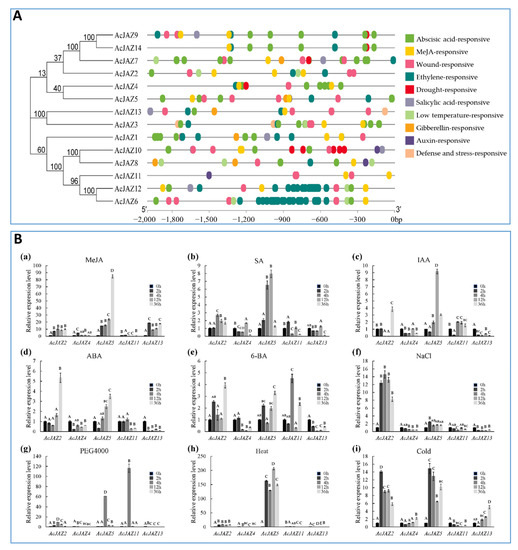
Figure 4.
The predicted cis-acting elements in the putative promoter regions of AcJAZs and the expression of AcJAZ genes in response to phytohormone treatments and abiotic stresses. (A) The phylogenetic tree of AcJAZs proteins is shown on the left. The abiotic stress-responsive cis-acting elements were predicted by the PlantCARE online tool in the 2000 bp sequences upstream of “ATG” of AcJAZ genes. The types of cis-acting elements were marked by different colors. (B) The expression analyses of AcJAZ genes under different treatments. One-month-old pineapple seedlings were treated with MeJA (100 μM (a)), SA (100 μM (b)), IAA (100 μM (c)), ABA (100 μM (d)), 6-BA (100 μM (e)), NaCl (150 mM (f)), PEG4000 (15% (g)), Heat (45 °C (h)), and Cold (4 °C (i)). The leaves of the treated seedlings were used for the analysis. AcPP2A gene was used for RT-qPCR normalization. Data in (B) were analyzed using one-way ANOVA and post hoc Tukey test, and significant differences are indicated by different letters (upper case) at p < 0.01.
We further investigated the expression of AcJAZ genes in response to other hormonal and stress conditions. AcJAZ2, AcJAZ4, AcJAZ5, AcJAZ11, and AcJAZ13, representatives of different groups, were selected for this analysis. The results showed that their expression is regulated by different phytohormones (IAA, ABA, SA, and 6-BA) and abiotic stress treatments (cold, heat, salt, and osmotic stresses) (Figure 4B). AcJAZ2 and AcJAZ5 were regulated significantly by SA, IAA, ABA, and 6-BA, and the expression of AcJAZ11 was induced considerably by 6-BA treatment. Under abiotic treatments, the expression of AcJAZ2 and AcJAZ5 was significantly induced by salt, osmotic, heat, and cold stresses. Furthermore, AcJAZ11 and AcJAZ13 were markedly induced by osmotic and cold treatments, respectively. Taken together, these results suggest that AcJAZs may play important roles in plant responses to phytohormone and abiotic stress, in addition to the JA signaling.
3.6. AcJAZs Respond to MeJA in Different Ways
JAZ proteins function as repressors of JA-responsive genes in JA signaling [51,52,53,54]. The cis-elements analysis showed that all of the JAZ genes have MeJA-responsive genes. Therefore, we investigated the expression levels of the 12 AcJAZs under MeJA treatment (Figure 5). The results indicated that AcJAZ6, AcJAZ8, and AcJAZ12 were insensitive to MeJA treatment and their expression levels did not change compared to control. AcJAZ3, AcJAZ4, AcJAZ7, AcJAZ11, and AcJAZ13 were the early-responsive genes, with a high expression level after 2 h of treatment. AcJAZ2 and AcJAZ5 could be considered constant MeJA-responsive genes as their expressions were continuously increased at different time points after treatment. Interestingly, two genes AcJAZ9 and AcJAZ14, were repressed and showed a decreasing expression tendency after the treatment (Figure 5).

Figure 5.
The expression analyses of AcJAZ genes under MeJA treatment. The expressions of most AcJAZ genes except AcJAZ1 and AcJAZ10 were analyzed by RT-qPCR (A–L). The leaves and roots from one-month-old pineapple seedlings treated with MeJA for 0 h, 2 h, 4 h, 12 h, and 36 h were used to prepare RNA samples. AcPP2A gene was used as a reference for RT-qPCR normalization. The upper case letters A, B, C, D indicated the significant difference (p < 0.01) among the samples of different time points. Any of the two samples containing the same letter belong to the same group, which would be regarded as not having a significant difference.
3.7. Protein–Protein Interaction (PPI) Analysis Revealed the Conserved Interactions among the Central JA-Signaling Regulators in Pineapple
The transcription factor MYC2 plays a crucial role in the JA-signaling pathway, which drives the expression of JA-responsive genes [24,55,56], whereas JAZs bind with MYC2 and inhibit its activity, thereby turning off the expression of JA-responsive genes. We identified the pineapple MYC2 gene (Aco018875.1), AcMYC2 (Table S5), and investigated its interactions with AcJAZ4, AcJAZ5, AcJAZ11, and AcJAZ13 by BiFC. We found that only AcJAZ5 and AcJAZ13 interacted with AcMYC2 in nuclei of tobacco leaf cells (Figure 6 and Figure S4, which confirmed the canonical function of JAZs in pineapple. In Arabidopsis, JAM1 is reported as a negative regulator of JA signaling that competitively binds to the promoters of JA-responsive genes and attenuates JA signaling [24,57,58,59,60]. Furthermore, several JAZs could interact with JAM proteins and repress the expression of their targets such as those of MYC2 transcription factors. We verified this regulatory mechanism in pineapple by identifying the JAM1 (Aco005839.1) gene, AcJAM1 (Table S6), and validating its interactions with AcJAZs using BiFC. AcJAM1 and AcJAZs (AcJAZ5 and AcJAZ13) showed a clear interaction in BiFC experiments, and the signal was detected in nuclei of tobacco leaf cells (Figure S4). Novel Interactor of JAZ (NINJA) is another protein that interacts with JAZs directly and recruits the co-repressor TOPLESS (TPL) [29]. Thus, the pineapple NINJA gene (Aco006735.1) AcNINJA was identified (Table S7), and the interactions between AcNINJA and AcJAZs were also investigated. The results showed that both AcJAZ5 and AcJAZ13 interacted with AcNINJA in nuclei of leaf cells, but no signal was detected from the interaction of AcNINJA with AcJAZ4 or AcJAZ11 (Figure 6 and Figure S4 and Table S8). In addition, homo- and heteromeric interactions between JAZ proteins widely exist in different species, including Arabidopsis, rubber tree, and cotton [61,62,63]. We analyzed and detected the heteromeric interactions of AcJAZ4 with AcJAZ5 and AcJAZ13, but not AcJAZ11 (Figure 6 and Table S8). The signal was localized in the cytoplasm and nuclei (Figure 6). Taken together, the PPI analysis revealed the interactions among the central JA-signaling regulators in pineapple and suggested the conservation of the JA-signaling pathway in plants.
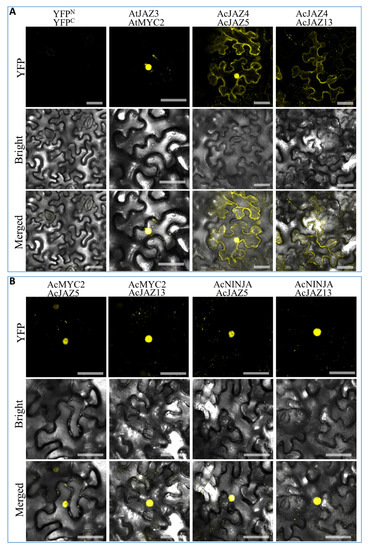
Figure 6.
The protein–protein interactions among central regulators of the JA-signaling pathway in pineapple. The images were taken under detecting YFP fluorescence (on the top), bright field (in the middle), and combining fluorescence and bright field (merged images, on the bottom). (A) Negative control (the empty vectors of YFPN and YFPC were transiently coexpressed in tobacco leaves), Positive control (construct pairs of YFPN-AtJAZ3 and YFPC-AtMYC2 were used as a positive control), and BiFC identification of the interactions of YFPN-AcJAZ4 with YFPC-AcJAZ5 and YFPC-AcJAZ13. (B) BiFC identification of the interactions of YFPN-AcMYC2 with YFPC-AcJAZ5 and YFPC-AcJAZ13, and YFPN-AcNINJA with YFPC-AcJAZ5 and YFPC-AcJAZ13. Scale bar = 50 μm.
4. Discussion
The JA-signaling pathway is one of the critical pathways involved in plant response to abiotic and biotic stresses. The basic helix–loop–helix transcription factors MYC2, MYC3, MYC4, and MYC5, are key regulators in this pathway which initiate the expression of JA-responsive genes, whereas the JAM1 transcription factor negatively regulates the pathway [24,55,56,57,58,59]. JAZ proteins repress MYC2 and JAM1 by interacting via the C-terminal Jas domain and N-terminal JIDS motif of MYC2 and JAM1. JAZ proteins also interact with NINJA and other JAZ proteins via the TIFY domain [61,62,63]. NINJA, a well-defined transcription co-repressor, interacts with TPL through its EAR domain to recruit the repressive CDK8 Mediator complex and Histone Deacetylase 19 (HD19), thus repressing the expression of target genes [59,64,65,66]. Although the pineapple genome has been deciphered [50], the JA-signaling pathway in pineapple has not been well characterized. In this study, the main regulators involved in JA signaling, including 9 MYCs, 8 NINJAs, 5 JAM1s, and 14 JAZs, were identified in pineapple (Tables S5–S7). Based on the gene-expression analysis of the AcJAZ gene family (Figure 4 and Figure 5) and protein-interaction results among the main regulators (Figure 6), we propose a hypothetical model of the JA-signaling pathway in pineapple, which is similar to that of Arabidopsis (Figure 7). However, it is worth mentioning that, AcJAZ2, AcJAZ4, AcJAZ5, AcJAZ11, and AcJAZ13 were the early JA-response genes in pineapple, and might play essential roles in pineapple JA signaling.
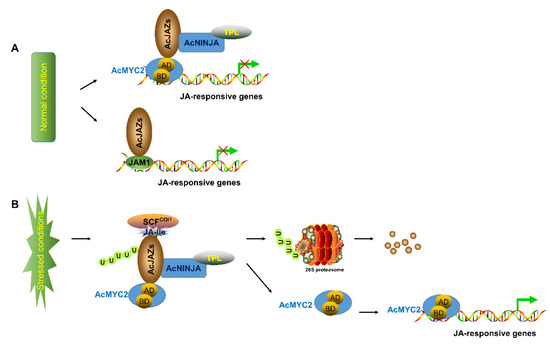
Figure 7.
Model of JA-signaling pathway in pineapple. The model indicates the core signal transduction mechanism of jasmonic acid (JA) signaling. (A) suggests that AcJAZ repressors bind to AcMYC2 or AcJAM1 to inhibit the expression of JA-responsive genes in pineapple under normal condition. (B) shows that the Skp1/Cullin1/F-box protein COI1 (SCFCOI1) complex mediates the transfer ubiquitin to AcJAZs for ubiquitination and 26S proteasome-mediated degradation, resulting in the release of transcription factors (TFs) such as AcMYC2 and the activation of JA-responsive genes in pineapple.
Based on the F153 genome data [50], we identified 14 gene members in the pineapple JAZ gene family (Table 1). A total of 42 JAZ proteins from pineapple and two species, rice (15) and Arabidopsis (13), representing monocot and dicot plants, were used for comparative and phylogenetic analyses [67,68]. The number of JAZ proteins in the pineapple genome was similar to those in rice and Arabidopsis, indicating that the JAZ gene family in those three species might encounter similar genome-wide duplication events. According to the Neighbor-Joining (NJ) phylogenetic tree, the 42 JAZ proteins from three species were clustered into six groups (Figure 1B). Group 6 only contained the JAZs from rice, while groups 1, 4, and 5 only had one JAZ from rice, reflecting the evolutionary differences among these three plants. There was a closer relationship between Arabidopsis and pineapple. Furthermore, the members in different pineapple JAZ gene family clusters showed significant divergences in exon–intron organization, sequence length, and motifs. Still, those in the same group were very similar for these characteristics (Figure 2A). In addition, the divergence in the motif composition and gene structure provided additional evidence to support phylogenetic groupings.
The JAZ proteins harbor two domains, Jas and TIFY, which interact either with MYC transcription factors when the JA-signaling pathway is not active or with the JA-Ile-SCFCOI complex when the pathway is activated. However, the TIFY domain mediates the interactions with NINJA and the formation of homo- and hetero- dimers within the JAZ subfamily [61,62,63]. Pfam results revealed that these AcJAZ proteins possessed a conserved TIFY domain and a Jas domain (Figure 2B). It has been reported that the TIFY domain has several deviations in plants [28,32]. Multiple sequence alignments indicated that the AcJAZ proteins also had several TIFY deviations (Figure S1A). The core sequence of “TIFY” was replaced with “TISF” in AcJAZ6, “TLSF” in AcJAZ11 and AcJAZ12, “TLLF” in AcJAZ8, and “TLLY” in AcJAZ10 (Figure S1A). The structural diversity of the core “TIFY” domain in pineapple JAZ proteins suggests functional diversity among the family members. In contrast, the sequences of the Jas domain among the JAZs in pineapple were more conserved (Figure S1B). Most of the JAZ genes are also JA-responsive genes in the JA-signaling pathway. For instance, AtJAZ10 was expressed immediately after JA treatment [69,70]. The results from MeJA treatment indicate that AcJAZs are JA-responsive genes, among which AcJAZ3, AcJAZ4, AcJAZ7, AcJAZ11, and AcJAZ13 are transiently responsive, while AcJAZ2, AcJAZ5, and JAZ13 are constantly JA-responsive. It has been reported that OsJAZ8 could be induced by the JA signal to confer resistance to bacterial blight [71]. Phylogenetic analysis showed that AcJAZ3, AcJAZ13, and OsJAZ8 are in the same subcluster, suggesting that MeJA-induced AcJAZ3 and AcJAZ13 might also respond to biotic stress. Under MeJA treatment, the expression of AcJAZ3, AcJAZ4, AcJAZ7, AcJAZ11, and AcJAZ1 were transiently up-regulated, and that of AcJAZ2 and AcJAZ5 constantly up-regulated (Figure 5). The JA-responsive expression of AcJAZ genes in pineapple also provides evidence for the negative feedback loop of the JA-signaling pathway.
The JA-signaling pathway has also been reported to be involved in various abiotic stresses. A previous study showed that OsJAZ1 is strongly up-regulated under drought stress, and its overexpression of OsJAZ1 negatively modulates the drought resistance [72]. Cis-acting regulatory elements in the promoter regions play a critical role in regulating gene expression by controlling the affinity of transcription factors. In this study, the cis-acting elements related to phytohormones (MeJA, SA, IAA, ABA, GA, and ETH) and stress responses (wound, osmotic, and low temperature) were discovered in the putative promoter regions of the pineapple JAZ genes (Figure 4). The expression profiles of AcJAZs under phytohormone (MeJA, IAA, ABA, SA, and 6-BA) and abiotic (cold, heat, salt, and osmotic) treatments (Figure 4) suggest that AcJAZ genes play a pivotal role in plant response to abiotic stresses.
5. Conclusions
In this study, 14 pineapple JAZ genes were identified and classified into five groups. The AcJAZ genes shared a similar gene structure and motif arrangement. The conservative motifs Jas and TIFY were present in all AcJAZs with their specialized functions. The putative promoter regions of AcJAZs contain between three and eight abiotic stress-responsive cis-acting elements. The expression patterns of AcJAZs in different tissues and in response to various abiotic stresses suggested that AcJAZs were associated with plant development and differentiation and significant response to different stresses and phytohormones. This finding indicates that AcJAZs may be involved in multiple biological processes during growth and development as well as resisting stresses. Moreover, the BiFC identification of protein interactions of AcJAZs with the central JA-signaling regulators suggested that AcJAZs and other vital players function in the JA-signaling pathway in response to abiotic stresses in pineapple. The results will be helpful in improving breeding for stress tolerance in pineapple.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/biology11030445/s1, Figure S1. The alignment of 14 AcJAZ proteins showed the tify motif and Jas domain; Figure S2. Motif logos of AcJAZs; Figure S3. Motifs in AcJAZ identified by Pfam; Figure S4. The protein–protein interactions among central regulators of the JA-signaling pathway in pineapple (continued). Table S1. The list of RT-PCR Primers; Table S2. Prediction of subcellular localization of AcJAZ proteins; Table S3. Prediction of subcellular localization of OsJAZ proteins. Table S4. The cis-acting abiotic responsive elements in Pineapple JAZ genes; Table S5. MYC2 genes in pineapple; Table S6. JAM1 genes in pineapple; Table S7. NINJA genes in pineapple; Table S8. BiFC identification of protein–protein interactions among central regulators of the JA-signaling pathway in pineapple; Supplemental File S1. All protein sequences used for phylogenetic analysis in this study; Supplemental File S2. The motif information of pineapple JAZ proteins.
Author Contributions
Conceptualization, Y.Q. and Y.C.; Funding acquisition, Y.Q. and Y.C.; Investigation, L.Y., L.C., X.Z., X.G., K.Y., S.J., Y.W., X.H., C.D., B.H. and F.D.; Methodology, L.C. and Y.C.; Project administration, Y.Q. and Y.C.; Visualization, K.Y.; Writing—original draft, L.Y. and Y.C.; Writing—review and editing, L.C., H.Z., P.Z., M.A., Y.Q. and Y.C.; All authors have read and agreed to the published version of the manuscript.
Funding
Y.C. was supported by a grant from the National Natural Science Foundation of China (Grant No. 32170380). Y.Q. was supported by a grant from the National Natural Science Foundation of China (Grant No. 31970333) and grants from the Science and Technology Program of Fujian Province (Grant No. 2019N51010021 and KJB19006A). The Funding bodies were not involved in the design of the study and collection, analysis, and interpretation of data and in writing the manuscript.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
All data have been provided in the manuscript as main figures and tables or as Supplementary Materials.
Acknowledgments
We thank the editors and reviewers for their revision of this manuscript.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Wasternack, C. Jasmonates: An Update on Biosynthesis, Signal Transduction and Action in Plant Stress Response, Growth and Development. Ann. Bot. 2007, 100, 681–697. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Turner, J. Wound-Induced Endogenous Jasmonates Stunt Plant Growth by Inhibiting Mitosis. PLoS ONE 2008, 3, e3699. [Google Scholar] [CrossRef] [PubMed]
- Wasternack, C.; Hause, B. Jasmonates: Biosynthesis, perception, signal transduction and action in plant stress response, growth and development. An update to the 2007 review in Annals of Botany. Ann. Bot. 2013, 111, 1021–1058. [Google Scholar] [CrossRef]
- Huang, H.; Liu, B.; Liu, L.; Song, S. Jasmonate action in plant growth and development. J. Exp. Bot. 2017, 68, 1349–1359. [Google Scholar] [CrossRef]
- Światek, A.; Lenjou, M.; Van Bockstaele, D.R.; Inze, D.; Van Onckelen, H. Differential Effect of Jasmonic Acid and Abscisic Acid on Cell Cycle Progression in Tobacco BY-2 Cells. Plant Physiol. 2002, 128, 201–211. [Google Scholar] [CrossRef]
- Noir, S.; Bomer, M.; Takahashi, N.; Ishida, T.; Tsui, T.; Balbi, V.; Shanahan, H.P.; Sugimoto, K.; Devoto, A. Jasmonate Controls Leaf Growth by Repressing Cell Proliferation and the Onset of Endoreduplication while Maintaining a Potential Stand-By Mode. Plant Physiol. 2013, 161, 1930–1951. [Google Scholar] [CrossRef] [PubMed]
- He, Y.; Fukushige, H.; Hildebrand, D.F.; Gan, S. Evidence supporting a role of jasmonic acid in Arabidopsis leaf senescence. Plant Physiol. 2002, 128, 876–884. [Google Scholar] [CrossRef]
- Kim, J.; Chang, C.; Tucker, M.L. To grow old: Regulatory role of ethylene and jasmonic acid in senescence. Front. Plant Sci. 2015, 6, 20. [Google Scholar] [CrossRef]
- Zhao, J.; Davis, L.C.; Verpoorte, R. Elicitor signal transduction leading to production of plant secondary metabolites. Biotechnol. Adv. 2005, 23, 283–333. [Google Scholar] [CrossRef]
- De Domenico, S.; Taurino, M.; Gallo, A.; Poltronieri, P.; Pastor, V.; Flors, V.; Santino, A. Oxylipin dynamics in Medicago truncatula in response to salt and wounding stresses. Physiol. Plant. 2019, 165, 198–208. [Google Scholar] [CrossRef]
- Bali, S.; Jamwal, V.L.; Kaur, P.; Kohli, S.K.; Ohri, P.; Gandhi, S.G.; Bhardwaj, R.; Al-Huqail, A.A.; Siddiqui, M.H.; Ahmad, P. Role of P-type ATPase metal transporters and plant immunity induced by jasmonic acid against Lead (Pb) toxicity in tomato. Ecotoxicol. Environ. Saf. 2019, 174, 283–294. [Google Scholar] [CrossRef] [PubMed]
- Hu, Y.; Jiang, Y.; Han, X.; Wang, H.; Pan, J.; Yu, D. Jasmonate regulates leaf senescence and tolerance to cold stress: Crosstalk with other phytohormones. J. Exp. Bot. 2017, 68, 1361–1369. [Google Scholar] [CrossRef] [PubMed]
- Kazan, K. Diverse roles of jasmonates and ethylene in abiotic stress tolerance. Trends Plant Sci. 2015, 20, 219–229. [Google Scholar] [CrossRef] [PubMed]
- Dar, T.A.; Uddin, M.; Khan, M.M.A.; Hakeem, K.; Jaleel, H. Jasmonates counter plant stress: A review. Environ. Exp. Bot. 2015, 115, 49–57. [Google Scholar] [CrossRef]
- Singh, I.; Shah, K. Exogenous application of methyl jasmonate lowers the effect of cadmium-induced oxidative injury in rice seedlings. Phytochemistry 2014, 108, 57–66. [Google Scholar] [CrossRef] [PubMed]
- Scholz, S.S.; Vadassery, J.; Heyer, M.; Reichelt, M.; Bender, K.W.; Snedden, W.A.; Boland, W.; Mithöfer, A. Mutation of the Arabidopsis calmodulin-like protein CML37 deregulates the jasmonate pathway and enhances susceptibility to herbivory. Mol. Plant 2014, 7, 1712–1726. [Google Scholar] [CrossRef] [PubMed]
- Brossa, R.; López-Carbonell, M.; Jubany-Marí, T.; Alegre, L. Interplay between abscisic acid and jasmonic acid and its role in water-oxidative stress in wild-type, ABA-deficient, JA-deficient, and ascorbate-deficient Arabidopsis plants. J. Plant Growth Regul. 2011, 30, 322–333. [Google Scholar] [CrossRef]
- Anderson, J.P.; Badruzsaufari, E.; Schenk, P.M.; Manners, J.M.; Desmond, O.J.; Ehlert, C.; Maclean, D.J.; Ebert, P.R.; Kazan, K. Antagonistic interaction between abscisic acid and jasmonate-ethylene signaling pathways modulates defense gene expression and disease resistance in Arabidopsis. Plant Cell 2004, 16, 3460–3479. [Google Scholar] [CrossRef] [PubMed]
- Feys, B.J.F.; Benedetti, C.E.; Penfold, C.N.; Turner, J. Arabidopsis Mutants Selected for Resistance to the Phytotoxin Coronatine Are Male Sterile, Insensitive to Methyl Jasmonate, and Resistant to a Bacterial Pathogen. Plant Cell 1994, 6, 751–759. [Google Scholar] [CrossRef]
- Fonseca, S.; Chini, A.; Hamberg, M.; Adie, B.; Porzel, A.; Kramell, R.; Miersch, O.; Wasternack, C.; Solano, R. (+)-7-iso-Jasmonoyl-L-isoleucine is the endogenous bioactive jasmonate. Nat. Chem. Biol. 2009, 5, 344–350. [Google Scholar] [CrossRef]
- Yan, J.; Li, S.; Gu, M.; Yao, R.; Li, Y.; Chen, J.; Yang, M.; Tong, J.; Xiao, L.; Nan, F. Endogenous Bioactive Jasmonate Is Composed of a Set of (+)-7-iso-JA-Amino Acid Conjugates. Plant Physiol. 2016, 172, 2154–2164. [Google Scholar] [CrossRef] [PubMed]
- Qi, T.; Huang, H.; Song, S.; Xie, D. Regulation of Jasmonate-Mediated Stamen Development and Seed Production by a bHLH-MYB Complex in Arabidopsis. Plant Cell 2015, 27, 1620–1633. [Google Scholar] [CrossRef]
- Gao, C.; Qi, S.; Liu, K.; Li, D.; Jin, C.; Li, Z.; Huang, G.; Hai, J.; Zhang, M.; Chen, M. MYC2, MYC3, and MYC4 function redundantly in seed storage protein accumulation in Arabidopsis. Plant Physiol. Biochem. 2016, 108, 63–70. [Google Scholar] [CrossRef] [PubMed]
- Goossens, J.; Mertens, J.; Goossens, A. Role and functioning of bHLH transcription factors in jasmonate signalling. J. Exp. Bot. 2017, 68, 1333–1347. [Google Scholar] [CrossRef] [PubMed]
- Devoto, A.; Nietorostro, M.; Xie, D.; Ellis, C.; Harmston, R.; Patrick, E.; Davis, J.; Sherratt, L.; Coleman, M.J.; Turner, J. COI1 links jasmonate signalling and fertility to the SCF ubiquitin–ligase complex in Arabidopsis. Plant J. 2002, 32, 457–466. [Google Scholar] [CrossRef] [PubMed]
- Mosblech, A.; Thurow, C.; Gatz, C.; Feussner, I.; Heilmann, I. Jasmonic acid perception by COI1 involves inositol polyphosphates in Arabidopsis thaliana. Plant J. 2011, 65, 949–957. [Google Scholar] [CrossRef] [PubMed]
- Major, I.T.; Yoshida, Y.; Campos, M.L.; Kapali, G.; Xin, X.F.; Sugimoto, K.; Ferreira, D.D.O.; He, S.Y.; Howe, G.A. Regulation of growth-defense balance by the JASMONATE ZIM-DOMAIN (JAZ)-MYC transcriptional module. New Phytol. 2017, 215, 1533–1547. [Google Scholar] [CrossRef] [PubMed]
- Vanholme, B.; Grunewald, W.; Bateman, A.; Kohchi, T.; Gheysen, G. The tify family previously known as ZIM. Trends Plant Sci. 2007, 12, 239–244. [Google Scholar] [CrossRef] [PubMed]
- Pauwels, L.; Barbero, G.F.; Geerinck, J.; Tilleman, S.; Grunewald, W.; Perez, A.C.; Chico, J.M.; Bossche, R.V.; Sewell, J.; Gil, E. NINJA connects the co-repressor TOPLESS to jasmonate signalling. Nature 2010, 464, 788–791. [Google Scholar] [CrossRef]
- Thines, B.; Katsir, L.; Melotto, M.; Niu, Y.; Mandaokar, A.; Liu, G.; Nomura, K.; He, S.; Howe, G.; Browse, J. JAZ repressor proteins are targets of the SCF(COI1) complex during JA signalling. Nature 2007, 448, 661–665. [Google Scholar] [CrossRef]
- Chini, A.; Fonseca, S.; Fernandez, G.; Adie, B.; Chico, J.; Lorenzo, O.; Garciacasado, G.; Lopezvidriero, I.; Lozano, F.M.; Ponce, M.R. The JAZ family of repressors is the missing link in jasmonate signalling. Nature 2007, 448, 666–671. [Google Scholar] [CrossRef] [PubMed]
- Tian, S.; Liu, S.; Wang, Y.; Wang, K.; Yin, C.; Yue, Y.; Hu, H. Genome-wide identification and characterization of JAZ protein family in two Petunia progenitors. Plants 2019, 8, 203. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Qiao, L.; Bai, J.; Wang, P.; Duan, W.; Yuan, S.; Yuan, G.; Zhang, F.; Zhang, L.; Zhao, C. Genome-wide characterization of JASMONATE-ZIM DOMAIN transcription repressors in wheat (Triticum aestivum L.). Bmc Genom. 2017, 18, 152. [Google Scholar] [CrossRef] [PubMed]
- Chini, A.; Ben-Romdhane, W.; Hassairi, A.; Aboul-Soud, M.A. Identification of TIFY/JAZ family genes in Solanum lycopersicum and their regulation in response to abiotic stresses. PLoS ONE 2017, 12, e0177381. [Google Scholar] [CrossRef] [PubMed]
- Garrido-Bigotes, A.; Figueroa, N.E.; Figueroa, P.M.; Figueroa, C.R. Jasmonate signalling pathway in strawberry: Genome-wide identification, molecular characterization and expression of JAZ s and MYC s during fruit development and ripening. PLoS ONE 2018, 13, e0197118. [Google Scholar] [CrossRef] [PubMed]
- Hong, H.; Xiao, H.; Yuan, H.; Zhai, J.; Huang, X. Cloning and characterisation of JAZ gene family in Hevea brasiliensis. Plant Biol. 2015, 17, 618–624. [Google Scholar] [CrossRef] [PubMed]
- Liu, F.; Sun, T.; Wang, L.; Su, W.; Gao, S.; Su, Y.; Xu, L.; Que, Y. Plant jasmonate ZIM domain genes: Shedding light on structure and expression patterns of JAZ gene family in sugarcane. BMC Genom. 2017, 18, 771. [Google Scholar] [CrossRef]
- Guo, Q.; Yoshida, Y.; Major, I.T.; Wang, K.; Sugimoto, K.; Kapali, G.; Havko, N.E.; Benning, C.; Howe, G.A. JAZ repressors of metabolic defense promote growth and reproductive fitness in Arabidopsis. Proc. Natl. Acad. Sci. USA 2018, 115, E10768–E10777. [Google Scholar] [CrossRef]
- Demianski, A.J.; Chung, K.M.; Kunkel, B.N. Analysis of Arabidopsis JAZ gene expression during Pseudomonas syringae pathogenesis. Mol. Plant Pathol. 2012, 13, 46–57. [Google Scholar] [CrossRef]
- Carlier, J.D.; d’Eeckenbrugge, G.C.; Leitão, J.M. Pineapple. Fruits Nuts 2007, 4, 331–342. [Google Scholar]
- Ouyang, S.; Zhu, W.; Hamilton, J.A.; Lin, H.; Campbell, M.; Childs, K.L.; Thibaudnissen, F.; Malek, R.L.; Lee, Y.; Zheng, L. The TIGR Rice Genome Annotation Resource: Improvements and new features. Nucleic Acids Res. 2007, 35, 883–887. [Google Scholar] [CrossRef] [PubMed]
- Letunic, I.; Bork, P. 20 years of the SMART protein domain annotation resource. Nucleic Acids Res. 2017, 46, D493–D496. [Google Scholar] [CrossRef] [PubMed]
- Voorrips, R.E. MapChart: Software for the Graphical Presentation of Linkage Maps and QTLs. J. Hered. 2002, 93, 77–78. [Google Scholar] [CrossRef] [PubMed]
- Zheng, Y.; Chen, X.; Wang, P.; Sun, Y.; Yue, C.; Ye, N. Genome-wide and expression pattern analysis of JAZ family involved in stress responses and postharvest processing treatments in Camellia sinensis. Sci. Rep. 2020, 10, 2792. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Li, G.; Cai, M.; Priyadarshani, S.V.G.N.; Aslam, M.; Zhou, Q.; Huang, X.; Wang, X.; Liu, Y.; Qin, Y. Genome-Wide Analysis of the YABBY Transcription Factor Family in Pineapple and Functional Identification of AcYABBY4 Involvement in Salt Stress. Int. J. Mol. Sci. 2019, 20, 5863. [Google Scholar] [CrossRef] [PubMed]
- Xie, T.; Chen, C.; Li, C.; Liu, J.; Liu, C.; He, Y. Genome-wide investigation of WRKY gene family in pineapple: Evolution and expression profiles during development and stress. BMC Genom. 2018, 19, 490. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Li, Y.; Jin, X.; Liu, L.; Dai, X.; Liu, Y.; Zhao, L.; Zheng, P.; Wang, X.; Liu, Y. Floral transcriptomes reveal gene networks in pineapple floral growth and fruit development. Commun. Biol. 2020, 3, 500. [Google Scholar] [CrossRef] [PubMed]
- Yuan, M.; Xu, C.Y. BiFC Assay for Detecting Protein-protein Interaction in Tobacco Leaves. Bio-Protocol. 2018, 101, e1010133. [Google Scholar]
- Jiang, Y.; Wang, D.; Yao, Y.; Eubel, H.; Künzler, P.; Møller, I.M.; Xu, D. MULocDeep: A deep-learning framework for protein subcellular and suborganellar localization prediction with residue-level interpretation. Comput. Struct. Biotechnol. J. 2021, 19, 4825–4839. [Google Scholar] [CrossRef]
- Ming, R.; VanBuren, R.; Wai, C.M.; Tang, H.; Schatz, M.C.; Bowers, J.E.; Lyons, E.; Wang, M.-L.; Chen, J.; Biggers, E. The pineapple genome and the evolution of CAM photosynthesis. Nat. Genet. 2015, 47, 1435–1442. [Google Scholar] [CrossRef]
- Turner, J.G.; Ellis, C.; Devoto, A. The Jasmonate Signal Pathway. Plant Cell 2002, 14 (Suppl. 1), S153–S164. [Google Scholar] [CrossRef] [PubMed]
- Kazan, K.; Manners, J.M. Jasmonate Signaling: Toward an Integrated View. Plant Physiol. 2008, 146, 1459–1468. [Google Scholar] [CrossRef] [PubMed]
- Gfeller, A.; Liechti, R.; Farmer, E.E. Jasmonate Signaling Pathway. Sci. Signal. 2010, 3, cm4. [Google Scholar]
- Ruan, J.; Zhou, Y.; Zhou, M.; Yan, J.; Khurshid, M.; Weng, W.; Cheng, J.; Zhang, K. Jasmonic Acid Signaling Pathway in Plants. Mol. Sci. 2019, 20, 2479. [Google Scholar] [CrossRef] [PubMed]
- Boter, M.; Ruíz-Rivero, O.; Abdeen, A.; Prat, S. Conserved MYC transcription factors play a key role in jasmonate signaling both in tomato and Arabidopsis. Genes Dev 2004, 18, 1577–1591. [Google Scholar] [CrossRef] [PubMed]
- Niu, Y.; Figueroa, P.; Browse, J. Characterization of JAZ-interacting bHLH transcription factors that regulate jasmonate responses in Arabidopsis. J. Exp. Bot. 2011, 62, 2143–2154. [Google Scholar] [CrossRef] [PubMed]
- Nakata, M.; Mitsuda, N.; Herde, M.; Koo, A.J.K.; Moreno, J.E.; Suzuki, K.; Howe, G.A.; Ohme-Takagi, M. A bHLH-Type Transcription Factor, ABA-INDUCIBLE BHLH-TYPE TRANSCRIPTION FACTOR/JA-ASSOCIATED MYC2-LIKE1, Acts as a Repressor to Negatively Regulate Jasmonate Signaling in Arabidopsis. Plant Cell 2013, 25, 1641–1656. [Google Scholar] [CrossRef] [PubMed]
- Sasaki-Sekimoto, Y.; Jikumaru, Y.; Obayashi, T.; Saito, H.; Masuda, S.; Kamiya, Y.; Ohta, H.; Shirasu, K. Basic Helix-Loop-Helix Transcription Factors JASMONATE-ASSOCIATED MYC2-LIKE1 (JAM1), JAM2, and JAM3 Are Negative Regulators of Jasmonate Responses in Arabidopsis. Plant Physiol. 2013, 163, 291–304. [Google Scholar] [CrossRef] [PubMed]
- Fonseca, S.; Fernández-Calvo, P.; Fernández, G.M.; Díez-Díaz, M.; Gimenez-Ibanez, S.; López-Vidriero, I.; Godoy, M.; Fernández-Barbero, G.; Van Leene, J.; De Jaeger, G.; et al. bHLH003, bHLH013 and bHLH017 Are New Targets of JAZ Repressors Negatively Regulating JA Responses. PLoS ONE 2014, 9, e86182. [Google Scholar] [CrossRef]
- Sasaki-Sekimoto, Y.; Saito, H.; Masuda, S.; Shirasu, K.; Ohta, H. Comprehensive analysis of protein interactions between JAZ proteins and bHLH transcription factors that negatively regulate jasmonate signaling. Plant Signal. Behav. 2014, 9, e27639. [Google Scholar] [CrossRef]
- Chini, A.; Fonseca, S.; Chico, J.M.; Fernandezcalvo, P.; Solano, R. The ZIM domain mediates homo- and heteromeric interactions between Arabidopsis JAZ proteins. Plant J. 2009, 59, 77–87. [Google Scholar] [CrossRef]
- Chao, J.; Zhao, Y.; Jin, J.; Wu, S.; Deng, X.; Chen, Y.; Tian, W.-M. Genome-Wide Identification and Characterization of the JAZ Gene Family in Rubber Tree (Hevea brasiliensis). Front. Genet. 2019, 10, 372. [Google Scholar] [CrossRef]
- Sun, H.; Chen, L.; Li, J.; Hu, M.; Ullah, A.; He, X.; Yang, X.; Zhang, X. The JASMONATE ZIM-Domain Gene Family Mediates JA Signaling and Stress Response in Cotton. Plant Cell Physiol. 2017, 58, 2139–2154. [Google Scholar] [CrossRef] [PubMed]
- Ito, J.; Fukaki, H.; Onoda, M.; Li, L.; Li, C.; Tasaka, M.; Furutani, M. Auxin-dependent compositional change in Mediator in ARF7- and ARF19-mediated transcription. Proc. Natl. Acad. Sci. USA 2016, 113, 6562–6567. [Google Scholar] [CrossRef] [PubMed]
- Long, J.A.; Ohno, C.; Smith, Z.R.; Meyerowitz, E.M. TOPLESS Regulates Apical Embryonic Fate in Arabidopsis. Science 2006, 312, 1520. [Google Scholar] [CrossRef] [PubMed]
- Leydon, A.R.; Wang, W.; Juarez-Solis, S.; Zemke, J.E.; Zahler, M.L.; Zheng, N.; Nemhauser, J.L. Stucture-function analysis of Arabidopsis TOPLESS reveals conservation of repression mechanisms across eukaryotes. Biotech. Week 2020, 4, 1474–1525. [Google Scholar]
- Ye, H.; Du, H.; Tang, N.; Li, X.; Xiong, L. Identification and expression profiling analysis of TIFY family genes involved in stress and phytohormone responses in rice. Plant Mol. Biol. 2009, 71, 291–305. [Google Scholar] [CrossRef]
- Thireault, C.; Shyu, C.; Yoshida, Y.; Aubin, B.S.; Campos, M.L.; Howe, G.A. Repression of jasmonate signaling by a non-TIFY JAZ protein in Arabidopsis. Plant J. 2015, 82, 669–679. [Google Scholar] [CrossRef] [PubMed]
- Moreno, J.E.; Shyu, C.; Campos, M.L.; Patel, L.C.; Chung, H.S.; Yao, J.; He, S.Y.; Howe, G.A. Negative Feedback Control of Jasmonate Signaling by an Alternative Splice Variant of JAZ10. Plant Physiol. 2013, 162, 1006–1017. [Google Scholar] [CrossRef] [PubMed]
- PFernández-Calvo, P.; Chini, A.; Fernández-Barbero, G.; Chico, J.M.; Gimenez-Ibanez, S.; Geerinck, J.; Eeckhout, D.; Schweizer, F.; Godoy, M.; Franco-Zorrilla, J.M.; et al. The Arabidopsis bHLH Transcription Factors MYC3 and MYC4 Are Targets of JAZ Repressors and Act Additively with MYC2 in the Activation of Jasmonate Responses. Plant Cell 2011, 23, 701–715. [Google Scholar] [CrossRef]
- Yamada, S.; Kano, A.; Tamaoki, D.; Miyamoto, A.; Shishido, H.; Miyoshi, S.; Taniguchi, S.; Akimitsu, K.; Gomi, K. Involvement of OsJAZ8 in Jasmonate-Induced Resistance to Bacterial Blight in Rice. Plant Cell Physiol. 2012, 53, 2060–2072. [Google Scholar] [CrossRef]
- Fu, J.; Wu, H.; Ma, S.; Xiang, D.; Liu, R.; Xiong, L. OsJAZ1 Attenuates Drought Resistance by Regulating JA and ABA Signaling in Rice. Front. Plant Sci. 2017, 8, 2108. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).