Maternal Relationships among Ancient and Modern Southern African Sheep: Newly Discovered Mitochondrial Haplogroups
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Modern Samples
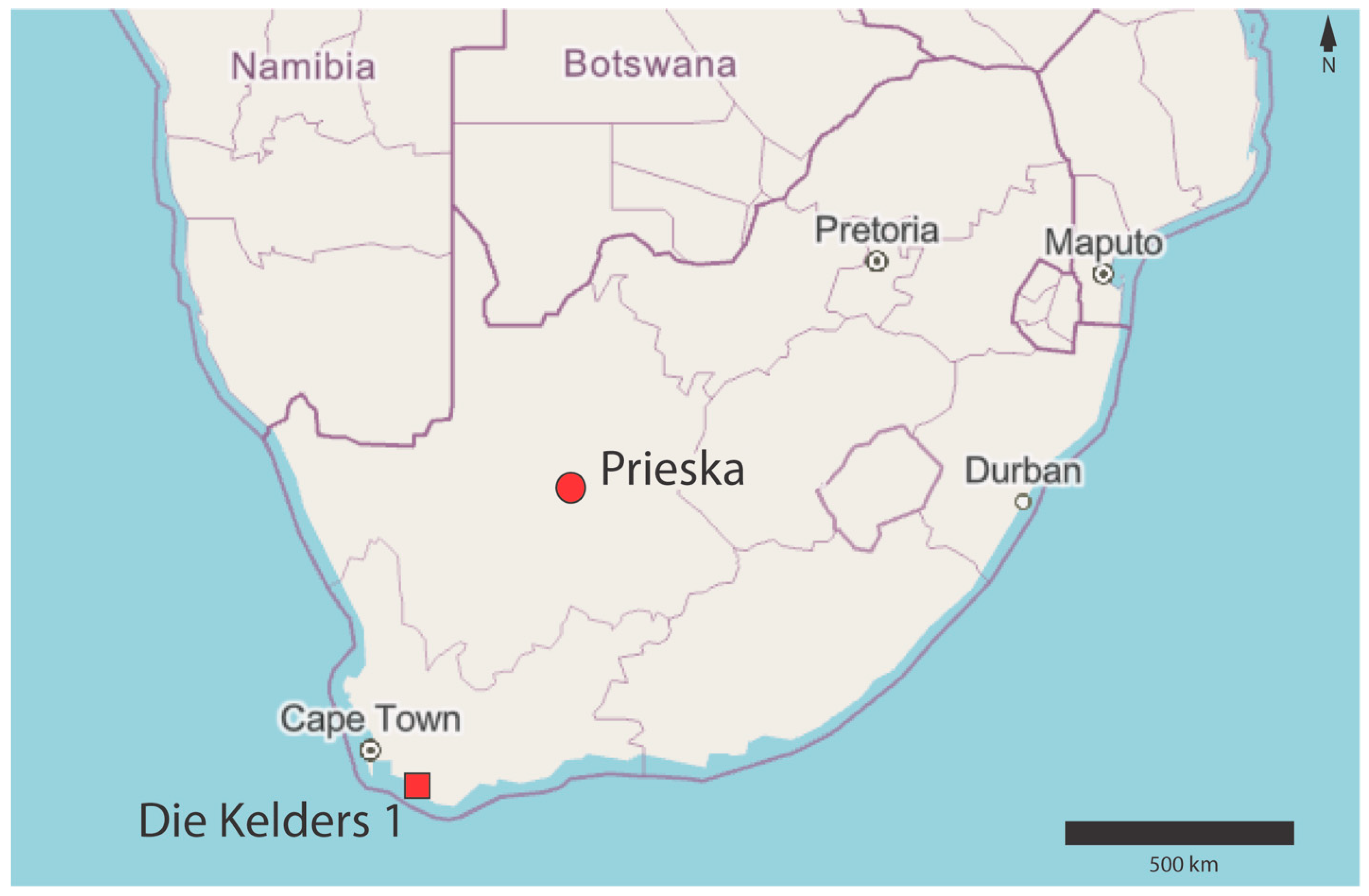
2.2. Archaeological Samples
2.3. Data Analysis
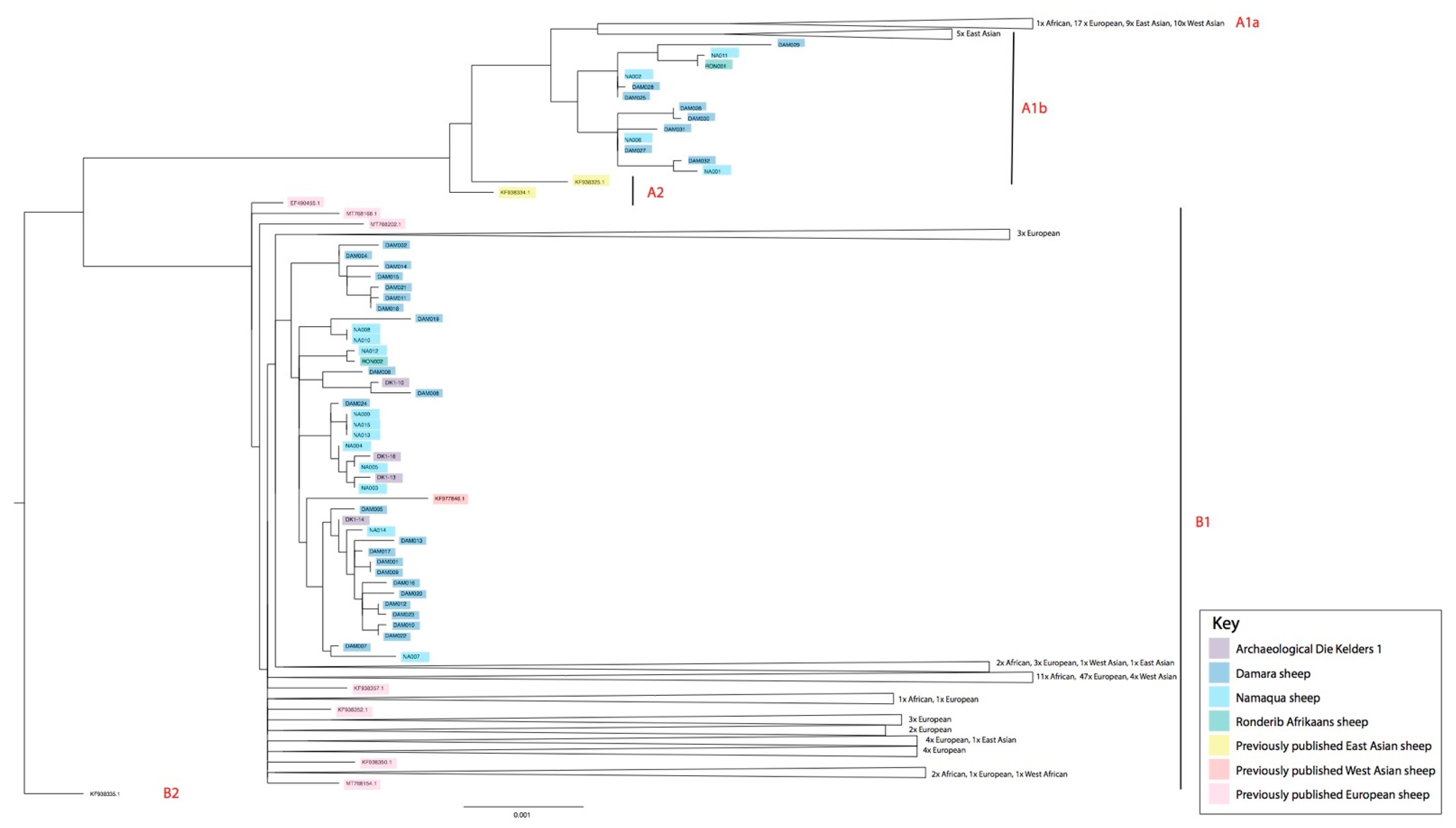
3. Results
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Kosgey, I.; Rowlands, G.; van Arendonk, J.; Baker, R. Small ruminant production in smallholder and pastoral/extensive farming systems in Kenya. Small Rumin. Res. 2008, 77, 11–24. [Google Scholar] [CrossRef]
- Legge, T. The beginning of caprine domestication in Southwest Asia. In The Origins and Spread of Agriculture and Pastoralism in Eurasia; Harris, D.R., Ed.; Institute of Archaeology, University College London: London, UK, 1996; pp. 238–262. [Google Scholar]
- Álvarez, I.; Capote, J.; Traoré, A.; Fonseca, N.; Pérez, K.; Cuervo, M.; Fernández, I.; Goyache, F. Mitochondrial analysis sheds light on the origin of hair sheep. Anim. Genet. 2013, 44, 344–347. [Google Scholar] [CrossRef] [PubMed]
- Demirci, S.; Baştanlar, E.K.; Dagtas, N.; Pişkin, E.; Engin, A.; Ozer, F.; Yüncü, E.; Dogan, S.A.; Togan, I. Mitochondrial DNA diversity of modern, ancient and wild sheep (Ovis gmelinii anatolica) from Turkey: New insights on the evolutionary history of sheep. PLoS ONE 2013, 8, e81952. [Google Scholar] [CrossRef] [PubMed]
- Finlay, E.; Tapio, M.; Kierstein, G.; Jianlin, H.; Hanotte, O. Assessment of Sheep Domestication History Based on Mitochondrial DNA Diversity of Contemporary Old World Sheep; European Society for Evolutionary Biology: Uppsala, Sweden, 2007. [Google Scholar]
- Muigai, A.W.T.; Hanotte, O. The origin of African sheep: Archaeological and genetic perspectives. Afr. Archaeol. Rev. 2013, 30, 39–50. [Google Scholar] [CrossRef]
- di Lernia, S. Cultural control over wild animals during the early Holocene: The case of Barbary sheep in the central Sahara. In Before Food Production in North Africa; di Lernia, S., Manzi, G., Eds.; A.B.A.C.O. Edizioni: Forli, Italy, 1998; pp. 113–126. [Google Scholar]
- di Lernia, S. The Uan Afuda Cave: Hunter-Gatherer Societies of the Central Sahara (Arid Zone Archaeology Monographs); Edizioni All’Insegna del Giglio: Rome, Italy, 1999. [Google Scholar]
- Klein, R.G.; Scott, K. Re-analysis of faunal assemblages from the Haua Fteah and other late quaternary archaeological sites in Cyrenaican Libya. J. Archaeol. Sci. 1986, 13, 515–542. [Google Scholar] [CrossRef]
- Saxon, E.C.; Close, A.E.; Cluzel, C.; Morse, V.; Shackleton, N.J. Results of recent investigations at Tamar Hat. Libyca 1974, 22, 49–91. [Google Scholar]
- Manwell, C.; Baker, C.M.A. Ammotragus lervia: Pregenitor of domesticated sheep or specialized offshoot of caprine evolution. Experientia 1975, 31, 1370–1371. [Google Scholar] [CrossRef]
- Barker, G.; Basell, L.; Brooks, I.; Burn, L.; Cartwright, C.; Cole, F.; Davison, J.; Farr, L.; Grün, R.; Hamilton, R.; et al. The Cyrenaican Prehistory Project 2008: The second season of investigations of the Haua Fteah cave and its landscape, and further results from the initial (2007) fieldwork. Libyan Stud. 2008, 39, 1–74. [Google Scholar] [CrossRef]
- Barthelme, J.W. Fisher-Hunters and Neolithic Pastoralists in East Turkana, Kenya; British Archaeological Reports; BAR International Series: Oxford, UK, 1985; Volume 254. [Google Scholar]
- Culley, C.; Janzen, A.; Brown, S.; Prendergast, M.E.; Shipton, C.; Ndiema, E.; Petraglia, M.D.; Boivin, N.; Crowther, A. Iron Age hunting and herding in coastal eastern Africa: ZooMS identification of domesticates and wild bovids at Panga ya Saidi, Kenya. J. Archaeol. Sci. 2021, 130, 105368. [Google Scholar] [CrossRef]
- Sadr, K. The first herders at the Cape of Good Hope. Afr. Archaeol. Rev. 1998, 15, 101–132. [Google Scholar] [CrossRef]
- Sadr, K. The Neolithic of southern Africa. J. Afr. Hist. 2003, 44, 195–209. [Google Scholar] [CrossRef]
- Sadr, K. Livestock first reached southern Africa in two separate events. PLoS ONE 2015, 10, e0134215. [Google Scholar] [CrossRef] [PubMed]
- Smith, A.B. Prehistoric pastoralism in the Southwestern Cape, South Africa. World Archaeol. 1983, 15, 79–89. [Google Scholar] [CrossRef]
- Smith, A.B. The Economics of Prehistoric Herding at the Cape. In Final Report to the Human Sciences Research Council; University of Cape Town: Cape Town, South Africa, 1987. [Google Scholar]
- Smith, A.B. Origins and spread of pastoralism in Africa. Annu. Rev. Anthropol. 1992, 21, 125–141. [Google Scholar] [CrossRef]
- Smith, A.B. Early domestic stock in southern Africa: A commentary. Afr. Archaeol. Rev. 1998, 15, 151–156. [Google Scholar] [CrossRef]
- Smith, A.B. The origins of the domesticated animals of southern Africa. In The Origins and Development of African Livestock: Archaeology, Genetics, Linguistics and Ethnography; Blench, R.M., MacDonald, K.C., Eds.; UCL Press: London, UK, 2000; pp. 222–238. [Google Scholar]
- Smith, A.B. Pastoral origins at the Cape, South Africa: Influences and arguments. South. Afr. Humanit. 2008, 20, 49–60. [Google Scholar]
- Lander, F.; Russell, T. The archaeological evidence for the appearance of pastoralism and farming in southern Africa. PLoS ONE 2018, 13, e0198941. [Google Scholar] [CrossRef]
- Russell, T. “Where goats connect people”: Cultural diffusion of livestock not food production amongst southern African hunter-gatherers during the Later Stone Age. J. Soc. Archaeol. 2017, 17, 115–137. [Google Scholar] [CrossRef]
- Russell, T.; Lander, F. “What is consumed is wasted”: From foraging to herding in the southern African Later Stone Age. Azania Archaeol. Res. Afr. 2015, 50, 267–317. [Google Scholar] [CrossRef]
- Russell, T.M. The Spatial Analysis of Radiocarbon Databases: The Spread of the First Farmers in Europe and of the Fat-Tailed Sheep in Southern Africa; BAR International Series/Archaeopress: Oxford, UK, 2004; Volume 1294. [Google Scholar]
- Sealy, J.; Yates, R. The chronology of the introduction of pastoralism to the Cape, South Africa. Antiquity 1994, 68, 58–67. [Google Scholar] [CrossRef]
- Sealy, J.; Yates, R. Direct radiocarbon dating of early sheep bones: Two further results. South Afr. Archaeol. Bull. 1996, 51, 109–110. [Google Scholar] [CrossRef]
- Orton, J.; Mitchell, P.; Klein, R.; Steele, T.; Horsburgh, K.A. An early date for cattle from Namaqualand, South Africa: Implications for the origins of herding in southern Africa. Antiquity 2013, 87, 108–120. [Google Scholar] [CrossRef][Green Version]
- Horsburgh, K.A. Molecular anthropology: The judicial use of genetic data in archaeology. J. Archaeol. Sci. 2015, 56, 141–145. [Google Scholar] [CrossRef]
- Horsburgh, K.A.; Moreno-Mayar, J.V. Molecular identification of sheep at Blydefontein Rock Shelter, South Africa. South. Afr. Humanit. 2015, 27, 65–90. [Google Scholar]
- Horsburgh, K.A.; Moreno-Mayar, J.V.; Gosling, A.L. Revisiting the Kalahari debate in the highlands: Ancient DNA provides new faunal identifications at Sehonghong, Lesotho. Azania Archaeol. Res. Afr. 2016, 51, 295–306. [Google Scholar] [CrossRef]
- Horsburgh, K.A.; Orton, J.; Klein, R.G. Beware the Springbok in sheep’s clothing: How secure are the faunal identifications upon which we build our models? Afr. Archaeol. Rev. 2016, 33, 353–361. [Google Scholar] [CrossRef]
- Horsburgh, K.A. Genetics and domestic fauna in southern Africa. In Oxford Research Encyclopedia of Anthropology; Oxford University Press: Oxford, UK, 2020. [Google Scholar]
- Horsburgh, K.A.; Gosling, A.L. Systematic ancient DNA species identification fails to find late Holocene domesticated cattle in southern Africa. Biology 2020, 9, 316. [Google Scholar] [CrossRef]
- Scott, K.; Plug, I. Osteomorphology and osteometry versus aDNA in taxonomic identification of fragmentary sheep and sheep/goat bones from archaeological deposits: Blydefontein Shelter, Karoo, South Africa. South. Afr. Humanit. 2016, 28, 61–79. [Google Scholar]
- Plug, I. Reply to Horsburgh et al. 2016: “Revisiting the Kalahari debate in the highlands”. Azania Archaeol. Res. Afr. 2017, 53, 98–113. [Google Scholar] [CrossRef]
- Bousman, B.C.; Mauldin, R.; Zoppi, U.; Higham, T.; Scott, L.; Brink, J. The quest for evidence of domestic stock at Blydefontein Rock Shelter. South. Afr. Humanit. 2016, 28, 39–60. [Google Scholar]
- Horsburgh, K.A. A reply to Plug 2017: Science requires self-correction. Azania Archaeol. Res. Afr. 2017, 53, 114–118. [Google Scholar] [CrossRef]
- Horsburgh, K.A.; Moreno-Mayar, J.V.; Klein, R.G. Counting and miscounting sheep: Genetic evidence for pervasive misclassification of wild fauna as domestic stock. South. Afr. Humanit. 2017, 30, 53–69. [Google Scholar]
- Le Meillour, L.; Zirah, S.; Zazzo, A.; Cersoy, S.; Détroit, F.; Imalwa, E.; Lebon, M.; Nankela, A.; Tombret, O.; Pleurdeau, D.; et al. Palaeoproteomics gives new insight into early southern African pastoralism. Sci. Rep. 2020, 10, 14427. [Google Scholar] [CrossRef] [PubMed]
- Coutu, A.N.; Taurozzi, A.J.; Mackie, M.; Jensen, T.Z.T.; Collins, M.J.; Sealy, J. Palaeoproteomics confirm earliest domesticated sheep in southern Africa ca. 2000 BP. Sci. Rep. 2021, 11, 6631. [Google Scholar] [CrossRef] [PubMed]
- Raven-Hart, R. Before Van Riebeeck: Callers at South Africa from 1488 to 1652; Struik Publishing: Cape Town, South Africa, 1967. [Google Scholar]
- Heinrich, A.R.; Schrire, C. Colonial fauna at the Cape of Good Hope: A proxy for colonial impact on indigenous people. In The Importance of Material Things; Schablitsky, J., Leone, M., Eds.; Society for Historical Archaeology: Germantown, MD, USA, 2011; Volume 2, pp. 121–141. [Google Scholar]
- Schrire, C. Historical Archaeology in South Africa: Material Culture of the Dutch East India Company at the Cape; Left Coast Press, Inc.: Walnut Creek, CA, USA, 2014. [Google Scholar]
- Thompson, L. A History of South Africa; Vail-Ballou Press: Binghamton, NY, USA, 1995. [Google Scholar]
- Mentzel, O.F.; Mandelbrote, H.J. A Geographical and Topographical Description of the Cape of Good Hope; Van Riebeeck Society: Cape Town, South Africa, 1921. [Google Scholar]
- Gordon, R.E.; Talbot, C.J. From Dias to Vorster: Source Material on South African History 1488–1975, 1st ed.; Nasou via Afrika Publishers: Cape Town, South Africa, 1977; 464p. [Google Scholar]
- du Toit, D. The Damara of southern Africa. Anim. Genet. Resour. Inf. 2007, 44, 89. [Google Scholar]
- Campbell, Q.P. The origin and description of southern Africa’s indigenous goats. South Afr. J. Anim. Sci. 2003, 4, 18–22. [Google Scholar]
- Almeida, A.M. The Damara in the context of southern Africa fat-tailed sheep breeds. Trop. Anim. Health Prod. 2011, 43, 1427–1441. [Google Scholar] [CrossRef]
- Kilminster, T.F.; Greeff, J.C. A note on the reproductive performance of Damara, Dorper and Merino sheep under optimum management and nutrition for Merino ewes in the eastern wheatbelt of western Australia. Trop. Anim. Health Prod. 2011, 43, 1459–1464. [Google Scholar] [CrossRef]
- Epstein, H., IV. The fat-tailed sheep of Africa. In The Origin of Domestic Animals of Africa; Epstein, H., Ed.; Africana Publishing Corporation: New York, NY, USA, 1971; pp. 110–174. [Google Scholar]
- Epstein, H. History and origin of the Ronderib and Namaqua Afrikaner sheep. Z. Tierzüchtung Züchtungsbiol. 1960, 74, 267–292. [Google Scholar] [CrossRef]
- Thutwa, K.; van Wyk, J.B.; Dzama, K.; Scholtz, A.J.; Cloete, S.W. Expression of cytokine genes at tick attachment and control sites of Namaqua Afrikaner, Dorper and South African Mutton Merino sheep. Vet. Parasitol. 2021, 291, 109384. [Google Scholar] [CrossRef]
- Moradi, M.H.; Nejati-Javaremi, A.; Moradi-Shahrbabak, M.; Dodds, K.G.; McEwan, J.C. Genomic scan of selective sweeps in thin and fat tail sheep breeds for identifying of candidate regions associated with fat deposition. BMC Genet. 2012, 13, 10. [Google Scholar] [CrossRef] [PubMed]
- Wood, N.J.; Phua, S.H. Variation in the control region sequence of the sheep mitochondrial genome. Anim. Genet. 1996, 27, 25–33. [Google Scholar] [CrossRef] [PubMed]
- Hiendleder, S.; Kaupe, B.; Wassmuth, R.; Janke, A. Molecular analysis of wild and domestic sheep questions current nomenclature and provides evidence for domestication from two different subspecies. Proc. R. Soc. B Boil. Sci. 2002, 269, 893–904. [Google Scholar] [CrossRef] [PubMed]
- Hiendleder, S.; Lewalski, H.; Wassmuth, R.; Janke, A. The complete mitochondrial DNA sequence of the domestic sheep (Ovis aries) and comparison with the other major ovine haplotype. J. Mol. Evol. 1998, 47, 441–448. [Google Scholar] [CrossRef]
- Hiendleder, S.; Mainz, K.; Plante, Y.; Lewalski, H. Analysis of mitochondrial DNA indicates that domestic sheep are derived from two different ancestral maternal sources: No evidence for contributions from urial and argali sheep. J. Hered. 1998, 89, 113–120. [Google Scholar] [CrossRef]
- Hiendleder, S.; Phua, S.H.; Hecht, W. A diagnostic assay discriminating between two major Ovis aries mitochondrial DNA haplotypes. Anim. Genet. 1999, 30, 211–213. [Google Scholar] [CrossRef]
- Meadows, J.R.S.; Hiendleder, S.; Kijas, J.W. Haplogroup relationships between domestic and wild sheep resolved using a mitogenome panel. Heredity 2011, 106, 700–706. [Google Scholar] [CrossRef]
- Guo, J.; Du, L.-X.; Ma, Y.-H.; Guan, W.-J.; Li, H.-B.; Zhao, Q.-J.; Li, X.; Rao, S.-Q. A novel maternal lineage revealed in sheep (Ovis aries). Anim. Genet. 2005, 36, 331–336. [Google Scholar] [CrossRef]
- Pedrosa, S.; Arranz, J.-J.; Brito, N.; Molina, A.; Primitivo, F.S.; Bayón, Y. Mitochondrial diversity and the origin of Iberian sheep. Genet. Sel. Evol. 2007, 39, 91–103. [Google Scholar] [CrossRef]
- Pedrosa, S.; Uzun, M.; Arranz, J.-J.; Gutiérrez-Gil, B.; Primitivo, F.S.; Bayón, Y. Evidence of three maternal lineages in near eastern sheep supporting multiple domestication events. Proc. R. Soc. B Boil. Sci. 2005, 272, 2211–2217. [Google Scholar] [CrossRef]
- Meadows, J.; Li, K.; Kantanen, J.; Tapio, M.; Sipos, W.; Pardeshi, V.; Gupta, V.; Calvo, J.; Whan, V.; Norris, B.; et al. Mitochondrial sequence reveals high levels of gene flow between breeds of domestic sheep from Asia and Europe. J. Hered. 2005, 96, 494–501. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Tapio, M.; Marzanov, N.; Ozerov, M.; Ćinkulov, M.; Gonzarenko, G.; Kiselyova, T.; Murawski, M.; Viinalass, H.; Kantanen, J. Sheep mitochondrial DNA variation in European, Caucasian, and central Asian areas. Mol. Biol. Evol. 2006, 23, 1776–1783. [Google Scholar] [CrossRef] [PubMed]
- Lv, F.-H.; Peng, W.-F.; Yang, J.; Zhao, Y.-X.; Li, W.-R.; Liu, M.-J.; Ma, Y.-H.; Zhao, Q.-J.; Yang, G.-L.; Wang, F.; et al. Mitogenomic meta-analysis identifies two phases of migration in the history of eastern Eurasian sheep. Mol. Biol. Evol. 2015, 32, 2515–2533. [Google Scholar] [CrossRef] [PubMed]
- Ghernouti, N.; Bodinier, M.; Ranebi, D.; Maftah, A.; Petit, D.; Gaouar, S. Control region of mtDNA identifies three migration events of sheep breeds in Algeria. Small Rumin. Res. 2017, 155, 66–71. [Google Scholar] [CrossRef]
- Othman, O.E.M.; Pariset, L.; Balabel, E.A.; Marioti, M. Genetic characterization of Egyptian and Italian sheep breeds using mitochondrial DNA. J. Genet. Eng. Biotechnol. 2015, 13, 79–86. [Google Scholar] [CrossRef]
- Othman, O.E.; Balabel, E.A.; Abdel-Samad, M.F. Mitochondrial DNA diversity in five Egyptian sheep breeds. Glob. Vet. 2014, 12, 369–375. [Google Scholar]
- Nigussie, H.; Mwacharo, J.M.; Osama, S.; Agaba, M.; Mekasha, Y.; Kebede, K.; Abegaz, S.; Pal, S.K. Correction to: Genetic diversity and matrilineal genetic origin of fat-rumped sheep in Ethiopia. Trop. Anim. Health Prod. 2020, 52, 3933. [Google Scholar] [CrossRef]
- Nigussie, H.; Mwacharo, J.M.; Osama, S.; Agaba, M.; Mekasha, Y.; Kebede, K.; Abegaz, S.; Pal, S.K. Genetic diversity and matrilineal genetic origin of fat-rumped sheep in Ethiopia. Trop. Anim. Health Prod. 2019, 51, 1393–1404. [Google Scholar] [CrossRef]
- Resende, A.; Gonçalves, J.; Muigai, A.W.T.; Pereira, F. Mitochondrial DNA variation of domestic sheep (Ovis aries) in Kenya. Anim. Genet. 2016, 47, 377–381. [Google Scholar] [CrossRef]
- Kandoussi, A.; Boujenane, I.; Auger, C.; Serranito, B.; Germot, A.; Piro, M.; Maftah, A.; Badaoui, B.; Petit, D. The origin of sheep settlement in western Mediterranean. Sci. Rep. 2020, 10, 10225. [Google Scholar] [CrossRef]
- Horsburgh, K.A.; Rhines, A. Genetic characterization of an archaeological sheep assemblage from South Africa’s Western Cape. J. Archaeol. Sci. 2010, 37, 2906–2910. [Google Scholar] [CrossRef]
- Schweitzer, F.R. Excavations at Die Kelders, Cape Province, South Africa: The Holocene deposits. Ann. South Afr. Mus. 1979, 78, 101–233. [Google Scholar]
- Avery, G.; Cruz-Uribe, K.; Goldberg, P.; Grine, F.E.; Klein, R.G.; Lenardi, M.J.; Marean, C.W.; Rink, W.J.; Schwarcz, H.P.; Thackeray, A.I.; et al. The 1992–1993 Excavations at the Die Kelders Middle and Later Stone Age Cave site, South Africa. J. Field Archaeol. 1997, 24, 263–291. [Google Scholar] [CrossRef]
- Goldberg, P. Micromorphology and site formation at Die Kelders Cave I., South Africa. J. Hum. Evol. 2000, 38, 43–90. [Google Scholar] [CrossRef] [PubMed]
- Klein, R.G.; Cruz-Uribe, K. Middle and Later Stone Age large mammal and tortoise remains from Die Kelders Cave 1, Western Cape Province, South Africa. J. Hum. Evol. 2000, 38, 169–195. [Google Scholar] [CrossRef]
- Klein, R.G.; Cruz-Uribe, K. Exploitation of large bovids and seals at Middle and Later Stone Age sites in South Africa. J. Hum. Evol. 1996, 31, 315–334. [Google Scholar] [CrossRef]
- Wilson, M.L. The late Holocene occupants of Die Kelders: Hunter-gatherers or herders? South. Afr. Field Archaeol. 1996, 5, 79–83. [Google Scholar]
- Schweitzer, F.R. Archaeological evidence for sheep at the Cape. South Afr. Archaeol. Bull. 1974, 29, 75–82. [Google Scholar] [CrossRef]
- Hogg, A.G.; Hua, Q.; Blackwell, P.G.; Niu, M.; Buck, C.; Guilderson, T.P.; Heaton, T.J.; Palmer, J.; Reimer, P.J.; Reimer, R.W.; et al. SHCal13 southern hemisphere calibration, 0–50,000 years cal BP. Radiocarbon 2013, 55, 1889–1903. [Google Scholar] [CrossRef]
- Allentoft, M.E.; Sikora, M.; Sjögren, K.G.; Rasmussen, S.; Rasmussen, M.; Stenderup, J.; Damgaard, P.B.; Schroeder, H.; Ahlström, T.; Vinner, L.; et al. Population genomics of Bronze Age Eurasia. Nature 2015, 522, 167–172. [Google Scholar] [CrossRef]
- Li, H.; Durbin, R. Fast and accurate short read alignment with Burrows–Wheeler transform. Bioinformatics 2009, 25, 1754–1760. [Google Scholar] [CrossRef] [PubMed]
- Jónsson, H.; Ginolhac, A.; Schubert, M.; Johnson, P.L.F.; Orlando, L. mapDamage2.0: Fast approximate Bayesian estimates of ancient DNA damage parameters. Bioinformatics 2013, 29, 1682–1684. [Google Scholar] [CrossRef] [PubMed]
- Lindgreen, S. AdapterRemoval: Easy cleaning of next-generation sequencing reads. BMC Res. Notes 2012, 5, 337. [Google Scholar] [CrossRef] [PubMed]
- Guindon, S.; Dufayard, J.-F.; Lefort, V.; Anisimova, M.; Hordijk, W.; Gascuel, O. New algorithms and methods to estimate maximum-likelihood phylogenies: Assessing the performance of PhyML 3.0. Syst. Biol. 2010, 59, 307–321. [Google Scholar] [CrossRef] [PubMed]
- Lefort, V.; Longueville, J.-E.; Gascuel, O. SMS: Smart Model Selection in PhyML. Mol. Biol. Evol. 2007, 34, 2422. [Google Scholar] [CrossRef] [PubMed]
- Horsburgh, K.A.; Prost, S.; Gosling, A.; Stanton, J.-A.; Rand, C.; Matisoo-Smith, E.A. The genetic diversity of the Nguni breed of African cattle (Bos spp.): Complete mitochondrial genomes of haplogroup T1. PLoS ONE 2013, 8, e71956. [Google Scholar] [CrossRef]
- Bonfiglio, S.; Ginja, C.; De Gaetano, A.; Achilli, A.; Olivieri, A.; Colli, L.; Tesfaye, K.; Agha, S.H.; Gama, L.T.; Cattonaro, F.; et al. Origin and spread of Bos taurus: New clues from mitochondrial genomes belonging to haplogroup T1. PLoS ONE 2012, 7, e38601. [Google Scholar] [CrossRef]

| Lab Identification | Element | Morphology | Provenience |
|---|---|---|---|
| DK1_02 | Right mandible | P2 P3 P4 M1 M2 M3 | Layer 2, AA2 |
| DK1_04 | Right mandible | M1 M2 | Layer 2b, B3 |
| DK1_05 | Right mandible | dp3 dp4 M1 M2 | Layer 2, A97 |
| DK1_06 | Right mandible | P2 M1 M2 M3 | Layer 2, A2 |
| DK1_09 | Right mandible | dp2 dp3 dp4 M1 | Layer 2, A3 |
| DK1_10 | Right mandible | M1 M2 | Layer 2, A98 |
| DK1_13 | Right mandible | P3 P4 | Layer 2, AA1 |
| DK1_14 | Right mandible | dp2 dp3 dp4 | Layer 2, D2 |
| DK1_16 | Left mandible | dp2 dp4 M1 | Layer 2, A98 |
| DK1_25 | Right mandible | dp3 dp4 | Layer 2, A9b |
| DK1_53 | Left mandible | dp2 dp3 dp4 | Layer 2, B3 |
| DK1_54 | Left mandible | P2 P3 P4 M1 M2 M3 | Layer 2, B3 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Horsburgh, K.A.; Beckett, D.B.; Gosling, A.L. Maternal Relationships among Ancient and Modern Southern African Sheep: Newly Discovered Mitochondrial Haplogroups. Biology 2022, 11, 428. https://doi.org/10.3390/biology11030428
Horsburgh KA, Beckett DB, Gosling AL. Maternal Relationships among Ancient and Modern Southern African Sheep: Newly Discovered Mitochondrial Haplogroups. Biology. 2022; 11(3):428. https://doi.org/10.3390/biology11030428
Chicago/Turabian StyleHorsburgh, K. Ann, Devri B. Beckett, and Anna L. Gosling. 2022. "Maternal Relationships among Ancient and Modern Southern African Sheep: Newly Discovered Mitochondrial Haplogroups" Biology 11, no. 3: 428. https://doi.org/10.3390/biology11030428
APA StyleHorsburgh, K. A., Beckett, D. B., & Gosling, A. L. (2022). Maternal Relationships among Ancient and Modern Southern African Sheep: Newly Discovered Mitochondrial Haplogroups. Biology, 11(3), 428. https://doi.org/10.3390/biology11030428






