Insights into the Divergence of Chinese Ips Bark Beetles during Evolutionary Adaptation
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Specimen Collection and DNA Extraction
2.2. Mitogenome De Novo Sequencing and Assembly
2.3. Phylogenetic Analysis
2.4. Estimation of Divergence Time
2.5. Comparative Mitogenome Analyses of Bark Beetles
2.6. Genetic Distance, Selection Pressure and Isolation by Distance (IBD) Analysis
2.7. SNP Sequencing and Analysis
3. Results
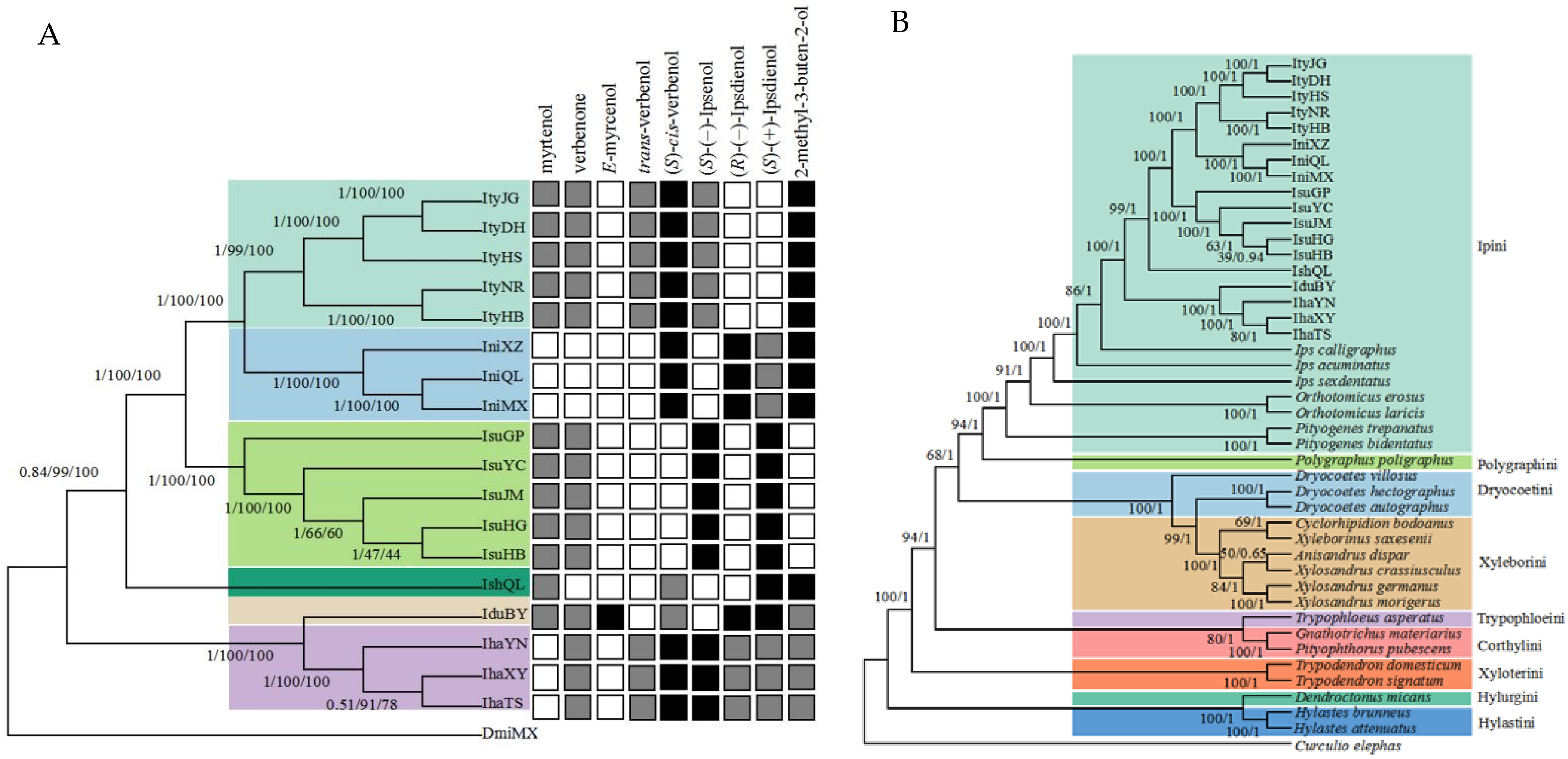
3.1. Phylogenetic Relationships, Divergence Times, and Pheromone Differences in Ips Bark Beetles
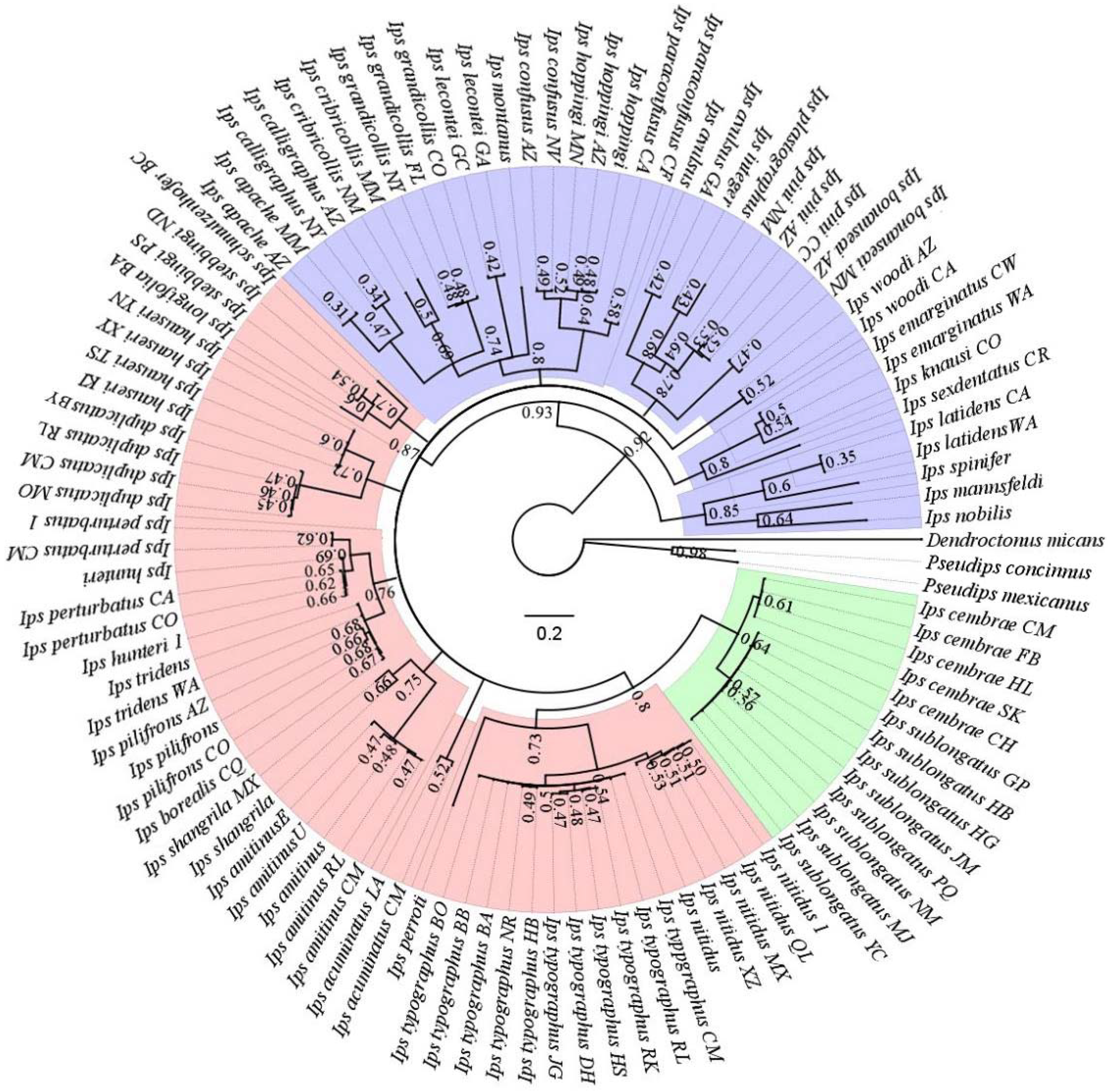
3.2. Phylogenetic Relationships among Ips Bark Beetles Worldwide
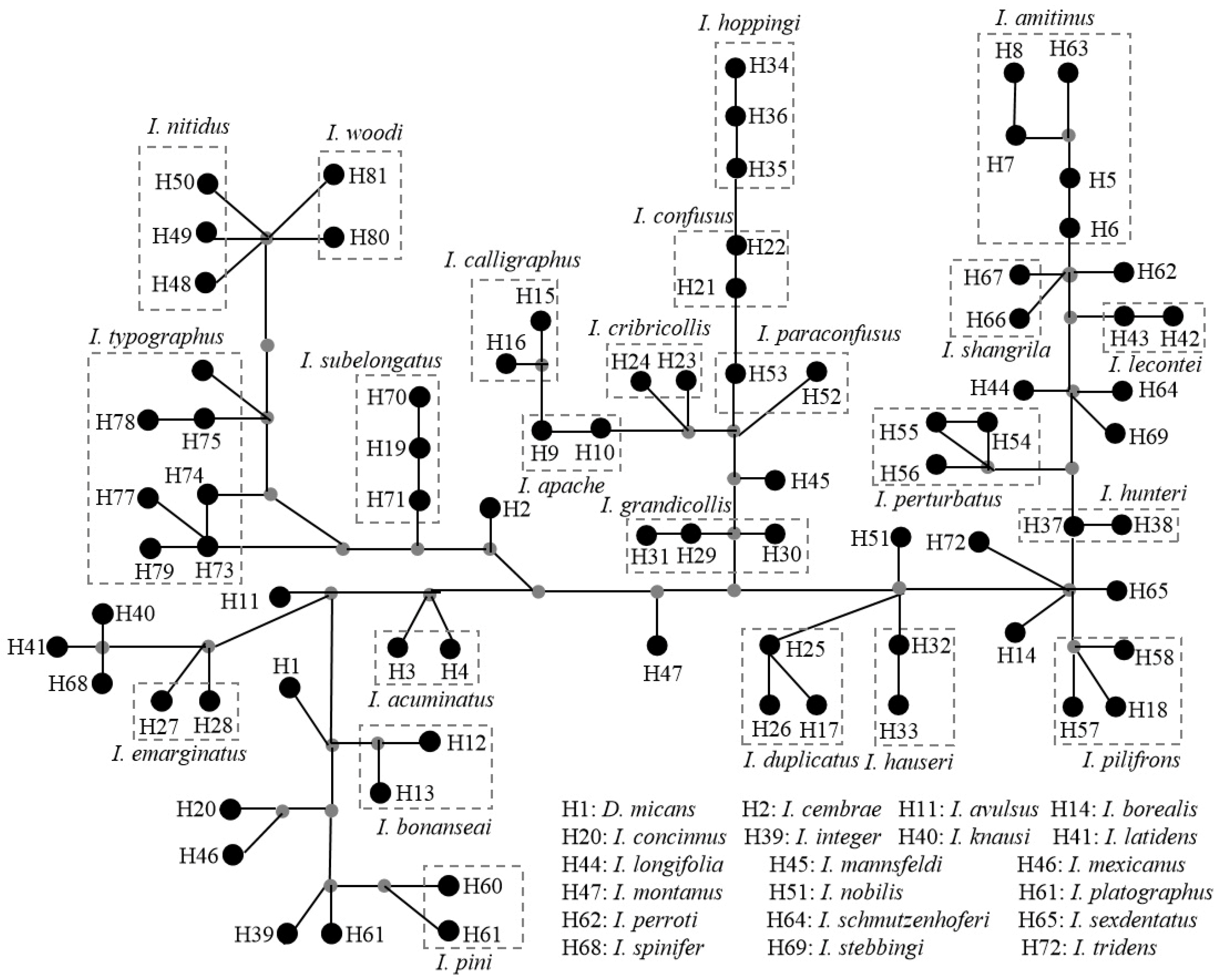
3.3. Analysis of Selection Pressure, Genetic Distance, and IBD Test
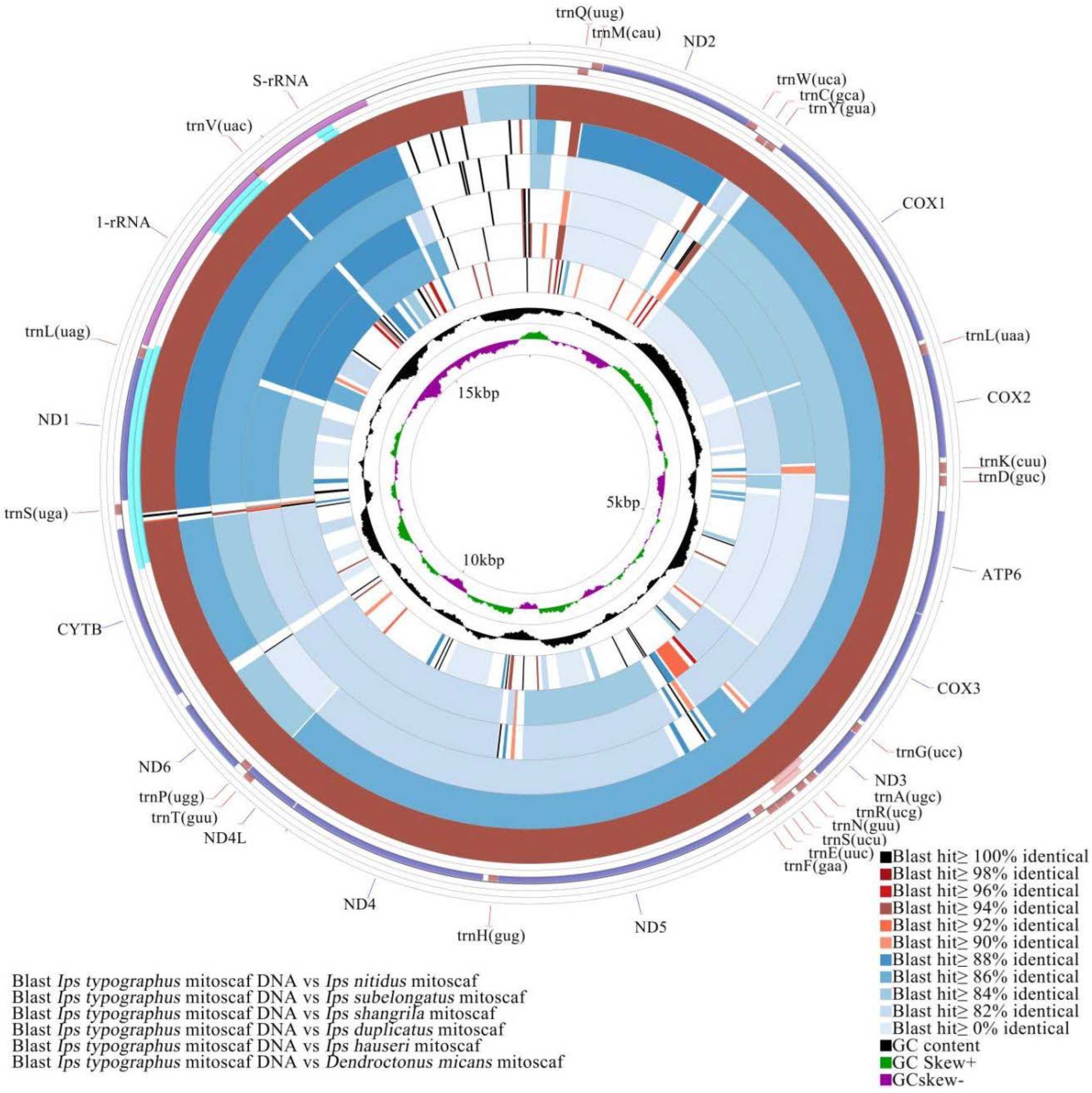
3.4. Comparative Mitochondrial Genome Analyses of Ips Bark Beetles
4. Discussion
4.1. Phylogenetic Relationships of Ips Species in Relation to Their Biological Characteristics
4.2. Adaptation of Ips Bark Beetles to a Complex Olfactory Environment
4.3. Evolutionary Lineages of the Ips Species in Relation to the Geographic Environment
4.4. Adaptive Evolution Inferred from Comparative Mitochondrial Genome ANALYSIS
4.5. Phylogenetic Status of Ips Species from China Based on COI Gene Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Fang, J.X.; Liu, M.; Zhang, S.F.; Liu, F.; Zhang, Z.; Zhang, Q.H.; Kong, X.B. Chemical signal interactions of the bark beetle with fungal symbionts, and host/non-host trees. J. Exp. Bot. 2020, 71, 6084–6091. [Google Scholar] [CrossRef] [PubMed]
- Byers, J.A. Bark beetle conversion of a plant compound to a sex-specific inhibitor of pheromone attraction. Science 1983, 220, 624–626. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fang, J.X.; Zhang, S.F.; Liu, F.; Zhang, Z.; Cheng, B.; Zhang, Q.H.; Kong, X.B. Functional investigation of monoterpenes for improved understanding of the relationship between hosts and bark beetles. J. Appl. Entomol. 2021, 145, 303–311. [Google Scholar] [CrossRef]
- Rosenberger, D.W.; Venette, R.C.; Aukema, B.H. Development of an aggressive bark beetle on novel hosts: Implications for outbreaks in an invaded range. J. Appl. Ecol. 2018, 55, 1526–1537. [Google Scholar] [CrossRef]
- Therrien, J.; Mason, C.J.; Cale, J.A.; Adams, A.; Aukema, B.H.; Currie, C.R.; Raffa, K.F.; Erbilgin, N. Bacteria influence mountain pine beetle brood development through interactions with symbiotic and antagonistic fungi: Implications for climate-driven host range expansion. Oecologia 2015, 179, 467–485. [Google Scholar] [CrossRef]
- Bracewell, R.R.; Vanderpool, D.; Good, J.M.; Six, D.L. Cascading speciation among mutualists and antagonists in a tree–beetle–fungi interaction. Proc. R. Soc. B-Biol. Sci. 2018, 285, 20180694. [Google Scholar] [CrossRef]
- Raffa, K.F.; Aukema, B.H.; Bentz, B.J.; Carroll, A.L.; Hicke, J.A.; Turner, M.G.; Romme, W.H. Cross-scale drivers of natural disturbances prone to anthropogenic amplification: The dynamics of bark beetle eruptions. BioScience 2008, 58, 501–517. [Google Scholar] [CrossRef] [Green Version]
- Biedermann, P.H.W.; Müller, J.; Grégoire, J.C.; Gruppe, A.; Hagge, J.; Hammerbacher, A.; Hofstetter, R.W.; Kandasamy, D.; Kolarik, M.; Kostovcik, M.; et al. Bark beetle population dynamics in the Anthropocene: Challenges and solution. Trends Ecol. Evol. 2019, 34, 914–924. [Google Scholar] [CrossRef] [Green Version]
- Cognato, A.I.; Felix, A. Phylogeny of Ips DeGeer species (Coleoptera: Scolytidae) inferred from Mitochondrial Cytochrome Oxidase I DNA sequence. Mol. Phylogenet. Evol. 2000, 14, 445–460. [Google Scholar] [CrossRef]
- Cognato, A.I.; Seybold, S.J.; Sperling, F.A.H. Incomplete barriers to mitochondrial gene flow between pheromone races of the North American pine engraver, Ips pini (Say) (Coleoptera: Scolytidae). Proc. R. Soc. B-Biol. Sci. 1999, 266, 1843–1850. [Google Scholar] [CrossRef] [Green Version]
- Cognato, A.I. Phylogenetic analysis reveals new genus of Ipini bark beetle (Scolytidae). Ann. Entomol. Soc. Am. 2000, 93, 362–366. [Google Scholar] [CrossRef]
- Bertheau, C.; Schuler, H.; Arthofer, W.; Avtzis, D.N.; Mayer, F.; Krumböck, S.; Moodley, Y.; Stauffer, C. Divergent evolutionary histories of two sympatric spruce bark beetle species. Mol. Ecol. 2013, 22, 3318–3332. [Google Scholar] [CrossRef] [PubMed]
- Stauffer, C.; Lakatos, F.; Hewitt, G.M. The phylogenetic relationships of seven European Ips (Scolytidae, Ipinae) species. Insect Mol. Biol. 1997, 6, 233–240. [Google Scholar] [CrossRef]
- Lin, C.P.; Danforth, B.N. How do insect nuclear and mitochondrial gene substitution patterns differ? Insights from Bayesian analyses of combined datasets. Mol. Phylogenet. Evol. 2004, 30, 686–702. [Google Scholar] [CrossRef]
- Gissi, C.; Iannelli, F.; Pesole, G. Evolution of the mitochondrial genome of Metazoa as exemplified by comparison of congeneric species. Heredity 2008, 101, 301–320. [Google Scholar] [CrossRef] [Green Version]
- Mayer, F.; Piel, F.B.; Cassel-Lundhagen, A.; Kirichenko, N.; Grumiau, L.; Økland, B.; Bertheau, C.; Grégoire, J.C.; Mardulyn, P. Comparative multi-locus phylogeography of two Palaearctic spruce bark beetles: Influence of contrasting ecological strategies on genetic variation. Mol. Ecol. 2015, 24, 1292–1310. [Google Scholar] [CrossRef]
- Du, H.C.; Fang, J.X.; Shi, X.; Zhang, S.F.; Liu, F.; Yu, C.M.; Zhang, Z.; Kong, X.B. Comparative analysis of eight mitogenomes of bark beetles and their phylogenetic implicaitons. Insects 2021, 12, 949. [Google Scholar] [CrossRef]
- Wang, S.; Meyer, E.; McKay, J.K.; Matz, M.V. 2b-RAD: A simple and flexible method for genome-wide genotyping. Nat. Methods 2012, 20, 9. [Google Scholar] [CrossRef]
- Seetharam, A.S.; Stuart, G.W. Whole genome phylogeny for 21 Drosophila species using predicted 2b-RAD fragments. PeerJ 2013, 1, e226. [Google Scholar] [CrossRef] [Green Version]
- Du, Z.Y.; Hasegawa, H.; Cooley, J.R.; Simon, C.; Yoshimura, J.; Cai, W.Z.; Li, H. Mitochondrial genomics reveals shared phylogeographic patterns and demographic history among three periodical cicada species groups. Mol. Biol. Evol. 2019, 36, 1187–1200. [Google Scholar] [CrossRef]
- Puritz, J.B.; Matz, M.V.; Toonen, R.J.; Weber, J.N.; Bolnick, D.I.; Bird, C.E. Demystifying the RAD fad. Mol. Ecol. 2014, 23, 5937–5942. [Google Scholar] [CrossRef] [PubMed]
- Arbizu, C.I.; Ellison, S.L.; Senalik, D.; Simon, P.W.; Spooner, D.M. Genotyping-by-sequencing provides the discriminating power to investigate the subspecies of Daucus carota (Apiaceae). BMC Evol. Biol. 2016, 16, 234. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Diaz-Arce, N.; Arrizabalaga, H.; Murua, H.; Irigoien, X.; Rodriguez, E.N. RAD-seq derived genome-wide nuclear markers resolve the phylogeny of tunas. Mol. Phylogenet. Evol. 2016, 102, 202–207. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kirkendall, L.R.; Biedermann, P.H.W.; Jordal, B.H. Evolution and Diversity of Bark and Ambrosia Beetles. In Bark Beetles: Biology and Ecology of Native and Invasive Species; Vega, F.E., Hofstetter, R.W., Eds.; Elsevier Inc.: New York, NY, USA, 2015; pp. 85–136. [Google Scholar]
- Hulcr, J.; Atkin Son, T.H.; Cognato, A.I.; Jordal, B.H.; McKenna, D.D. Morphology, taxonomy an phylogenetics of bark beetles. In Bark Beetles: Biology and Ecology of Native and Invasive Species; Vega, F.E., Hofstetter, R.W., Eds.; Elsevier Inc.: New York, NY, USA, 2015; pp. 41–84. [Google Scholar]
- Hou, X.Q.; Yuvaraj, J.K.; Roberts, R.E.; Zhang, D.D.; Unelius, C.R.; Lofstedt, C.; Andersson, M.N. Functional evolution of a bark beetle odorant receptor clade detecting monoterpenoids of different ecological origins. Mol. Biol. Evol. 2021, 38, 4934–4937. [Google Scholar] [CrossRef]
- Roelofs, W.L.; Brown, R.L. Pheromones and evolutionary relationships of Tortricidae. Annu. Rev. Ecol. Syst. 1982, 13, 395–422. [Google Scholar] [CrossRef]
- Symonds, M.R.E.; Elgar, M.A. The mode of pheromone evolution: Evidence from bark beetle. Proc. R. Soc. B-Biol. Sci. 2004, 271, 839–846. [Google Scholar] [CrossRef] [Green Version]
- Guo, H.; Lackus, N.; Köllner, T.G.; Li, R.; Bing, J.; Wang, Y.; Baldwin, I.T.; Xu, S. Evolution of a novel and adaptive floral scent in wild tobacco. Mol. Biol. Evol. 2020, 37, 1090–1099. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Q.H.; Birgersson, G.; Schlyter, F.; Chen, G.F. Pheromone components in the larch bark beetle, Ips cembrae, from China: Quantitative variation among attack phases and individuals. J. Chem. Ecol. 2000, 26, 841–858. [Google Scholar] [CrossRef]
- Zhang, Q.H.; Schlyter, F.; Chen, G.; Wang, Y. Electrophysiological and behavioral responses of Ips subelongatus to semiochemicals from its hosts, non-hosts, and conspecifics in China. J. Chem. Ecol. 2007, 33, 391–404. [Google Scholar] [CrossRef]
- Zhang, Q.H.; Schlyter, F.; Liu, G.T.; Sheng, M.L.; Birgersson, G. Electrophysiological and behavioral responses of Ips duplicatus to aggregation pheromone in Inner Mongolia, China: Amitinol as a potential pheromone component. J. Chem. Ecol. 2007, 33, 1303–1315. [Google Scholar] [CrossRef]
- Zhang, Q.H.; Ma, J.H.; Zhao, F.Y.; Song, L.W.; Sun, J.H. Aggregation pheromone of the Qinghai spruce bark beetle, Ips nitidus eggers. J. Chem. Ecol. 2009, 35, 610–617. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Q.H.; Song, L.W.; Ma, J.H.; Han, F.Z.; Sun, J.H. Aggregation pheromone of a newly described spruce bark beetle, Ips shangrila Cognato and Sun from China. Chemoecology 2009, 19, 203–210. [Google Scholar] [CrossRef]
- Bakke, A. Spruce bark beetle, Ips typographus: Pheromone production and field response to synthetic pheromones. Naturwissenschafren 1976, 62, 92. [Google Scholar] [CrossRef]
- Fang, J.X.; Du, H.C.; Shi, X.; Zhang, S.F.; Liu, F.; Zhang, Z.; Zu, P.J.; Kong, X.B. Monoterpenoid signals and their transcriptional responses to feeding and juvenile hormone regulation in bark beetle Ips huaseri. J. Exp. Biol. 2021, 224, jeb238030. [Google Scholar] [CrossRef]
- Eidmann, H.H.; Birgersson, G. Semiochemicals in the East Himalaya spruce bark beetle. Anz. Schädlingskunde Pflanzenschutz Umweltschutz 1988, 61, 147–148. [Google Scholar] [CrossRef]
- Vité, J.; Bakke, A.; Renwick, J.A.A. Pheromones in Ips (Coleoptera: Scolytidae): Occurrence and production. Can. Entomol. 1972, 104, 1967–1975. [Google Scholar] [CrossRef]
- Tittiger, C.; Blomquist, G.J. Pheromone biosynthesis in bark beetles. Curr. Opin. Insect Sci. 2017, 24, 68–74. [Google Scholar] [CrossRef]
- Ivarsson, P.; Schlyter, F.; Birgersson, G. Demonstration of de novo pheromone biosynthesis in Ips duplicatus (Coleoptera: Scolytidae): Inhibition of ipsdienol and E-myrcenol production by compaction. Insect Biochem. Mol. Biol. 1993, 23, 655–662. [Google Scholar] [CrossRef]
- Seybold, S.J.; Quilici, D.R.; Tillman, J.A.; Vanderwel, D.; Wood, D.L.; Blomquist, G.J. De novo biosynthesis of the aggregation pheromone components ipsenol and ipsdienol by the pine bark beetles Ips paraconfusus Lanier and Ips pini (Say) (Coleoptera:Scolytidae). Proc. Natl. Acad. Sci. USA 1995, 92, 8393–8397. [Google Scholar] [CrossRef] [Green Version]
- Cane, J.H.; Stock, M.W.; Wood, D.L.; Gast, S.J. Phylogenetic relationship of Ips bark beetles (Coleoptera: Scolytidae): Electrophoretic and morphometric analyses of the grandicollis group. Biochem. Syst. Ecol. 1990, 18, 359–368. [Google Scholar] [CrossRef]
- Cognato, A.I.; Seybold, S.J.; Wood, D.L.; Teale, S.A. A cladistic analysis of pheromone evolution in Ips bark beetles (Coleoptera: Scolytidae). Evolution 1997, 51, 313–318. [Google Scholar] [CrossRef] [PubMed]
- Huang, F.S.; Lu, J. The Classification Outline of Scolytidae from China; Tongji University Press: Shanghai, China, 2015. [Google Scholar]
- Dmitry, A.; Anton, K.; Jeffrey, S.; McLean, P.A.; Pevzner, A.N. HYBRIDSPADES: An algorithm for hybrid assembly of short and long reads. Bioinformatics 2016, 7, 1009–1015. [Google Scholar]
- Meng, G.; Li, Y.; Yang, C.; Liu, S. MitoZ: A toolkit for animal mitochondrial genome assembly, annotation and visualization. Nucleic Acid Res. 2019, 47, e63. [Google Scholar] [CrossRef]
- Guindon, S.; Gascuel, O. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst. Biol. 2003, 52, 696–704. [Google Scholar] [CrossRef] [Green Version]
- Ronquist, F.; Huelsenbeck, J.P. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 2003, 19, 1572–1574. [Google Scholar] [CrossRef] [Green Version]
- Huelsenbeck, J.P.; Ronquist, F.; Nielsen, R.; Bollback, J.P. Bayesian inference of phylogeny and its impact on evolutionary biology. Science 2011, 294, 2310–2314. [Google Scholar] [CrossRef] [Green Version]
- Bouckaert, R.; Heled, J.; Kühnert, D.; Vaughan, T.; Wu, C.H.; Xie, D.; Suchard, M.; Rambaut, A.; Drummond, A.J. BEAST 2: A software platform for bayesian evolutionary analysis. PLoS Comput. Biol. 2014, 10, e1003537. [Google Scholar] [CrossRef] [Green Version]
- Papadopoulou, A.; Anastasiou, I.; Vogler, A.P. Revisiting the insect mitochondrial molecular clock: The mid-Aegean trench Calibration. Mol. Biol. Evol. 2010, 27, 1659–1672. [Google Scholar] [CrossRef] [Green Version]
- Perna, N.T.; Kocher, T.D. Patterns of nucleotide composition at fourfold degenerate sites of animal mitochondrial genomes. J. Mol. Evol. 1995, 41, 353–358. [Google Scholar] [CrossRef] [PubMed]
- Tamura, K. MEGA5: Molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 2011, 28, 2731–2739. [Google Scholar] [CrossRef] [Green Version]
- Thompson, J.D.; Gibson, T.J.; Plewniak, F.; Jeanmougin, F.; Higgins, D.G. The CLUSTAL X Windows interface: Flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 1997, 25, 4876–4882. [Google Scholar] [CrossRef] [Green Version]
- Kimura, M. A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J. Mol. Evol. 1980, 16, 111–120. [Google Scholar] [CrossRef] [PubMed]
- Yang, Z. PAML 4: Phylogenetic analysis by maximum likelihood. Mol. Biol. Evol. 2007, 24, 1586–1591. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bandelt, H.; Forster, P.; Röhl, A. Median-joining networks for inferring intraspecifific phylogenies. Mol. Biol. Evol. 1999, 16, 37–48. [Google Scholar] [CrossRef] [PubMed]
- Bohonak, A.J. IBD (Isolation by Distance): A program for analyses of isolation by distance. J. Hered. 2002, 93, 153–154. [Google Scholar] [CrossRef]
- Catchen, J.; Hohenlohe, P.A.; Bassham, S. Stacks: An analysis tool set for population genomics. Mol. Ecol. 2013, 22, 3124–3140. [Google Scholar] [CrossRef] [Green Version]
- Li, R.; Yu, C.; Li, Y.; Lam, T.W.; Yiu, S.M.; Kristiansen, K.; Wang, J. SOAP2: An improved ultrafast tool for short read alignment. Bioinformatics 2009, 25, 1966–1967. [Google Scholar] [CrossRef] [Green Version]
- Pritchard, J.K.; Stephens, M.J.; Donnelly, P.J. Inference of population structure using multilocus genotype data. Genetics 2000, 155, 945–959. [Google Scholar] [CrossRef]
- Clary, D.O.; Wolstenholme, D.R. The ribosomal RNA genes of Drosophila mitochondrial DNA. Nucleic Acids Res. 1985, 13, 4029–4045. [Google Scholar] [CrossRef] [Green Version]
- Cognato, A.I.; Sun, J.H. DNA based cladograms augment the discovery of a new Ips species from China (Coleoptera: Curculionidae: Scolytinae). Cladistics 2007, 23, 539–551. [Google Scholar] [CrossRef]
- Feng, S.; Ru, D.; Sun, Y.; Mao, K.S.; Milne, R.; Liu, J.Q. Trans-lineage polymorphism and nonbifrucating diversification of the genus Picea. New Phytol. 2019, 222, 576–587. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shi, X.; Shen, J.C.; Zhang, S.F.; Liu, F.; Xu, F.Y.; Wang, G.L.; Zhang, Z.; Kong, X.B. Comparative analysis of the type and number of larval sensilla on the antennae and mouthparts of Ips typographus and Ips subelongatus using SEM. Zool. Anz. 2020, 289, 18–25. [Google Scholar] [CrossRef]
- Shi, X.; Zhang, S.F.; Liu, F.; Xu, F.Y.; Zhang, F.B.; Guo, X.B.; Zhang, Z.; Kong, X.B. SEM analysis of sensilla on the mouthparts and antennae of Asian larch bark beetle Ips subelongatus. Micron 2020, 1, 140. [Google Scholar] [CrossRef] [PubMed]
- Stauffer, C.; Kirisits, T.; Nussbaumer, C.; Pavlin, R.; Wingfield, M.J. Phylogenetic relationships between the European and Asian eight spined larch bark beetle populations (Coleoptera, Scolytidae) inferred from DNA sequences and fungal associates. Eur. J. Entomol. 2001, 98, 99–105. [Google Scholar] [CrossRef]
- Kohnle, U.; Vité, J.; Erbacher, C.; Bartels, J.; Francke, W. Aggregation response of European engraver beetles of the genus Ips mediated by terpenoid pheromones. Entomol. Exp. Appl. 1988, 49, 43–53. [Google Scholar] [CrossRef]
- Song, L.W.; Zhang, Q.H.; Chen, Y.Q.; Zuo, T.T.; Ren, B.Z. Field responses of the Asian larch bark beetle, Ips subelongatus, to potential aggregation pheromone components: Disparity between two populations in northeastern China. Insect Sci. 2011, 18, 311–319. [Google Scholar] [CrossRef]
- Chen, D.F.; Li, Y.J.; Zhang, Q.H.; Zhang, S.F.; Wang, H.B.; Zhang, Z.; Zhao, L.L.; Kong, X.B. Population divergence of aggregation pheromone responses in Ips subelongatus in Northeastern China. Insect Sci. 2016, 23, 728–738. [Google Scholar] [CrossRef]
- Byers, J.A.; Birgersson, G. Pheromone production in a bark beetle independent of myrcene precursor in host pine species. Naturwissenschaften 1990, 77, 385–387. [Google Scholar] [CrossRef]
- Schlyter, F.; Birgersson, G.; Byers, J.A.; Bakke, A. The aggregation pheromone of Ips duplicatus and its role in competitive interactions with I. typographus. Chemoecology 1992, 3, 103–112. [Google Scholar] [CrossRef]
- Renwick, J.; Hughes, P.; Krull, I. Selective production of cis- and trans-verbenol from (–)-and (+)-alpha by a bark beetle. Science 1976, 191, 199–201. [Google Scholar] [CrossRef]
- Schlyter, F.; Jakus, R.; Han, F.Z.; Ma, J.H.; Kalinová, B.; Mezei, P.; Sun, J.H.; Ujhelyiová, L.; Zhang, Q.H. Reproductive Isolation of Ips nitidus and I. shangrila in Mountain Forests of Western China: Responses to Chiral and Achiral Candidate Pheromone Components. J. Chem. Ecol. 2015, 41, 678–688. [Google Scholar] [CrossRef] [PubMed]
- Roelofs, W.L.; Liu, W.T.; Hao, G.X.; Jiao, H.M.; Rooney, A.P.; Linn, C.E. Evolution of moth sex pheromones via ancestral genes. Proc. Natl. Acad. Sci. USA 2002, 99, 13621–13626. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Baker, T.C. Mechanism for saltational shifts in pheromone communication systems. Proc. Natl. Acad. Sci. USA 2002, 99, 13368–13370. [Google Scholar] [CrossRef] [Green Version]
- Sun, Y.; Abbott, R.; Li, L.; Zou, J.; Liu, J. Evolutionary history of purple cone spruce (Picea purpurea) in the Qinghai-Tibet Plateau: Homoploidhybrid origin and pleistocene expansion. Mol. Ecol. 2014, 23, 343–359. [Google Scholar] [CrossRef] [PubMed]
- An, Z.S.; Kutzbach, J.E.; Prell, W.L.; Porter, S.C. Evolution of Asian monsoons an phased uplift of the Himalaya Tibetan Plateau since late miocene times. Nature 2001, 411, 62–66. [Google Scholar]
- Li, L.L.; Sun, Y.S.; Zou, J.B.; Wei, Y.; Wang, X.; Liu, J.Q. Origin and speciation of Picea schrenkiana and Picea smithiana in the center Asian highlands and himalayas. Plant Mol. Biol. Rep. 2015, 33, 661–672. [Google Scholar] [CrossRef] [Green Version]
- Santiago, M.; Ken, T.; Matthew, J.C.; Anna, O.; Carles, V.; Andreas, W.; Matthew, T.W. The genomic basis of adaptation to high-altitude habitats in the eastern honey bee (Apis cerana). Mol. Ecol. 2019, 28, 1–15. [Google Scholar]
- Sun, J.T.; Duan, X.Z.; Hoffmann, A.A.; Liu, Y.; Garvin, M.R.; Chen, L.; Hu, G.; Zhou, J.C.; Huang, H.J.; Xue, X.F.; et al. Mitochondrial variation in small brown planthoppers linked to multiple traits and probably reflecting a complex evolutionary trajectory. Mol. Ecol. 2019, 28, 3306–3323. [Google Scholar] [CrossRef]
- Boore, J.L. Animal mitochondrial genomes. Nucleic Acids Res. 1999, 27, 1767–1780. [Google Scholar] [CrossRef] [Green Version]
- Shen, Y.Y.; Liang, L.; Zhu, Z.H.; Zhou, W.P.; Irwin, D.; Zhang, Y.P. Adaptive evolution of energy metabolism genes and the origin of flight in bats. Proc. Natl. Acad. Sci. USA 2010, 107, 8666–8671. [Google Scholar] [CrossRef] [Green Version]
- Sun, Y.B.; Shen, Y.Y.; Irwin, D.M.; Zhang, Y.P. Evaluating the roles of energetic functional constraints on teleost mitochondrial encoded protein evolution. Mol. Biol. Evol. 2011, 28, 39–44. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, Z.; Yonezawa, T.; Liu, B.; Ma, T.; Shen, X.; Su, J.P.; Guo, S.C.; Hasegawa, M.; Liu, J.Q. Domestication relaxed selective constraints on the yak mitochondrial genome. Mol. Biol. Evol. 2011, 28, 1553–1556. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gu, M.L.; Dong, X.Q.; Shi, L.I.; Shi, L.; Lin, K.; Huang, X.Q.; Chu, J.Y. Differences in mtDNA whole sequence between Tibetan and Han populations suggesting adaptive selection to high altitude. Gene 2012, 496, 37–44. [Google Scholar] [CrossRef]
- Li, Y.; Ren, Z.M.; Shedlock, A.M.; Wu, J.Q.; Sang, L.; Tersing, T.; Hasegawa, M.; Yonezawa, T.; Zhong, Y. High altitude adaptation of the schizothoracine fishes (Cyprinidae) revealed by the mitochondrial genome analyses. Gene 2013, 517, 169–178. [Google Scholar] [CrossRef] [PubMed]
- Lv, F.; Yang, W.Y.; Chen, Z.T.; Xu, Q.; Zhou, Y.J.; Du, Y.Z. Three partial mitochondrial genomes from Ips (Coleoptera: Cruculionidae, Scolytinae) contribute to the phylogeny of Scolytinae. J. Asia-Pac. Enotomol. 2017, 20, 1007–1013. [Google Scholar]
- Wolfe, J.A. An analysis of Neogene climates in Beringia. Palaeogeogr. Palaeoclimatol. Palaeoecol. 1994, 108, 207–216. [Google Scholar] [CrossRef]
- Lanier, G.N.; Burkholder, W.E. Pheromones in speciation of Coleoptera. In Pheromones; Birch, M.C., Ed.; North-Holland: London, UK, 1974; pp. 161–189. [Google Scholar]
- Mckenna, D.D.; Shin, S.; Ahrens, D.; Balke, M.; Beza-Beza, C.; Clarke, D.J.; Donath, A.; Escalona, H.E.; Friedrich, F.; Letsch, H.; et al. The evolution and genomic basis of beetle diversity. Proc. Natl. Acad. Sci. USA 2019, 12, 24729–24737. [Google Scholar] [CrossRef] [Green Version]
- Meseguer, A.S.; Coeur, A.; Genson, G.; Jousselin, E. Unravelling the historical biogeography and diversifification dynamics of a highly diverse conifer-feeding aphid genus. J. Biogeogr. 2015. 42, 1482–1492.
- Godefroid, M.; Meseguer, A.S.; Saune, L.; Genson, G.; Streito, J.; Rossi, J.; Riveron, A.Z.; Mayer, F.; Cruaud, A.; Rasplus, J. Restriction-site associated DNA markers provide new insights into the evolutionary history of the bark beetle genus Dendroctonus. Mol. Phylogenet. Evol. 2019, 139, 106528. [Google Scholar] [CrossRef] [Green Version]


| Species | Code | Location (District, Province) | Latitude | Longitude | Altitude | Hosts | Accession Number |
|---|---|---|---|---|---|---|---|
| Ips typographus | ItyJG | Yanbian, Jilin | 43.35 | 130.18 | 660 | Picea koraiensis Nakai | OK110249 |
| ItyHB | Habahe, Xinjiang | 48.47 | 86.68 | 994 | P. obovata Ledeb. | OK147100 | |
| ItyHS | Hengshan, Jilin | 46.24 | 128.95 | 175 | P. koraiensis Nakai | OK147101 | |
| ItyDH | Dunhua, Jilin | 43.21 | 127.65 | 575 | P. koraiensis Nakai | OK147111 | |
| ItyNR | Naren, Xinjiang | 48.47 | 86.69 | 1024 | P. obovata Ledeb. | OK147109 | |
| Ips subelongatus | IsuHB | Habahe, Xinjiang | 48.39 | 86.79 | 1329 | Larix sibirica Ledeb. | OK110250 |
| IsuGP | Helong, Jilin | 42.04 | 128.8 | 572 | L. olgensis Henry | OK086802 | |
| IsuJM | Jiamusi, Heilongjiang | 46.43 | 130.65 | 192 | L. olgensis Henry | OK147102 | |
| IsuHG | Chifeng, Inner Mongolia | 43.57 | 117.52 | 1447 | L. principis-rupprechtii Mayr | OK147107 | |
| IsuYC | Yichun, Heilongjiang | 48.65 | 126.63 | 273 | L. gmelinii Rupr. | OK147112 | |
| Ips hauseri | IhaTS | Tienshan, Xinjiang | 43.18 | 82.85 | 1516 | P. schrenkiana Fisch. et Mey. | OK147104 |
| IhaYN | Yining, Xinjiang | 44.2 | 81.67 | 2016 | P. schrenkiana Fisch. et Mey. | OK147105 | |
| IhaXY | Xinyuan, Xinjiang | 43.27 | 84.3 | 1654 | P. schrenkiana Fisch. et Mey. | OK147106 | |
| Ips nitidus | IniMX | Maixiu, Qinghai | 35.26 | 101.89 | 3156 | P. crassifolia Kom. | OK147110 |
| IniXZ | Leiwuqi, Xizang | 31.25 | 96.49 | 3947 | P. likiangensis Pritz. | OK147108 | |
| IniQL | Qilian, Qinghai | 38.16 | 100.49 | 3407 | P. crassifolia Kom. | OK147114 | |
| Ips shangrila | IshQL | Qilian, Qinghai | 35.26 | 101.89 | 3156 | P. crassifolia Kom. | OK147103 |
| Ips duplicatus | IduBY | Chifeng, Inner Mongolia | 43.53 | 117.22 | 1357 | P. meyeri Rehd. et Wils. | OK147102 |
| Dendroctonus micans | DmiMX | Maixiu, Qinghai | 35.27 | 101.91 | 3050 | P. purpurea Mast. | OL147113 |
| Species | ATP6 | ATP8 | COI | COII | COIII | Cytb | ND1 | ND2 | ND3 | ND4 | ND4L | ND5 | ND6 | Average |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| I. nitidus | 0.0304 | 0.0219 | 0.0302 | 0.0473 | 0.0356 | 0.0603 | 0.0651 | 0.0693 | 0.0298 | 0.0220 | 0.0096 | 0.0621 | 0.2278 | 0.0547 |
| I. shangrila | 0.0334 | 0.0209 | 0.0159 | 0.0387 | 0.0894 | 0.0298 | 0.0414 | 0.0719 | 0.0530 | 0.0202 | 0.0203 | 0.0604 | 0.0353 | 0.0408 |
| I. duplicatus | 0.0315 | 0.0245 | 0.0050 | 0.0636 | 0.0322 | 0.0310 | 0.0507 | 0.1258 | 0.0461 | 0.0326 | 0.0540 | 0.0757 | 0.0485 | 0.0478 |
| I. hauseri | 0.0220 | 0.0196 | 0.0129 | 0.0128 | 0.0111 | 0.0118 | 0.0531 | 0.0566 | 0.0259 | 0.0339 | 0.0116 | 0.0521 | 0.0633 | 0.0297 |
| I. typgraphus | 0.0272 | 0.0001 | 0.0122 | 0.0001 | 0.0001 | 0.0240 | 0.0261 | 0.0897 | 0.0001 | 0.0001 | 0.0001 | 0.0216 | 0.1252 | 0.0251 |
| I. subelongatus | 0.0101 | 0.0001 | 0.0001 | 0.0001 | 0.0163 | 0.0287 | 0.0150 | 0.0381 | 0.0060 | 0.0001 | 0.0012 | 0.0362 | 0.0899 | 0.0186 |
| Formula Mode | r (Correlation Coefficient) | p-Value | |
|---|---|---|---|
| Ips (between all species) | genetic distance & geographic distance | 0.0526 | 0.198 |
| genetic distance & log(geographic distance) | 0.105 | 0.097 | |
| log(genetic distance) & geographic distance | 0.1383 | 0.07 | |
| log(genetic distance) & log(geographic distance) | 0.2282 | 0.017 | |
| I. typographus | genetic distance & geographic distance | 0.9943 | 0.019 |
| genetic distance & log(geographic distance) | 0.8111 | 0.019 | |
| log(genetic distance) & geographic distance | 0.8441 | 0.047 | |
| log(genetic distance) & log(geographic distance) | 0.8402 | 0.001 | |
| I. subelongatus | genetic distance & geographic distance | −0.1942 | 0.357 |
| genetic distance & log(geographic distance) | −0.2108 | 0.559 | |
| log(genetic distance) & geographic distance | −0.1508 | 0.384 | |
| log(genetic distance) & log(geographic distance) | −0.1938 | 0.487 | |
| I. nitidus | genetic distance & geographic distance | 0.9528 | 0.015 |
| genetic distance & log(geographic distance) | 0.9786 | 0.001 | |
| log(genetic distance) & geographic distance | 0.9432 | 0.001 | |
| log(genetic distance) & log(geographic distance) | 0.9720 | 0.001 | |
| I. hauseri | genetic distance & geographic distance | −0.0857 | 0.508 |
| genetic distance & log(geographic distance) | −0.0068 | 0.346 | |
| log(genetic distance) & geographic distance | −0.0414 | 0.508 | |
| log(genetic distance) & log(geographic distance) | 0.0376 | 0.508 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Du, H.; Fang, J.; Shi, X.; Yu, C.; Deng, M.; Zhang, S.; Liu, F.; Zhang, Z.; Han, F.; Kong, X. Insights into the Divergence of Chinese Ips Bark Beetles during Evolutionary Adaptation. Biology 2022, 11, 384. https://doi.org/10.3390/biology11030384
Du H, Fang J, Shi X, Yu C, Deng M, Zhang S, Liu F, Zhang Z, Han F, Kong X. Insights into the Divergence of Chinese Ips Bark Beetles during Evolutionary Adaptation. Biology. 2022; 11(3):384. https://doi.org/10.3390/biology11030384
Chicago/Turabian StyleDu, Huicong, Jiaxing Fang, Xia Shi, Chunmei Yu, Mei Deng, Sufang Zhang, Fu Liu, Zhen Zhang, Fuzhong Han, and Xiangbo Kong. 2022. "Insights into the Divergence of Chinese Ips Bark Beetles during Evolutionary Adaptation" Biology 11, no. 3: 384. https://doi.org/10.3390/biology11030384
APA StyleDu, H., Fang, J., Shi, X., Yu, C., Deng, M., Zhang, S., Liu, F., Zhang, Z., Han, F., & Kong, X. (2022). Insights into the Divergence of Chinese Ips Bark Beetles during Evolutionary Adaptation. Biology, 11(3), 384. https://doi.org/10.3390/biology11030384







