Can Model Experiments Give Insight into the Response of the Soil Environment to Flooding? A Comparison of Microcosm and Natural Event
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Soil Sampling Site
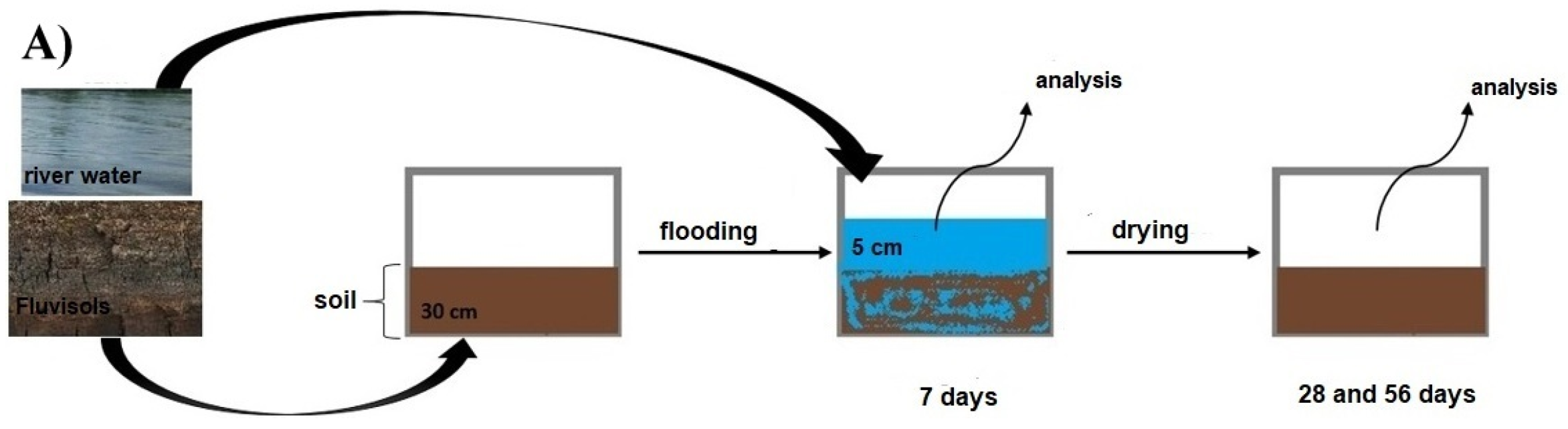
2.2. The Microcosm Experiment
2.3. The Natural Hydrological Event
2.4. Soil Sampling
2.5. pH Values and Moisture Contents
2.6. Enzymatic Activity of Soil
2.7. Community Level Physiological Profiling (CLPP)
2.8. Statistical Analysis and Visualization
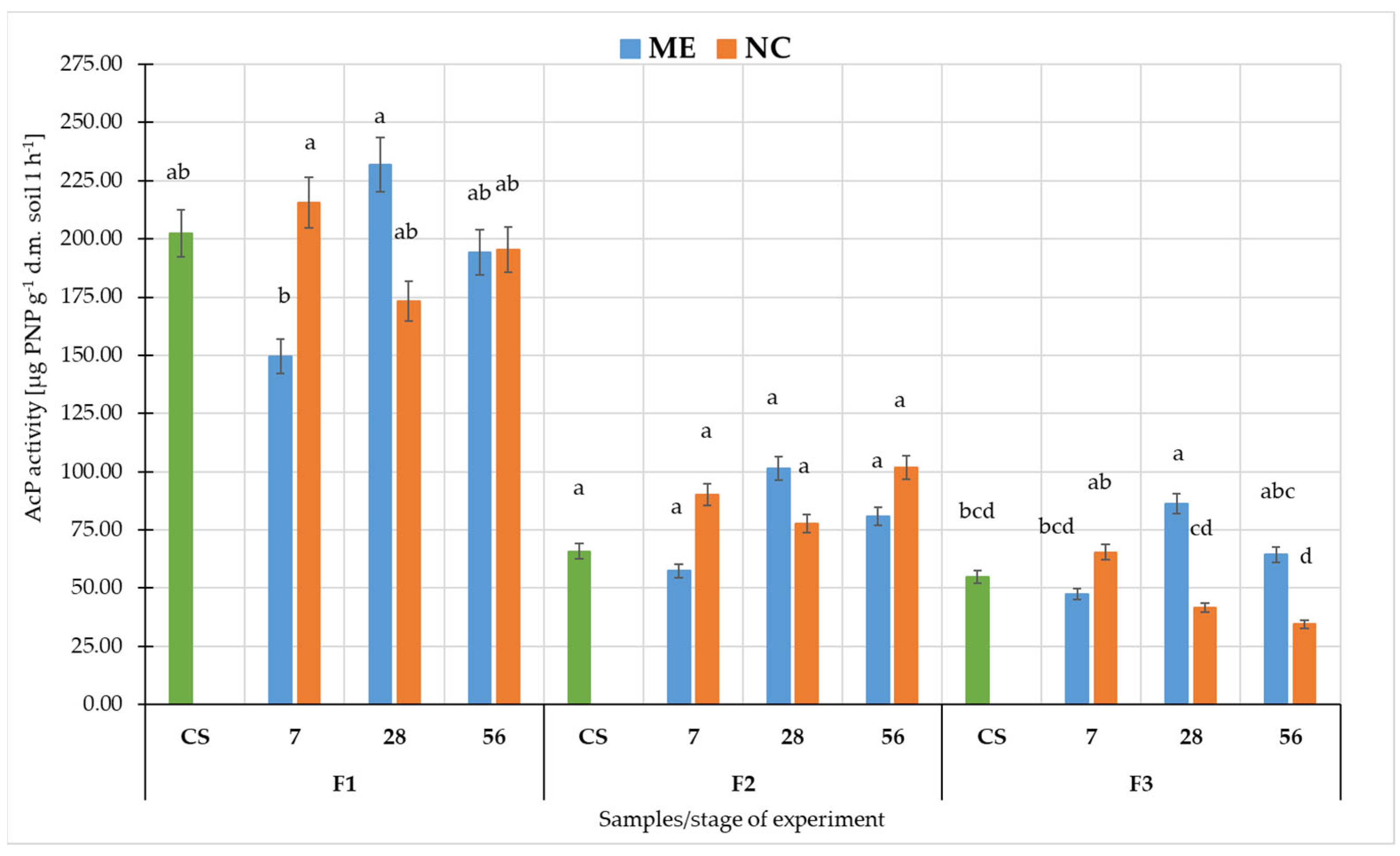
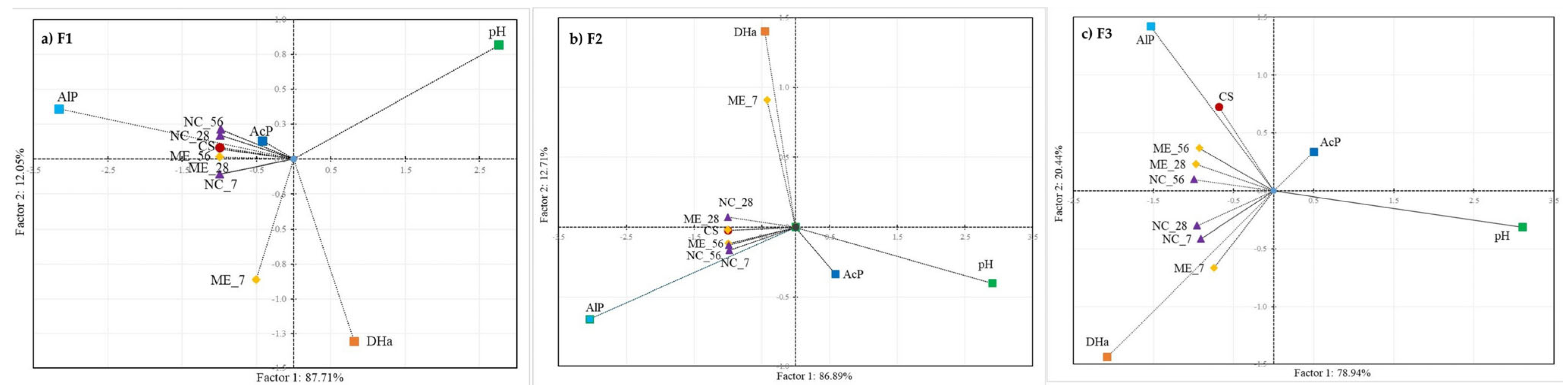
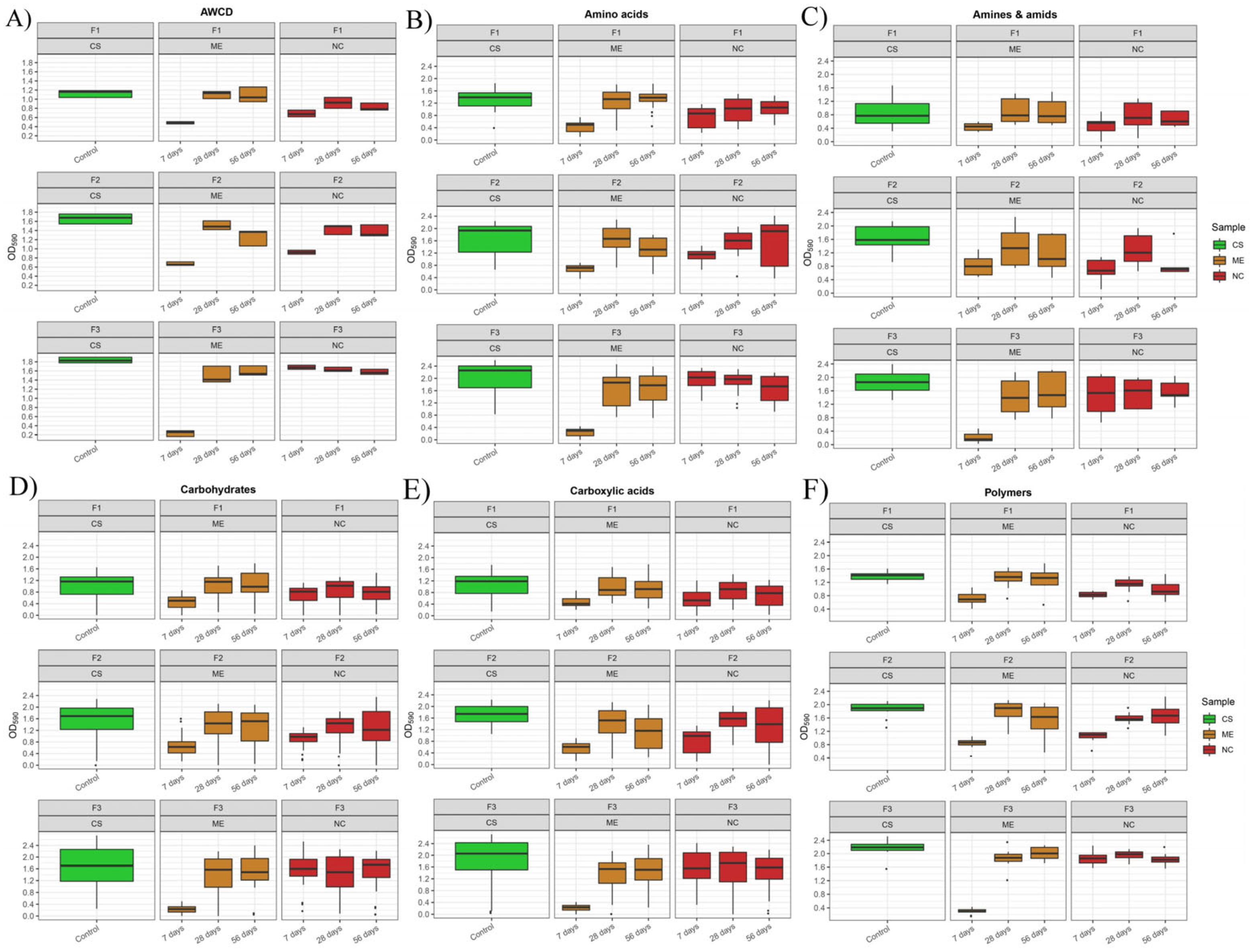
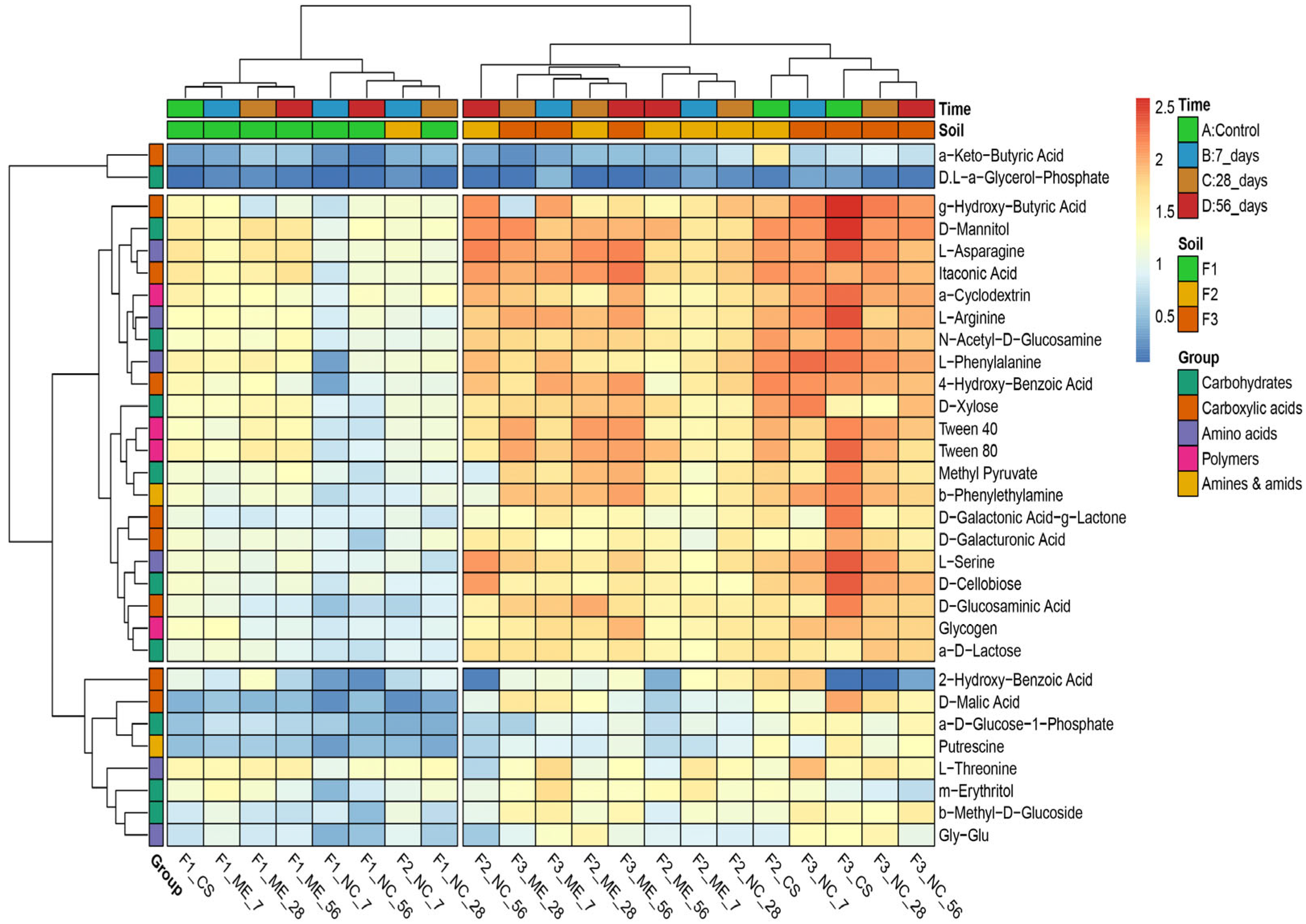
3. Results
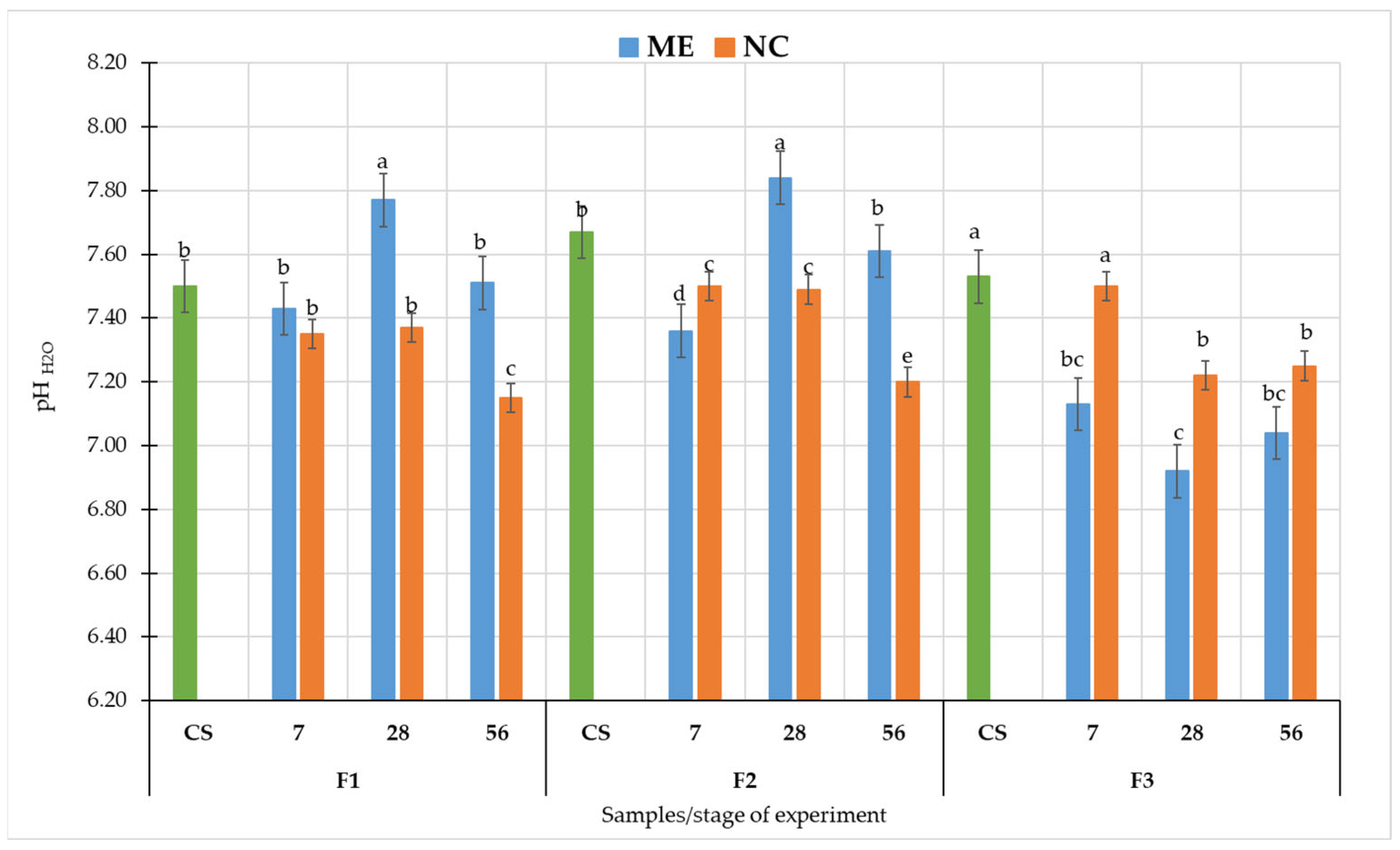
3.1. Effect of Simulated and Natural Flooding on Soil pH
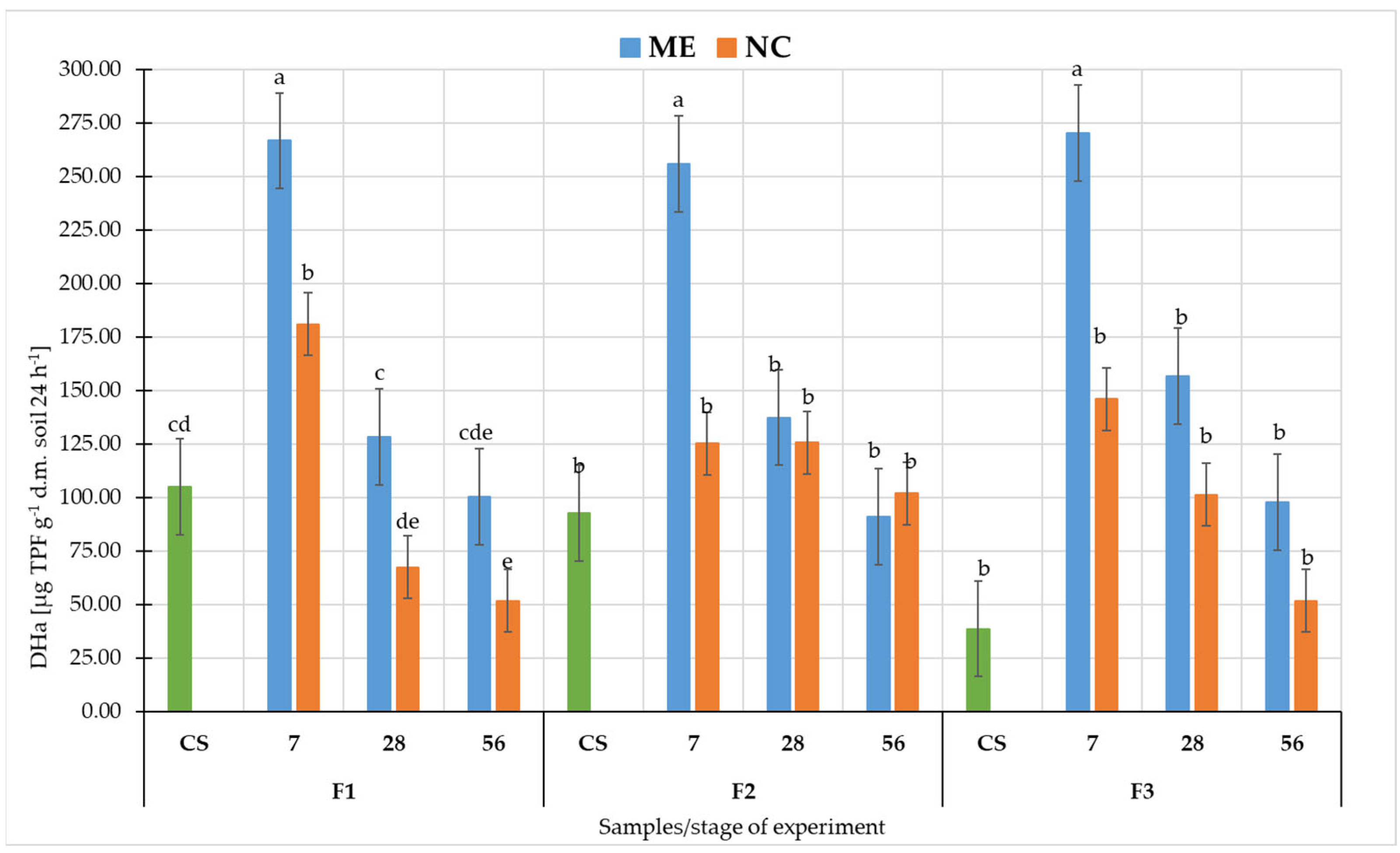
3.2. Metabolic Potential of Soil Microorganisms under Simulated and Natural Flooding
4. Discussion
4.1. Impact of Flooding on Soil Environment
4.2. Microcosm vs. Natural Condition
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- European Commission. Directive 2007/60/EC on the Assessment and Management of Flood Risks; Journal of the European Union Official: Brussels, Belgium, 2007. [Google Scholar]
- European Academies Science Advisory Council. Trends in Extreme Weather Events in Europe: Implications for National and European Union Adaptation strategies; German National Academy of Sciences Leopoldina: Halle, Germany, 2013; Volume 22, ISBN 978-3-8047-3239-1. [Google Scholar]
- Banach, A.M.; Banach, K.; Visser, E.J.W.; Stȩpniewska, Z.; Smits, A.J.M.; Roelofs, J.G.M.; Lamers, L.P.M. Effects of summer flooding on floodplain biogeochemistry in Poland; Implications for increased flooding frequency. Biogeochemistry 2009, 92, 247–262. [Google Scholar] [CrossRef]
- Blöschl, G.; Hall, J.; Parajka, J.; Perdigão, R.A.; Merz, B.; Arheimer, B.; Aronica, G.T.; Bilibashi, A.; Bonacci, O.; Borga, M.; et al. Changing climate shifts timing of European floods. Science 2017, 357, 588–590. [Google Scholar] [CrossRef]
- Vousdoukas, M.I.; Mentaschi, L.; Voukouvalas, E.; Verlaan, M.; Feyen, L. Extreme sea levels on the rise along Europe’s coasts. Earths Future 2017, 5, 304–323. [Google Scholar] [CrossRef]
- Mentaschi, L.; Vousdoukas, M.I.; Voukouvalas, E.; Dosio, A.; Feyen, L. Global changes of extreme coastal wave energy fluxes triggered by intensified teleconnection patterns. Geophys. Res. Lett. 2017, 44, 2416–2426. [Google Scholar] [CrossRef]
- Wolińska, A. Dehydrogenase activity of soil microorganisms and oxygen availability during reoxidation process of selected mineral soils from Poland. Acta Agrophys. Rozpr. Monogr. 2010, 3, 12–30. [Google Scholar]
- Banach, A.M.; Banach, K.; Ransen, R.H.M.P.; Jansen, R.H.M.; Visser, E.J.W.; Stȩpniewska, Z.; Roelofs, J.G.M.; Lamers, L.P.M. Effects of long-term flooding on biogeochemistry and vegetation development in floodplains; a mesocosm experiment to study interacting effects of land use and water quality. Biogeosciences 2009, 6, 1325–1339. [Google Scholar] [CrossRef]
- Young, I.; Ritz, K. Tillage, habitat space and function of soil microbes. Soil Tillage Res. 2000, 53, 201–213. [Google Scholar] [CrossRef]
- Gu, Y.; Wang, P.; Kong, C.H. Urease, invertase, dehydrogenase and polyphenoloxidase activities in paddy soil influenced by allelopathic rice variety. Eur. J. Soil Biol. 2009, 45, 436–441. [Google Scholar] [CrossRef]
- Mhuantong, W.; Wongwilaiwalin, S.; Laothanachareon, T.; Eurwilaichitr, L.; Tangphatsornruang, S.; Boonchayaanant, B.; Limpiyakorn, T.; Pattaragulwanit, K.; Punmatharith, T.; McEvoy, J.; et al. Survey of microbial diversity in flood areas during Thailand 2011 flood crisis using high-throughput tagged amplicon pyrosequencing. PLoS ONE 2015, 10, e0128043. [Google Scholar] [CrossRef]
- Unger, I.M.; Motavalli, P.P.; Muzika, R.-M. Changes in soil chemical properties with flooding: A field laboratory approach. Agric. Ecosyst. Environ. 2009, 131, 105–110. [Google Scholar] [CrossRef]
- Argiroff, W.A.; Zak, D.R.; Lanser, C.M.; Wiley, M.J. Microbial Community Functional Potential and Composition Are Shaped by Hydrologic Connectivity in Riverine Floodplain Soils. Microb. Ecol. 2017, 73, 630–644. [Google Scholar] [CrossRef] [PubMed]
- Maliszewska-Kordybach, B.; Klimkowicz-Pawlas, A.; Smreczak, B.; Gała̧zka, R. Effect of flooding on contamination of agricultural soils with metals and PAHs: The middle Vistula Gap case study. Water Air Soil Pollut. 2012, 223, 687–697. [Google Scholar] [CrossRef]
- Liu, B.; Li, Y.; Zhang, X.; Wang, J.; Gao, M. Effects of chlortetracycline on soil microbial communities: Comparisons of enzyme activities to the functional diversity via Biolog EcoPlatesTM. Eur. J. Soil Biol. 2015, 68, 69–76. [Google Scholar] [CrossRef]
- Floch, C.; Chevremont, A.C.; Joanico, K.; Capowiez, Y.; Criquet, S. Indicators of pesticide contamination: Soil enzyme compared to functional diversity of bacterial communities via Biolog® Ecoplates. Eur. J. Soil Biol. 2011, 47, 256–263. [Google Scholar] [CrossRef]
- Roeselers, G.; Zippel, B.; Staal, M.; Van Loosdrecht, M.; Muyzer, G. On the reproducibility of microcosm experiments—Different community composition in parallel phototrophic biofilm microcosms. FEMS Microbiol. Ecol. 2006, 58, 169–178. [Google Scholar] [CrossRef]
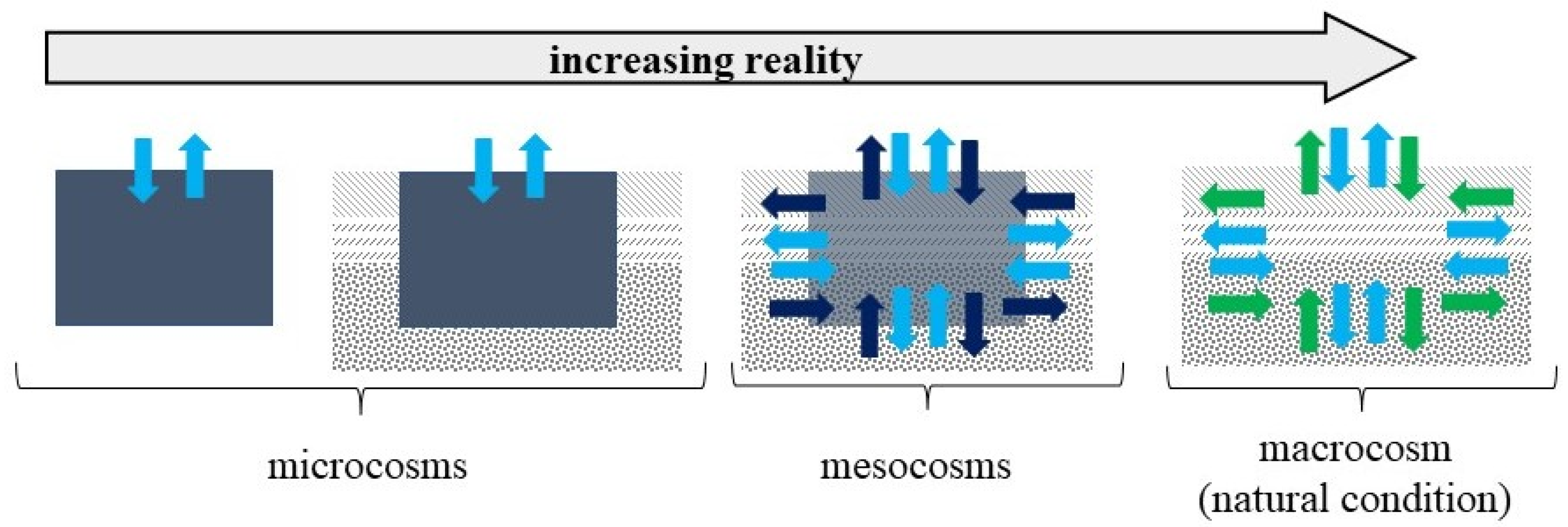
- Carpenter, S.R. Microcosm Experiments have Limited Relevance for Community and Ecosystem Ecology. Ecology 1996, 77, 677–680. [Google Scholar] [CrossRef]
- Kampichler, C.; Bruckner, A.; Kandeler, E. Use of enclosed model ecosystems in soil ecology: A bias towards laboratory research. Soil Biol. Biochem. 2001, 33, 269–275. [Google Scholar] [CrossRef]
- Beyers, R.J. The Microcosm Approach to Ecosystem Biology. Am. Biol. Teach. 1964, 26, 491–498. [Google Scholar] [CrossRef]
- Schindler, D.W. Replication versus Realism: The Need for Ecosystem-Scale Experiments on JSTOR. Ecosystems 1998, 1, 323–334. [Google Scholar] [CrossRef]
- Bergholz, P.W.; Strawn, L.K.; Ryan, G.T.; Warchocki, S.; Wiedmann, M. Spatiotemporal Analysis of Microbiological Contamination in New York State Produce Fields following Extensive Flooding from Hurricane Irene, August 2011. J. Food Prot. 2016, 79, 384–391. [Google Scholar] [CrossRef]
- Unger, I.M.; Kennedy, A.C.; Muzika, R.-M. Flooding effects on soil microbial communities. Appl. Soil Ecol. 2009, 42, 1–8. [Google Scholar] [CrossRef]
- Zasada, A.A.; Formińska, K.; Ogrodnik, A.; Gierczyński, R.; Jagielski, M. Screening for anthrax occurrence in soil of flooded rural areas in Poland after rainfalls in spring 2010. Ann. Agric. Environ. Med. 2014, 21, 460–463. [Google Scholar] [CrossRef][Green Version]
- Anjos, L.; Gaistardo, C.; Deckers, J.; Dondeyne, S.; Eberhardt, E.; Gerasimova, M.; Harms, B.; Jones, A.; Krasilnikov, P.; Reinsch, T.; et al. World Reference Base for Soil Resources 2014. International Soil Classification System for Naming Soils and Creating Legends for Soil Maps; Schad, P., Van Huyssteen, C., Micheli, E., Eds.; FAO: Rome, Italy, 2014; ISBN 9789251083697. [Google Scholar]
- Furtak, K.; Grządziel, J.; Gałązka, A.; Niedźwiecki, J. Analysis of soil properties, bacterial community composition, and metabolic diversity in fluvisols of a floodplain area. Sustainability 2019, 11, 3929. [Google Scholar] [CrossRef]
- Furtak, K.; Grządziel, J.; Gałązka, A.; Niedźwiecki, J. Prevalence of unclassified bacteria in the soil bacterial community from floodplain meadows (fluvisols) under simulated flood conditions revealed by a metataxonomic approachss. Catena 2020, 188, 104448. [Google Scholar] [CrossRef]
- The Institute of Meteorology and Water Management—State Research Institute Internetowy Serwis Pogodowy IMGW-PIB. Available online: https://danepubliczne.imgw.pl/data/dane_pomiarowo_obserwacyjne/dane_hydrologiczne/miesieczne/2019/ (accessed on 6 January 2020).
- Kabała, C.; Musztyfaga, E.; Gałka, B.; Łabuńska, D.; Mańczyńska, P. Conversion of soil pH 1:2.5 KCl and 1:2.5 H2O to 1:5 H2O: Conclusions for soil management, environmental monitoring, and international soil databases. Pol. J. Environ. Stud. 2016, 25, 647–653. [Google Scholar] [CrossRef]
- Reynolds, S.G. The gravimetric method of soil moisture determination Part I A study of equipment, and methodological problems. J. Hydrol. 1970, 11, 258–273. [Google Scholar] [CrossRef]
- Casida, L.; Klein, D.; Santoro, T. Soil dehydrogenase activity. Soil Sci. 1964, 98, 371–376. [Google Scholar] [CrossRef]
- Tabatabai, M.A. Soil enzymes. In Methods of Soil Analysis; Page, A.L., Miller, R.H., Keeney, D.R., Eds.; American Society of Agronomy Inc. and Soil Science Society of America Inc.: Madison, WI, USA, 1982; pp. 903–948. [Google Scholar]
- Frąc, M.; Oszust, K.; Lipiec, J. Community Level Physiological Profiles (CLPP), Characterization and Microbial Activity of Soil Amended with Dairy Sewage Sludge. Sensors 2012, 12, 3253. [Google Scholar] [CrossRef]
- Gryta, A.; Frąc, M.; Oszust, K. The Application of the Biolog EcoPlate Approach in Ecotoxicological Evaluation of Dairy Sewage Sludge. Appl. Biochem. Biotechnol. 2014, 174, 1434–1443. [Google Scholar] [CrossRef]
- Weber, K.P.; Legge, R.L. One-dimensional metric for tracking bacterial community divergence using sole carbon source utilization patterns. J. Microbiol. Methods 2009, 79, 55–61. [Google Scholar] [CrossRef]
- Insam, H.; Goberna, M. Use of Biolog® for the Community Level Physiological Profiling (CLPP) of environmental samples. In Molecular Microbial Ecology Manual; Kowalchuk, G.A., de Brujin, F.J., Head, I.M., Akkermans, A.D., van Elsas, J.D., Eds.; Kluwer Academic Publishers: Dordrecht, The Netherlands, 2004; pp. 853–860. [Google Scholar]
- Garland, J.L.; Mills, A.L. Classification and characterization of heterotrophic microbial communities on the basis of patterns of community-level sole-carbon-source utilization. Appl. Environ. Microbiol. 1991, 57, 2351–2359. [Google Scholar] [CrossRef] [PubMed]
- Zak, J.C.; Willig, M.R.; Moorhead, D.L.; Wildman, H.G. Functional diversity of microbial communities: A quantitative approach. Soil Biol. Biochem. 1994, 26, 1101–1108. [Google Scholar] [CrossRef]
- Cartwright, J.M. Hydrologic and Soil Data Collected in Limestone Cedar Glades at Stones River National Battlefield, Tennessee; U.S. Geological Survey: Washington, DC, USA, 2015. [Google Scholar] [CrossRef]
- Gomez, E.; Garland, J.; Conti, M. Reproducibility in the response of soil bacterial community-level physiological profiles from a land use intensification gradient. Appl. Soil Ecol. 2004, 26, 21–30. [Google Scholar] [CrossRef]
- R Core Team. R: A Language and Environment for Statistical Computing; R Core Team: Vienna, Austria, 2019; Available online: https://www.R-project.org/ (accessed on 2 January 2022).
- R Core Team. Raivo Kolde Pheatmap: Pretty Heatmaps. R Package Version 1.0.12; R Core Team: Vienna, Austria, 2019. [Google Scholar]
- Ginestet, C. ggplot2: Elegant Graphics for Data Analysis. J. R. Stat. Soc. Ser. A Stat. Soc. 2011, 174, 245–246. [Google Scholar] [CrossRef]
- Jollife, I.T.; Cadima, J. Principal component analysis: A review and recent developments. Philos. Trans. R. Soc. A Math. Phys. Eng. Sci. 2016, 374, 20150202. [Google Scholar] [CrossRef]
- Ponnamperuma, F.N. Effects of flooding on soils. In Flooding and Plant Growth, Physiological Ecology; Kozlowski, T.T., Ed.; Academic Press, Elsevier Inc.: Cambridge, MA, USA, 1984; pp. 9–45. ISBN 9780124241206. [Google Scholar]
- Włodarczyk, T.; Szarlip, P.; Brzezińska, M.; Kotowska, U. Redox potential, nitrate content and pH in flooded Eutric Cambisol during nitrate reduction. Res. Agric. Eng. 2007, 53, 20–28. [Google Scholar] [CrossRef]
- Liu, Y.; Cheng, J.; Liu, X.; Zhong, H.; Wang, B.; Kong, Z.; Wu, L. Tracking the changes of wetland soil bacterial community and metabolic potentials under drought and flooding conditions in experimental microcosms. J. Soils Sediments 2021, 21, 2404–2417. [Google Scholar] [CrossRef]
- Bednarek, W.; Dresler, S.; Tkaczyk, P.; Hanaka, A. Physicochemical properties of surface soil layer after the flood in the middle Vistula River valley. J. Elemntol. 2014, 19, 17–29. [Google Scholar] [CrossRef]
- Sessitsch, A.; Weilharter, A.; Gerzabek, M.H.; Kirchmann, H.; Kandeler, E. Microbial population structures in soil particle size fractions of a long-term fertilizer field experiment. Appl. Environ. Microbiol. 2001, 67, 4215–4224. [Google Scholar] [CrossRef]
- Grządziel, J.; Gałązka, A. Microplot long-term experiment reveals strong soil type influence on bacteria composition and its functional diversity. Appl. Soil Ecol. 2018, 124, 117–123. [Google Scholar] [CrossRef]
- Furtak, K.; Gałązka, A. Edaphic factors and their influence on microbiological biodiversity of the soil environment. Postępy Mikrobiol. Adv. Microbiol. 2019, 58, 375–384. [Google Scholar] [CrossRef]
- Bastida, F.; Kandeler, E.; Moreno, J.L.; Ros, M.; García, C.; Hernández, T. Application of fresh and composted organic wastes modifies structure, size and activity of soil microbial community under semiarid climate. Appl. Soil Ecol. 2008, 40, 318–329. [Google Scholar] [CrossRef]
- Mocek-Płóciniak, A. Wykorzystanie aktywności enzymatycznej do oceny wpływu antropogenicznych zmian wywołanych przez metale ciężkie w środowisku glebowym. Nauk. Przyr. Technol. 2010, 4, 86. [Google Scholar]
- Yan, N.; Marschner, P.; Cao, W.; Zuo, C.; Qin, W. Influence of salinity and water content on soil microorganisms. Int. Soil Water Conserv. Res. 2015, 3, 316–323. [Google Scholar] [CrossRef]
- Weaver, M.A.; Zablotowicz, R.M.; Krutz, L.J.; Bryson, C.T.; Locke, M.A. Microbial and vegetative changes associated with development of a constructed wetland. Ecol. Indic. 2012, 13, 37–45. [Google Scholar] [CrossRef]
- Subhani, A.; Changyong, H.; Zhebgmiao, X.; Min, L.; El-ghamry, A.M. Impact of soil environment and agronomic practices on microbial/dehydrogenase enzyme activity in soil. Pak. J. Biol. Sci. 2001, 4, 333–338. [Google Scholar] [CrossRef]
- Wolinska, A.; Stepniewsk, Z. Dehydrogenase Activity in the Soil Environment. In Dehydrogenases; Canuto, R.A., Ed.; BoD—Books on Demand: Norderstedt, Germany, 2012; pp. 183–210. ISBN 9789533070193. [Google Scholar]
- Bielinska, E.J. Oznaczenie Aktywnosci Fosfataz. Acta Agrophys. Rozpr. I Monogr. 2005, 2005, 63–74. [Google Scholar]
- Banerjee, A.; Sanyal, S.; Sen, S. Soil phosphatase activity of agricultural land: A possible index of soil fertility. Agric. Sci. Res. J. 2012, 2, 412–419. [Google Scholar]
- Krämer, S.; Green, D.M. Acid and alkaline phosphatase dynamics and their relationship to soil microclimate in a semiarid woodland. Soil Biol. Biochem. 2000, 32, 179–188. [Google Scholar] [CrossRef]
- Furtak, K.; Gałązka, A. Enzymatic activity as a popular parameter used to determine the quality of the soil environment. Pol. J. Agron. 2019, 37, 22–30. [Google Scholar] [CrossRef]
- Sardans, J.; Peñuelas, J. Drought decreases soil enzyme activity in a Mediterranean Quercus ilex L. forest. Soil Biol. Biochem. 2005, 37, 455–461. [Google Scholar] [CrossRef]
- Schnecker, J.; Meeden, D.B.; Calderon, F.; Cavigelli, M.; Lehman, R.M.; Tiemann, L.K.; Grandy, A.S. Microbial activity responses to water stress in agricultural soils from simple and complex crop rotations. SOIL 2021, 7, 547–561. [Google Scholar] [CrossRef]
- Cardozo Junior, F.M.; Carneiro, R.F.V.; Rocha, S.M.B.; Nunes, L.A.P.L.; dos Santos, V.M.; de Lima Feitoza, L.; de Araújo, A.S.F. The Impact of Pasture Systems on Soil Microbial Biomass and Community-level Physiological Profiles. Land Degrad. Dev. 2018, 29, 284–291. [Google Scholar] [CrossRef]
- Bhattacharyya, P.; Roy, K.S.; Neogi, S.; Adhya, T.K.; Rao, K.S.; Manna, M.C. Effects of rice straw and nitrogen fertilization on greenhouse gas emissions and carbon storage in tropical flooded soil planted with rice. Soil Tillage Res. 2012, 124, 119–130. [Google Scholar] [CrossRef]
- Dinter, T.; Geihser, S.; Gube, M.; Daniel, R.; Kuzyakov, Y. Impact of sea level change on coastal soil organic matter, priming effects and prokaryotic community assembly. FEMS Microbiol. Ecol. 2019, 95, fiz129. [Google Scholar] [CrossRef]
- Oest, A.; Alsaffar, A.; Fenner, M.; Azzopardi, D.; Tiquia-Arashiro, S.M. Patterns of change in metabolic capabilities of sediment microbial communities in river and lake ecosystems. Int. J. Microbiol. 2018, 2018, 6234931. [Google Scholar] [CrossRef]
- Borowik, A.; Wyszkowska, J. Soil moisture as a factor affecting the microbiological and biochemical activity of soil. Plant Soil Environ. 2016, 62, 250–255. [Google Scholar] [CrossRef]
- Gliński, J.; Stępniewska, Z.; Kasiak, A. Changes of an enzymatic activity in soils with respect to their water content and oxygen status. Rocz. Glebozn. 1983, 34, 53–59. [Google Scholar]
- Wagner, D.; Eisenhauer, N.; Cesarz, S. Plant species richness does not attenuate responses of soil microbial and nematode communities to a flood event. Soil Biol. Biochem. 2015, 89, 135–149. [Google Scholar] [CrossRef]
- Fierer, N.; Schmiel, J.P. A proposed mechanism for the pulse in carbon dioxide production commonly observed following the rapid rewetting of a dry soil. Soil Sci. Soc. Am. J. 2003, 67, 798–805. [Google Scholar] [CrossRef]
- Kołwzan, B.; Adamiak, W.; Grabas, K.; Pawełczyk, A. Basics of Microbiology in Environmental Protection, 2nd ed.; Oficyna Wydawnicza Politechniki Wrocławskiej: Wrocław, Poland, 2006. [Google Scholar]
- Libudzisz, Z.; Kowal, K.; Żakowska, Z. Technical Microbiology, Volume 1: Microorganisms and Their Environment of Occurrence; Wydawnictwo Naukowe PWN: Warszawa, Poland, 2007. [Google Scholar]
- Dassonville, F.; Renault, P. Interactions between microbial processes and geochemical transformations under anaerobic conditions: A review. Agronomie 2002, 22, 51–68. [Google Scholar] [CrossRef]
- An, Q.; Cheng, J.R.; Wang, Y.T.; Zhu, M.J. Performance and energy recovery of single and two stage biogas production from paper sludge: Clostridium thermocellum augmentation and microbial community analysis. Renew. Energy 2020, 148, 214–222. [Google Scholar] [CrossRef]
- Datta, S.; Saha, D.; Chattopadhyay, L.; Majumdar, B. Genome Comparison Identifies Different Bacillus Species in a Bast Fibre-Retting Bacterial Consortium and Provides Insights into Pectin Degrading Genes. Sci. Rep. 2020, 10, 8169. [Google Scholar] [CrossRef]
- Neumann, A.P.; Suen, G. The Phylogenomic Diversity of Herbivore-Associated Fibrobacter spp. is Correlated to Lignocellulose-Degrading Potential. mSphere 2018, 3, e00593-18. [Google Scholar] [CrossRef] [PubMed]
- Eller, G.; Krüger, M.; Frenzel, P. Comparing field and microcosm experiments: A case study on methano- and methylo-trophic bacteria in paddy soil. FEMS Microbiol. Ecol. 2005, 51, 279–291. [Google Scholar] [CrossRef]
- Wynn, G.; Paradise, C.J. Effects of microcosm scaling and food resources on growth and survival of larval Culex pipiens. BMC Ecol. 2001, 1, 3. [Google Scholar] [CrossRef]
- Turner, B. Soil as an Archetype of Complexity: A Systems Approach to Improve Insights, Learning, and Management of Coupled Biogeochemical Processes and Environmental Externalities. Soil Syst. 2021, 5, 39. [Google Scholar] [CrossRef]
- Al-Kaisi, M.M.; Lal, R.; Olson, K.R.; Lowery, B. Fundamentals and Functions of Soil Environment. In Soil Health and Intensification of Agroecosystems; Elsevier Inc.: Amsterdam, The Netherlands, 2017; pp. 1–23. ISBN 9780128054017. [Google Scholar]
- Kumar, S.; Dutta, V. Constructed wetland microcosms as sustainable technology for domestic wastewater treatment: An overview. Environ. Sci. Pollut. Res. 2019, 26, 11662–11673. [Google Scholar] [CrossRef]
- Heck, K.; Coltman, E.; Schneider, J.; Helmig, R. Influence of Radiation on Evaporation Rates: A Numerical Analysis. Water Resour. Res. 2020, 56, e2020WR027332. [Google Scholar] [CrossRef]
- Palta, J.A.; Chen, X.; Milroy, S.P.; Rebetzke, G.J.; Dreccer, M.F.; Watt, M. Large root systems: Are they useful in adapting wheat to dry environments? Funct. Plant Biol. 2011, 38, 347. [Google Scholar] [CrossRef]
- Albiach, R.; Canet, R.; Pomares, F.; Ingelmo, F. Microbial biomass content and enzymatic activities after the application of organic amendments to a horticultural soil. Bioresour. Technol. 2000, 75, 43–48. [Google Scholar] [CrossRef]
- Polyak, Y.M.; Bakina, L.G.; Chugunova, M.V.; Mayachkina, N.V.; Gerasimov, A.O.; Bure, V.M. Effect of remediation strategies on biological activity of oil-contaminated soil—A field study. Int. Biodeterior. Biodegrad. 2018, 126, 57–68. [Google Scholar] [CrossRef]
- Balota, E.L.; Chaves, J.C.D. Enzymatic activity and mineralization of carbon and nitrogen in soil cultivated with coffee and green manures. Rev. Bras. Ciênc. Solo 2010, 34, 1573–1583. [Google Scholar] [CrossRef]
- Cayuela, M.L.; Sinicco, T.; Mondini, C. Mineralization dynamics and biochemical properties during initial decomposition of plant and animal residues in soil. Appl. Soil Ecol. 2009, 41, 118–127. [Google Scholar] [CrossRef]
- Mundra, S.; Kjønaas, O.J.; Morgado, L.N.; Krabberød, A.K.; Ransedokken, Y.; Kauserud, H. Soil depth matters: Shift in composition and inter-kingdom co-occurrence patterns of microorganisms in forest soils. FEMS Microbiol. Ecol. 2021, 97, fiab022. [Google Scholar] [CrossRef] [PubMed]
- Spohn, M.; Klaus, K.; Wanek, W.; Richter, A. Microbial carbon use efficiency and biomass turnover times depending on soil depth—Implications for carbon cycling. Soil Biol. Biochem. 2016, 96, 74–81. [Google Scholar] [CrossRef]
- Liu, L.; Zhu, K.; Wurzburger, N.; Zhang, J. Relationships between plant diversity and soil microbial diversity vary across taxonomic groups and spatial scales. Ecosphere 2020, 11, e02999. [Google Scholar] [CrossRef]
- Hovatter, S.R.; Dejelo, C.; Case, A.L.; Blackwood, C.B. Metacommunity organization of soil microorganisms depends on habitat defined by presence of Lobelia siphilitica plants. Ecology 2011, 92, 57–65. [Google Scholar] [CrossRef]
- Höhener, P.; Dakhel, N.; Christophersen, M.; Broholm, M.; Kjeldsen, P. Biodegradation of hydrocarbons vapors: Comparison of laboratory studies and field investigations in the vadose zone at the emplaced fuel source experiment, Airbase Værløse, Denmark. J. Contam. Hydrol. 2006, 88, 337–358. [Google Scholar] [CrossRef]
- Luckinbill, L.S. Regulation, Stability, and Diversity in A Model Experimental Microcosm. Ecology 1979, 60, 1098. [Google Scholar] [CrossRef]
- Ranjard, L.; Echairi, A.; Nowak, V.; Lejon, D.P.H.; Nouaïm, R.; Chaussod, R. Field and microcosm experiments to evaluate the effects of agricultural Cu treatment on the density and genetic structure of microbial communities in two different soils. FEMS Microbiol. Ecol. 2006, 58, 303–315. [Google Scholar] [CrossRef] [PubMed]
- Renella, G.; Chaudri, A.M.; Brookes, P.C. Fresh additions of heavy metals do not model long-term effects on microbial biomass and activity. Soil Biol. Biochem. 2002, 34, 121–124. [Google Scholar] [CrossRef]
- Teuben, A.; Verhoef, H.A. Relevance of micro- and mesocosm experiments for studying soil ecosystem processes. Soil Biol. Biochem. 1992, 24, 1179–1183. [Google Scholar] [CrossRef]
- Burrows, L.A.; Edwards, C.A. The Use of Integrated Soil Microcosms to Assess the Impact of Carbendazim on Soil Ecosystems. Ecotoxicology 2004, 13, 143–161. [Google Scholar] [CrossRef]
- Tolle, D.A.; Arthur, M.F.; Van Voris, P. Microcosm/field comparison of trace element uptake in crops grown in fly ash-amended soil. Sci. Total Environ. 1983, 31, 243–261. [Google Scholar] [CrossRef]
- Benton, T.G.; Solan, M.; Travis, J.M.J.; Sait, S.M. Microcosm experiments can inform global ecological problems. Trends Ecol. Evol. 2007, 22, 516–521. [Google Scholar] [CrossRef]



| Abbreviation | Fluvisol | Location | Condition | Day of Flooding | Day of Drying |
|---|---|---|---|---|---|
| CS | F1 | Wojszyn; 51°20′03.4″ N 21°56′43.2″ E | Control sample 1 | - | - |
| ME | Microcosm experiment | 7 | 28, 56 | ||
| NC | Natural condition | 7 | 28, 56 | ||
| CS | F2 | Janowiec; 51°19′29.9″ N 21°55′19.2″ E | Control sample 1 | - | - |
| ME | Microcosm experiment | 7 | 28, 56 | ||
| NC | Natural condition | 7 | 28, 56 | ||
| CS | F3 | Janowiec; 51°19′14.4″ N 21°54′42.9″ E | Control sample 1 | - | - |
| ME | Microcosm experiment | 7 | 28, 56 | ||
| NC | Natural condition | 7 | 28, 56 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Furtak, K.; Grządziel, J.; Gałązka, A. Can Model Experiments Give Insight into the Response of the Soil Environment to Flooding? A Comparison of Microcosm and Natural Event. Biology 2022, 11, 386. https://doi.org/10.3390/biology11030386
Furtak K, Grządziel J, Gałązka A. Can Model Experiments Give Insight into the Response of the Soil Environment to Flooding? A Comparison of Microcosm and Natural Event. Biology. 2022; 11(3):386. https://doi.org/10.3390/biology11030386
Chicago/Turabian StyleFurtak, Karolina, Jarosław Grządziel, and Anna Gałązka. 2022. "Can Model Experiments Give Insight into the Response of the Soil Environment to Flooding? A Comparison of Microcosm and Natural Event" Biology 11, no. 3: 386. https://doi.org/10.3390/biology11030386
APA StyleFurtak, K., Grządziel, J., & Gałązka, A. (2022). Can Model Experiments Give Insight into the Response of the Soil Environment to Flooding? A Comparison of Microcosm and Natural Event. Biology, 11(3), 386. https://doi.org/10.3390/biology11030386








