Simple Summary
We sequenced the complete chloroplast genomes of three Ceriops species (C. decandra, C. zippeliana, and C. tagal) and Avicennia lanata and performed comparative analyses among them. All chloroplast genomes have a circular quadripartite structure containing LSC, SSC, and two IR regions. The rpl32 gene was lost in C. zippeliana, and the infA gene was present in only A. lanata. Comparative genome analysis showed that the IR contraction or expansion events resulted in the differentiation of three genes and pseudogenes. Additionally, repeats and SSRs were identified and compared among them and other relative mangrove species. The phylogenetic analysis strongly supports that C. decandra is evolutionarily closer to C. zippeliana and A. lanata is closer to A. marina. In addition, two primer pairs were developed for species identification unique to the three Ceriops species.
Abstract
Ceriops and Avicennia are true mangroves in the middle and seaward zones of mangrove forests, respectively. The chloroplast genomes of Ceriops decandra, Ceriops zippeliana, and Ceriops tagal were assembled into lengths of 166,650, 166,083 and 164,432 bp, respectively, whereas Avicennia lanata was 148,264 bp in length. The gene content and gene order are highly conserved among these species. The chloroplast genome contains 125 genes in A. lanata and 129 genes in Ceriops species. Three duplicate genes (rpl2, rpl23, and trnM-CAU) were found in the IR regions of the three Ceriops species, resulting in expansion of the IR regions. The rpl32 gene was lost in C. zippeliana, whereas the infA gene was present in A. lanata. Short repeats (<40 bp) and a lower number of SSRs were found in A. lanata but not in Ceriops species. The phylogenetic analysis supports that all Ceriops species are clustered in Rhizophoraceae and A. lanata is in Acanthaceae. In a search for genes under selective pressures of coastal environments, the rps7 gene was under positive selection compared with non-mangrove species. Finally, two specific primer sets were developed for species identification of the three Ceriops species. Thus, this finding provides insightful genetic information for evolutionary relationships and molecular markers in Ceriops and Avicennia species.
1. Introduction
Mangroves are extremely important plants to coastal ecosystems. They protect shorelines from erosion and provide marine nursery areas and breeding sites for a variety of marine and terrestrial organisms (e.g., fish, crustaceans, reptiles, birds, and mammals) [,]. For human beings, they are used for food, fuels, timber, and medicines [,]. Mangroves grow in the intertidal zones of tropical and subtropical regions with extreme environmental conditions such as frequent tidal inundation, oxygen-poor soil, and high salinity [,]. There are roughly 70 mangrove species in 28 genera in 16–19 families [,]. Indeed, a few mangrove species in the families Rhizophoraceae and Acanthaceae occupy most areas of mangrove forests [,]. In the past decades, mangrove forest areas have been dramatically decreasing due to anthropogenic impacts and climate change [,,]. Therefore, the genetic information of mangroves is crucial for understanding their genetic conservation, population structure, evolution history, and species identification [,,,]. Recently, a number of whole mangrove genomes have been reported [,,,,,,,,].
Ceriops (Rhizophoraceae, Rosids) and Avicennia (Acanthaceae, Asterids) are classified as true mangroves and the most dominant species in the middle and seaward zones of mangrove forests, respectively [,,]. Both species have adapted to extreme conditions in mangrove habitats. For example, Ceriops is a viviparous mangrove species that has seeds producing propagules or beginning to germinate on the mother plants and is a salt excluder by filtering salt out at the roots []. Avicennia specially adapted with pneumatophores (pencil-like aerial roots) and salt glands on the upper and lower leaf surfaces, which secrete excess salt from the leaves [,]. The genus Ceriops contains five species, including Ceriops australis, Ceriops decandra, Ceriops pseudodecandra, Ceriops tagal, and Ceriops zippeliana [,,,]. In the past, C. zippeliana was believed to be a synonym of C. decandra [,]; however, differences in morphology and a trnL intron of chloroplast DNA between them were reported and they were suggested as different species []. C. decandra and C. tagal are widespread species in a large geographical range from Eastern Africa and throughout tropical Asia and Northern Australia to Melanesia, Micronesia, and Southern China [,,], while C. pseudodecandra and C. australis are endemic to Australia []. C. zippeliana is found in Southeast Asia (Thailand, Malaysia, Singapore, Indonesia, and the Philippines) []. Based on the International Union for Conservation of Nature (IUCN) Red List, C. decandra is classified as a near threatened species due to habitat loss []. In addition to Ceriops, Avicennia, which is the pioneer of the mangrove swamp, comprises at least eight species [,,]. The distribution of Avicennia species is separated into two geographic parts: the Indo-West Pacific (IWP) and Atlantic-East Pacific (AEP) regions. At least six species (A. alba, A. integra, A. lanata, A. marina, A. officinalis, and A. rumphiana) are distributed in the IWP region, whereas three species (A. germinans, A. schaueriana, and A. bicolor) are found in the AEP region [,,]. Notably, A. lanata is distributed only in Southeast Asia []. Recently, A. bicolor, A. integra, A. lanata, and A. rumphiana have been listed as vulnerable species on the IUCN Red List [].
Chloroplasts are photosynthetic organelles in algae and land plants that have their own genomes. Chloroplast genomes are highly conserved because of uni-parent inheritance or maternal inheritance []. The sizes of chloroplast genomes in mangrove species are around 145–168 kb [,,,]. Chloroplast genomes in mangrove species usually contain four regions, including one large single-copy region (LSC), two inverted repeats (IRA and IRB), and one single small-copy region (SSC) [,,,]. To date, several chloroplast genomes have been reported because of the development of DNA sequencing technology and bioinformatics methods [,,]. For Ceriops and Avicennia species, only the chloroplast genomes of C. tagal and A. marina are available [,]. Therefore, both the Ceriops and Avicennia genera suffer from a lack of chloroplast genomes to compare their genomes and to understand phylogenetic relationships among them.
Mangroves present very special ecological characteristics, and understanding the genome structure through this molecular finding will further provide valuable genetic information regarding the evolutionary trends in plants according to harsh climatic conditions. In this study, we investigated the chloroplast genomes of four mangrove species that are commonly distributed in the middle (Ceriops decandra, C. zippeliana, and C. tagal) and seaward (Avicennia lanata) zones of the coastal region of Southeast Asia to understand the evolutionary relationships under different coastal environments and to identify genetic markers for species identification and candidate genes under selective pressures. The four mangrove species were sequenced, assembled, and annotated. Comparisons of chloroplast genomes among the three Ceriops species and between the Ceriops and Avicennia species were performed to reveal their evolutionary relationships. Different numbers of SSRs and short repeats were identified among them. Genes under positive selection were identified and might correlate with adaptive selection that could be used for further studies on the response to stress conditions in mangroves. Finally, two sets of species-specific primers were developed for species identification of the three Ceriops species based on SSRs. These chloroplast genomes provide valuable genetic information and potential molecular markers for mangrove species in the southeast coastal regions.
2. Materials and Methods
2.1. Samples, DNA Isolation, and Sequencing
Four mangrove species (Ceriops decandra, Ceriops zippeliana, Ceriops tagal, and Avicennia lanata) were used in this study. C. decandra is a near threatened species, C. zippeliana was formerly recognized as C. decandra, and C. tagal is a widespread species [,]. A. lanata is listed as a vulnerable species [].
Fresh leaves of C. decandra, C. zippeliana, C. tagal, and A. lanata were collected from the Ranong, Chanthaburi, Ranong, and Prachuap Khiri Khan provinces in Thailand, respectively (Table S1). The leaf samples were frozen in liquid nitrogen for DNA isolation. Genomic DNA was extracted using the standard cetyltrimethylammonium bromide (CTAB) method []. Each sample was sequenced using the Illumina HiSeqX ten platform with paired-end reads of 150 bp.
2.2. Chloroplast Genome Assembly and Annotation
The chloroplast genome of C. decandra was assembled using NOVOPlasty version 4.2 []. The chloroplast rbcL sequence of C. tagal (NCBI accession number: MH240830) was used as a seed sequence. The chloroplast genomes of C. tagal, C. zippeliana, and A. lanata were assembled using GetOrganelle [] with the reference genome-based strategy based on the C. tagal chloroplast genome (MH240830) for Ceriops species and the A. marina chloroplast genome (MT012822) for Avicennia species. Notably, GetOrganelle was used mainly for assembling the four mangrove chloroplast genomes due to the highly accurate results of organelle genomes [,]. However, it was not fit to complete the chloroplast genome of C. decandra; thus, NOVOPlasty was used instead.
All four chloroplast genome sequences were annotated using GeSeq with default settings []. The start–stop loci and intron–exon borders of coding genes were edited manually after comparation with reported mangrove chloroplast genes. All transfer RNAs (tRNAs) were predicted using ARAGORN v1.2.36 [] implemented in the GeSeq software. The circular structures of the chloroplast genomes were illustrated using OGDRAW v1.3. []. Finally, the sequences and annotated genes of the four chloroplast genomes were deposited in GenBank (NCBI accession numbers OK258321 (A. lanata), OK258322 (C. tagal), OK272497 (C. decandra), and OK272496 (C. zippeliana)).
2.3. Comparative Genome Analysis
Comparative genome analysis for Ceriops and Avicennia was carried out using mVISTA with the Shuffle-LAGAN mode []. The species in this analysis included three Ceriops species (in this study), A. lanata (in this study), and four previously reported mangrove species, including Kandelia obovata (NC_042718), Rhizophora stylosa (NC_042819), and Bruguiera parviflora (MW836113) in the family Rhizophoraceae and A. marina (MT012822) in the family Acanthaceae. The previously reported chloroplast genome of C. tagal (MH240830.1, China) was used as a reference for comparison []. In addition, the junctions and borders of the IR regions were illustrated using IRscope [].
2.4. Repeat and SSR Identification
REPuter [] was used to identify repeat sequences in the four chloroplast genomes. Furthermore, simple sequence repeats (SSRs) in the chloroplast genome sequences of the three Ceriops species and A. lanata in this study as well as the previously reported chloroplast genome sequences of C. tagal (NCBI: MH240380; CNSA: CNS0105415) and A. marina (MT012822 and CNS0105414) were identified using MISA []. The thresholds for mononucleotide, dinucleotide, trinucleotide, tetranucleotide, pentanucleotide, and hexanucleotide SSRs were set to 10, 5, 4, 3, 3, and 3, respectively []. The minimum distance of compound SSRs was ≤100 bp (default).
2.5. Phylogenetic Analysis
To assess the phylogenetic relationships of Ceriops and Avicennia species, phylogenetic analyses were performed using the maximum likelihood (ML) method based on 50 conserved chloroplast protein-coding genes in 59 plant species, including the 4 species in this study, 19 other mangrove species, 35 relative land plant species, and 1 outgroup species as Ranunculus macranthus (NC_008796) (Table S2). The 50 conserved genes are atpA, atpB, atpE, atpF, atpI, ccsA, matK, ndhA, ndhD, ndhE, ndhG, ndhH, ndhI, ndhK, petA, petD, petG, petL, petN, psaA, psaB, psaC, psaJ, psbA, psbC, psbD, psbF, psbH, psbJ, psbL, psbM, psbN, psbT, rbcL, rpl2, rpl14, rpl23, rpl33, rpl36, rpoB, rpoC1, rps2, rps3, rps4, rps8, rps11, rps12, rps14, rps15, and rps18. Each gene sequence was aligned individually using MUSCLE with default settings implemented in MEGA X []. All gaps in the aligned sequences were removed. The aligned sequences were concatenated in each species. The GTR+I+G model was predicted to be the best fit model for the dataset using the find best DNA/protein model tool in MEGA X. ML analysis was used to construct a phylogenetic tree based on the nucleotide substitution matrix using RAxML version 8.2.10 [] with the GTRGAMMAI (GTR+I+G) model. Node supports were estimated by performing 1000 bootstrap replicates. Finally, the phylogenetic tree was visualized using FigTree v1.4.3 (http://tree.bio.ed.ac.uk/software/figtree/; accessed on 15 November 2021). Furthermore, gene gain and loss of rpl32, rps16, and infA in mangrove and other non-mangrove species were plotted on the phylogenetic tree.
2.6. Gene Selective Pressure Analysis
A total of 61 shared chloroplast protein-coding genes were used to investigate selection pressures for two mangrove genera, Ceriops and Avicennia (Table S3). We compared species pairs contained between the three Ceriops species and six relative mangrove and non-mangrove species (Kandelia obovata (NC_042718), Rhizophora apiculata (MW387538), Bruguiera parviflora (MW836113), Pellacalyx yunnanensis (NC_048998), Erythroxylum novogranatense (NC_030601), and Ranunculus macranthus (NC_008796)) as well as between A. lanata and six relative mangrove and non-mangrove species (Avicennia marina (NC_047414), Coffea arabica (NC_008535), Nicotiana tabacum (NC_001879), Eucommia ulmoides (NC_037948), Lonicera japonica (NC_026839), and R. macranthus). Notably, R. macranthus was used as an assumed ancestor for the two mangrove genera. Pairwise sequence alignments for each gene in each species pair were generated using MUSCLE with default settings in MEGA X [,]. Then, the values of non-synonymous (Ka) and synonymous (Ks) nucleotide substitutions and Ka/Ks (substitution ratio) in all aligned genes were calculated using KaKs Calculator version 2.0 []. Notably, the Ka/Ks ratios were not available (NA) and ~50, indicating no substitution and extremely low Ks values that were replaced to be zero []. The Ka/Ks ratios were then visualized using R with the heatmap function [].
2.7. Development of Species-Specific Molecular Markers for Ceriops Species
Two primer pairs were designed from the IR region of the three Ceriops chloroplast genome sequences using Primer3 []. PCR amplifications were carried out in 20 µL volumes containing 1 µg genomic DNA, 2 µL dNTPs (2.5 mM each), 2 µL of Taq PCR buffer, 0.2 µL of Taq DNA polymerase, and 1.0 µL of each primer. The amplification conditions were 94 °C for 2 min; followed by 30 cycles of 94 °C for 20 s (denaturation), 55 °C for 30 s (annealing), and 72 °C for 30 s (extension); and a final extension of 72 °C for 5 min. PCR products and the DNA ladder were analyzed using a 1% agarose gel to reveal PCR product sizes.
3. Results
3.1. Chloroplast Genome Features
A total of 66.32 million reads (150 bp) were generated for the three Ceriops species and Avicennia lanata by the Illumina HiseqX ten platform (Table S1). These data were used to assemble the four chloroplast genomes with over 300× coverage. The sizes of the complete chloroplast genomes of C. decandra, C. zippeliana, C. tagal, and A. lanata were 166,650, 166,083, 164,432, and 148,264 bp in length, respectively (Figure 1 and Table 1). All four species exhibit a typical quadripartite structure, which consists of one large single copy (LSC), one small single copy (SSC), and a pair of inverted repeats (IRs). All four regions of A. lanata (LSC: 87,995 bp; SSC: 17,949 bp; IRs: 21,160 bp) were shorter than those of the three Ceriops species (92,660–95,217 bp; 18,054–19,158 bp; 26,307–26,535 bp). The overall GC content in the whole chloroplast genomes of the three Ceriops species and A. lanata was 35% and 38%, respectively. The GC content in the IR regions (~42–44%) was greater than that in the LSC (~32–37%) and SSC (~29–33%) regions.
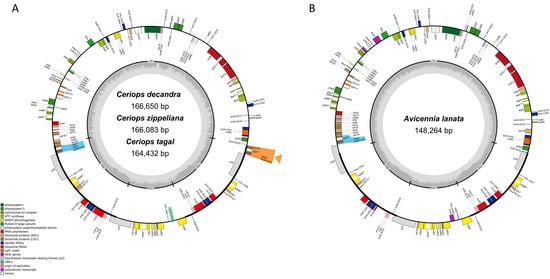
Figure 1.
The chloroplast features of four mangrove species. (A) Complete chloroplast maps of Ceriops decandra, Ceriops zippeliana, and Ceriops tagal. (B) Complete chloroplast map of Avicennia lanata. Genes located outside and inside the circle are transcribed clockwise and counter-clockwise, respectively. The grey bar area in the inner circle indicates GC content of the genome, whereas the lighter grey area indicates AT content of the genome. LSC, SSC, and IRs (IRA and IRB) represent large single copy, small single copy, and inverted repeats, respectively. Genes based on different functional groups are shown in different colors. Green rectangle indicates a loss region (rpl32) of C. zippeliana. Labeling in blue color indicates the same region of three genes (rpl2, rpl23, and trnM-CAU) in both Ceriops and Avicennia species, whereas labeling in orange color with an orange arrow indicates a unique region of three duplicate genes (rpl2, rpl23, and trnM-CAU) in three Ceriops species compared to A. lanata. ** indicates genes containing introns.

Table 1.
Summary of the chloroplast genomes of three Ceriops species and Avicennia lanata.
A total of 125 (A. lanata)–129 (Ceriops species) genes, including 81–84 protein-coding genes, 36–38 transfer RNA (tRNA) genes, and 8 ribosomal RNA (rRNA) genes, were identified (Table 1 and Table 2). Among them, 14 and 17 genes were duplicated in the IR regions of A. lanata and the three Ceriops species, respectively (Figure 1 and Table 1 and Table 2). The seventeen genes in the IR regions of the Ceriops species were ndhB, rpl2, rpl23, rps7, rps12, ycf2, rrn4.5, rrn5, rrn16, rrn23, trnA-UGC, trnE-UUC, trnL-CAA, trnM-CAU, trnN-GUU, trnR-ACG, and trnV-GAC. In A. lanata, rpl2, rpl23, and trnM-CAU were not found in the IR regions; however, they were located in the LSC region. All photosynthesis genes (45 genes), small subunits of ribosomal proteins (13 genes), DNA-dependent RNA polymerase (4 genes), ribosomal RNA genes (8 genes), other metabolic genes (matK, accD, cemA, clpP, and ccsA), and conserved open reading frames (ycf1, ycf2, ycf3, and ycf4) were found in all four chloroplast genomes. Notably, the rpl32 gene was lost in C. zippeliana and the infA gene was only present in A. lanata. In C. zippeliana, a novel tRNA gene, trnY-AUA, was predicted to be located between trnS-GCU and trnT-CGU of the LSC region. Three genes, rps16, rpl16, and ycf2, were found to be pseudogenes in A. lanata, whereas the rps19 gene was a pseudogene in the Ceriops species. Among all the annotated genes, eight protein-coding genes (atpF, ndhA, ndhB, petB, petD, rpl2, rpl16, and rpoC1) and six tRNA genes (trnA-UGC, trnC-ACA, trnE-UUC, trnK-UUU, trnL-UAA, and trnT-CGU) contain a single intron, and two genes (rps12 and ycf3) contain two introns (Table 2). The largest intron in all species was found in the trnK-UUU gene (2504–2585 bp), which contains the matK gene.

Table 2.
List of annotated genes in the chloroplast genomes of three Ceriops species and Avicennia lanata.
3.2. Comparative Analysis of Chloroplast Genomes
The comparison of eight mangrove chloroplast genomes (three Ceriops species, three relative mangrove species in the family Rhizophoraceae, and two Avicennia species) showed similar gene organization and variation regions (Figure 2). Gene orientation was assessed among the mangrove species, revealing a conserved gene structure in the chloroplast genomes. Coding regions were more conserved than the non-coding regions. Additionally, IR regions were more conserved than the LSC and SSC regions, suggesting low divergence in the IR regions. The IR regions were highly conserved between C. tagal and the Rhizophoraceae species and between C. tagal and the Avicennia species at over 98% and 90%, respectively. Low similarity (<80%) of nine protein-coding gene sequences (trnK-UUU, trnT-CGU, trnL-AAA, trnC-ACA, rps3, rpl22, ycf1, rps15, and rpl32) was observed between C. tagal and the Avicennia species. The highly divergent regions were also found in most intergenic regions, especially between the mangrove species in the family Rhizophoraceae and Avicennia species.
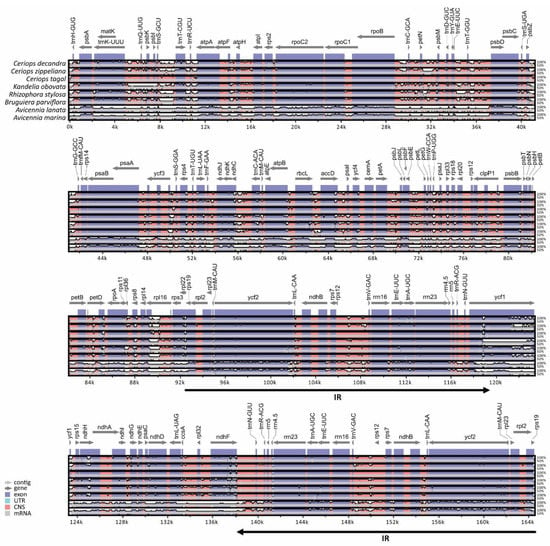
Figure 2.
Alignment map of 8 mangrove chloroplast genomes in Rhizophoraceae and Acanthaceae. Horizontal axis is genome position, whereas vertical axis shows sequence identity. Grey arrows indicate genes and transcriptional directions, and black arrows indicate IR regions. Exons and conserved non-coding sequences (CNS) are shown in blue and red, respectively.
3.3. Chloroplast Boundary Structures
The chloroplast boundary structures of the LSC, SSC, and IRs were compared among the three Ceriops species and two Avicennia species (Figure 3). In all species, the ycf1 and ndhF genes are located at the boundary of SSC/IRb and SSC/IRa, respectively. The size of ycf1 is approximately 5800 bp for the Ceriops species and 5500 bp for the Avicennia species. The ycf1 gene is ~1400 bp away from the SSC/IRb border in the Ceriops species, whereas it is ~800 bp away in the Avicennia species. Additionally, the size of the ndhF gene is similar in all species (2231 bp in C. tagal, A. lanata, and A. marina; 2237 bp in C. decandra; and 2243 bp in C. zippeliana). The LSC/IRb and LSC/IRa junctions in the three Ceriops species positioned the rps19 gene and rps19 pseudogene, respectively (Figure 3A). In contrast, the LSC/IRb junction in the two Avicennia species positioned the ycf2 gene (Figure 3B). Notably, no gene stretches across the boundary between the LSC and IRa regions of the two Avicennia species. These results reveal that the contraction and expansion of both LSC/IRa and LSC/IRb boundary regions occurred in Avicennia and Ceriops species, respectively, during their evolution.
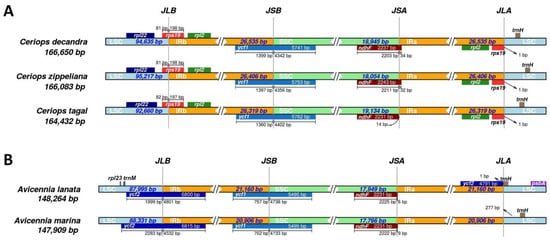
Figure 3.
Comparison of IR boundaries of chloroplast genomes. (A) IR boundaries among three Ceriops species. (B) IR boundaries among two Avicennia species.
3.4. Chloroplast Repeats and SSRs
Repeats in the chloroplast genomes of the three Ceriops species and A. lanata were identified (Figure 4A–C and Table S4). The number of forward, reverse, palindromic, and complement repeats was different in each species. For example, 23, 35, 25, and 17 forward repeats were found in C. decandra, C. zippeliana, C. tagal, and A. lanata, respectively (Figure 4A). The number of forward repeats in C. zippeliana (35) was the highest, while the number of palindromic repeats in A. lanata (20) was the highest (Figure 4A). There was no complement repeat in C. zippeliana. Interestingly, all Ceriops species contained long repeats (>30 bp), whereas A. lanata species consisted of short repeats (<40 bp) (Figure 4B). Usually, most repeats in all species were observed in the LSC region (Figure 4C).
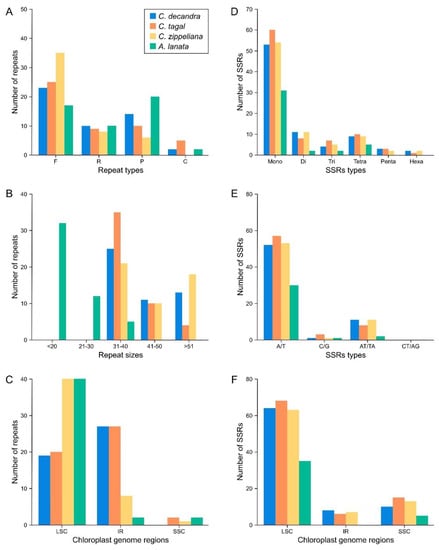
Figure 4.
Statistical analysis of repeats and SSRs in four mangrove chloroplast genomes. (A) Sorted by type of repeat. (B) Frequency by repeat types. (C) Sorted by repeat region of genome. (D) Sorted by type of SSR. (E) Frequency by SSR type. (F) Sorted by SSR region of genome.
SSRs in the chloroplast genomes of Ceriops species, A. lanata, and related mangrove species were analyzed (Figure 4D–F and Table 3 and Table S5). Mononucleotide SSRs were the most prevalent in all species (Figure 4D), consisting predominantly of A/T repeats, at over 90% (Figure 4E). Most SSRs were found in the LSC region (Figure 4F). Some SSRs were unique in each Ceriops species.

Table 3.
Number of SSRs in the chloroplast genomes of three Ceriops and two Avicennia species.
3.5. Ceriops Species Identification Based on Species-Specific Molecular Markers
Two pairs of primers were designed and tested to identify the differences between Ceriops species. PCR products of the chloroplast genomes exhibited different sizes among the three Ceriops species based on one molecular marker using two primer pairs (Figure 5). The PCR product of the two primer pairs confirmed the same variation. For the first one, the PCR product sizes of C. tagal, C. decandra, and C. zippeliana were 167, 207, and 226 bp, respectively (Figure 5 and Table S6). For the other one, the PCR product sizes of C. tagal, C. decandra, and C. zippeliana were 323, 363, and 382 bp, respectively (Figure 5 and Table S6). The difference in PCR product sizes occurred from indels and SSRs in the IR regions. For example, the chloroplast sequence of these regions of C. tagal and C. zippeliana contained 18 and 5 dinucleotide (AT/TA) repeats, respectively (102,313–102,348 and 154,744–154,779 bp: C. tagal; 120,115–120,124 and 156,400–156,409 bp: C. zippeliana) (Table S5), whereas there were no SSRs in these regions of C. decandra based on the SSR analysis criteria in this study due to short (AT/TA) repeats (<4 repeats) (Table S6).
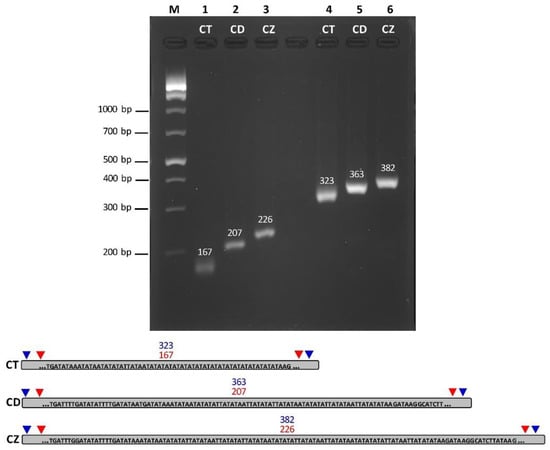
Figure 5.
Analysis of PCR products by 1% agarose gel electrophoresis with graphical genomes positing primer pairs. PCR products were amplified with two primer pairs in three Ceriops chloroplasts. Lane M: DNA size marker; Lane 1: PCR product amplified with the first primer set in C. tagal (CT: 167 bp); Lane 2: PCR product amplified with the first primer set in C. decandra (CD: 207 bp); Lane 3: PCR product amplified with the first primer set in C. zippeliana (CZ: 226 bp); Lane 4: PCR product amplified with the second primer set in C. tagal (323 bp); Lane 5: PCR product amplified with the second primer set in C. decandra (363 bp); Lane 6: PCR product amplified with the second primer set in C. zippeliana (382 bp). Graphical genomes show a part of the IR region used for designing specific primers of three Ceriops species based on different SSRs. Red arrows indicate the position of the first primer set, whereas blue arrows indicate the position of the second primer set.
3.6. Phylogenetic Relationships
The maximum likelihood (ML) analysis, based on 50 conserved chloroplast genes in 59 plant species, resulted in the best single tree (Figure 6). The ML tree shows two major clades corresponding to Rosids and Asterids. This tree highly supports that all Ceriops species are in the family Rhizophoraceae (Rosids), whereas A. lanata is in the family Acanthaceae (Asterids). C. decandra is closely related to C. zippeliana with a monophyletic branch supported by 100% bootstrap values. C. tagal is a sister species of the other two Ceriops species. For other mangrove species in the family Rhizophoraceae, Kandelia obovata is closer to the Ceriops species than Rhizophora and Bruguiera species. In addition, A. lanata and A. marina are grouped together in the family Acanthaceae (Asterids), with a bootstrap value of 100%.
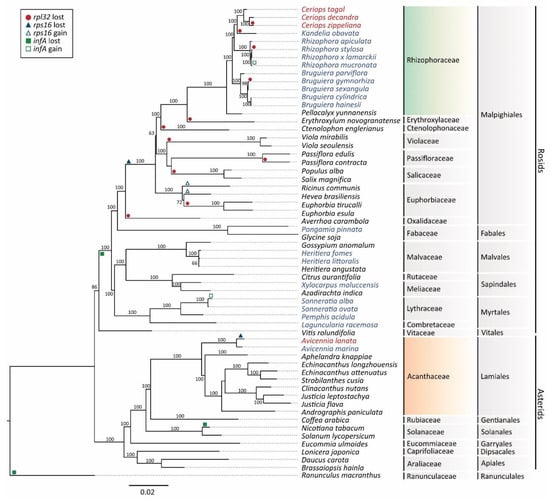
Figure 6.
Maximum likelihood (ML) tree for 50 chloroplast protein-coding genes in 59 plant species. Values above the branches represent bootstrap with 1000 replicates. The mangrove species in this study are indicated in red text, whereas other mangrove species are indicated in blue text. The Rhizophoraceae lineage is indicated in gradient green, and the Acanthaceae lineage is indicated in gradient orange. Gain and loss of the rpl32, rps16, and infA genes are shown in different symbols and colors.
The gain and loss of the rpl32, rps16, and infA genes in mangrove and non-mangrove species were plotted in the phylogenetic tree (Figure 6). For example, the rpl32 gene was lost in four mangrove species in Rhizophoraceae, namely C. zippeliana, K. obovata, Rhizophora stylosa, and Bruguiera gymnorhiza. The rps6 gene was lost in all mangrove species in Rhizophoraceae and most land plant species in Malpighiales, but not in Acanthaceae (Lamiales). The infA gene was also lost in all mangrove species in Rhizophoraceae and most land plant species in Rosids.
3.7. Chloroplast Genes under Positive Selection
To identify candidate genes under positive selection, the values of Ka/Ks (non-synonymous/synonymous) were estimated for 61 conserved chloroplast protein-coding genes in Ceriops and Avicennia species to relative mangrove species and non-mangrove species (assumed ancestors) (Figure 7 and Table S7). Most Ka/Ks ratios were lower than 1.0. However, there were two genes, rps7 and rps15, in which the Ka/Ks ratios were greater than 1.0 in several compared species pairs, suggesting positive selection during their evolution. The rps7 gene was under positive selection in both Ceriops species and A. lanata compared with relative non-mangrove species. The average Ka/Ks ratio of the rps7 gene between the Ceriops species compared with Ctenolophon englerianus, Averrhoa carambola, and Vitis rolundifolia was 1.06, 1.11, and 1.81, respectively. The Ka/Ks ratio of the rps7 gene between A. lanata compared with Eucommia ulmoides and Lonicera japonica was 1.03 and 1.13, respectively. In addition, the rps15 gene was positively selected in C. decandra and C. zippeliana. The Ka/Ks average ratio of the rps15 gene between the two Ceriops species compared with Pellacalyx yunnanensis, B. parviflora, and R. apiculata was 1.16, 1.17, and 1.42, respectively.
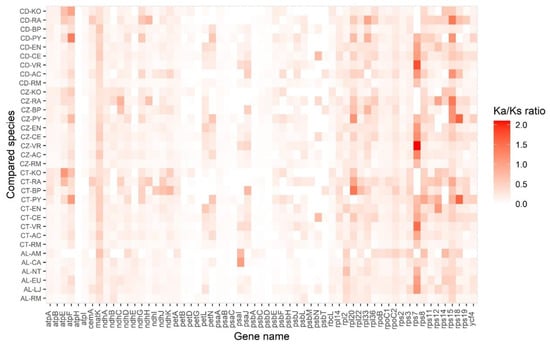
Figure 7.
Heatmap of Ka/Ks ratios between every compared species in 61 chloroplast genes. The scale ratios associated with each value are shown in the key beside the figure. AL (Avicennia lanata), AM (Avicennia marina), CA (Coffea arabica), EU (Eucommia ulmoides), LJ (Lonicera japonica), NT (Nicotiana tabacum), RM (Ranunculus macranthus), CD (Ceriops decandra), CZ (Ceriops zippeliana), CT (Ceriops tagal), AC (Averrhoa carambola), BP (Bruguiera parviflora), CE (Ctenolophon englerianus), EN (Erythroxylum novogranatense), KO (Kandelia obovata), PY (Pellacalyx yunnanensis), and RA (Rhizophora apiculata).
4. Discussion
Diverse chloroplast genome sequences have been used to study the evolution of mangrove species and to identify different mangrove species [,,,]. In the current study, we reported the chloroplast genomes of four mangrove species, including three Ceriops species (C. decandra, C. zippeliana, and C. tagal) and Avicennia lanata. Based on morphological characteristics, Ceriops is classified to the family Rhizophoraceae of the order Rosids (polypetalous), whereas Avicennia belongs to the family Acanthaceae of the order Asterids (sympetalous) [,]. Ceriops and Avicennia have a convergent evolution and are the most dominant species in the middle and seaward zones of mangrove forests, respectively [,,]. The three Ceriops chloroplast genomes (164.4–166.7 kb) were slightly different, consistent with published chloroplast genomes of mangrove species (middle zone) in Rhizophoraceae such as C. tagal, Kandelia obovata, Rhizophora species, and Bruguiera species (160.3–164.6 kb) [,,,,,]. In contrast, the smaller chloroplast genome of A. lanata was 148.2 kb, which is similar to the previously reported chloroplast genome of Avicennia marina (147.9–152.3 kb) [,]. In addition, the chloroplast genomes of Sonneratia alba and Sonneratia apetala, which are true mangroves in the family Lythraceae of the order Rosids in the seaward zone, were approximately 153.1 kb [,]. This finding suggests that the size of mangrove chloroplast genomes in the seaward zone may be compact compared with mangrove species in the middle zone, which is caused by adaptation under coastal stress conditions, especially salt stress. Salinity can affect plants in several ways, such as by changing the chloroplast size, number, lamellar organization, and lipid and starch accumulation and interfering with cross-membrane transportation [].
Chloroplast genomes are usually conserved in genome organization, gene order, and gene content []. Nevertheless, gene gain and loss have been found among the four mangrove species. The infA gene (translation initiation factor 1) was found in A. lanata but not in the Ceriops species and other mangrove species in Rhizophoraceae [,,]. The loss of the infA gene from the chloroplast to the nucleus occurred independently in multiple angiosperm lineages, especially in Rosids [,]. The rpl16 and rps16 genes became pseudogenes in A. lanata but not in A. marina []. The rpl16 gene has been independently pseudogenized in several angiosperm lineages across eudicots and monocots [,,]. Notably, the rps16 gene was not found in the three Ceriops species, consistent with other mangrove and land plant species in the order Malpighiales [,,]. The rps16 gene has been a pseudogene or lost by the nuclear encoded rps16 in many higher plants [,,,]. Three genes, namely rpl2, rpl23, and trnM-CAU, retained one copy in the LSC region of A. lanata and were found in only a single copy in the LSC region of A. marina []. In contrast, the three genes are located in the IR regions in the Ceriops species; thus, they have two copies, concordant with other mangrove species in Rhizophoraceae [,,]. Contraction at the LSC/IR junction, which was observed in several land plants, might result in the deletion of rpl2 and rpl23 from one of the IR regions [,]. Remarkably, rpl32 was lost in C. zippeliana but not the other Ceriops species and A. lanata. The loss of rpl32 has occurred in many mangrove species in the family Rhizophoraceae, such as K. obovata, R. stylosa, and B. gymnorhiza [,]. Transfer of chloroplast rpl32 to the nucleus DNA occurred independently in several families of Malpighiales plants, such as Rhizophoraceae, Erythroxylaceae, Ctenolophonaceae, Violaceae, Passifloraceae, Salicaceae, and Euphorbiaceae [,,,,,,]. These reveal gene evolution in Ceriops, Avicennia, and other mangrove species.
The border positions of the LSC, SSC, and IR regions were compared among the Ceriops and Avicennia chloroplast genomes. The boundaries of the LSC/IRa and LSC/IRb regions between the Ceriops species and A. lanata had different gene positioning. In the Ceriops species, the rps9 gene was located at the LSC/IRb border and the rps9 pseudogene was located at the LSC/IRa border, concordant with other mangrove species such as Bruguiera species []. Meanwhile, the ycf2 gene was located at the LSC/IRb border in A. lanata and no gene was located at the LSC/IRa border, which was similar with A. marina and some non-mangrove species in the Acanthaceae, such as Ruellia breedlovei (KP300014) [,]. One of the reasons for chloroplast genome variation among angiosperms is the contraction or expansion of the IR regions []. These indicated that the contraction of the IR regions in A. lanata and the expansion of the IR regions in the Ceriops species may be mainly caused by decreasing and increasing gene duplications in the IR regions, respectively, during their evolution.
Repeats of the four mangrove species varied among them. The occurrence of short repeats (<40 bp) and a small number of SSRs was found in A. lanata due to a compact chloroplast genome containing small non-coding regions. The mangrove species carry mostly forward repeats in their chloroplast genomes that are similar in other mangrove chloroplast genomes [,,]. For SSRs, most consist of repetitions of an A/T mononucleotide in all four mangrove species, concordant with other mangrove species [,,]. SSRs with repeat length differences occur from the process of mutation [], which could be used for identifying related species. In general, differentiation between C. decandra and C. zippeliana based on morphology can be difficult; therefore, we designed two species-specific primer sets based on one different SSR in the IR regions among the three Ceriops species. The specific primer sets were tested and could be used to identify C. decandra and C. zippeliana as well as C. tagal.
C. decandra is more closely related to C. zippeliana than to C. tagal, concordant with the results based on morphological and molecular evidence as well as the phylogenetic tree based on the trnL intron sequence of chloroplast genomes []. The Ceriops species are more closely related to K. obovata than other mangrove species in Rhizophoraceae, consistent with the results based on 44 conserved genes in 71 species (14 mangrove species and 57 land plant species) using Bayesian inference (BI) and ML []. In addition, the chloroplast genome of two Avicennia species was shown to be closely related to Acanthaceae, with a bootstrap value of 100%. These two lineages (Ceriops and Avicennia) had paraphyletic clades of the phylogeny, indicating convergent evolution.
The low average Ka/Ks ratios of most conserved genes in the four mangrove species suggest that the whole-chloroplast protein level of the species has been subjected to strong purifying selections. In general, synonymous changes (Ks) occur more often than non-synonymous substitutions (Ka); as a result, the ratios of Ka/Ks are commonly lower than 1.0 []. Remarkably, the Ka/Ks ratios of two chloroplast genes (rps7 and rps15) were greater than 1.0, suggesting positive selection pressure. The rps7 gene encodes ribosomal protein S7 involved in the regulation of chloroplast translation []. Positive selection on the rps7 gene has also been observed in many mangrove species (such as K. obovata, Rhizophora species, and Bruguiera species) and some land plants (such as Ananas comosus (pineapple)) [,,]. Moreover, the rps15 gene encoding ribosomal protein S15 was under positive selection in C. decandra and C. zippeliana. The multiple sequence alignment result showed co-variation of three sites in the rps15 amino acid sequence that occurs in C. decandra and C. zippeliana but not in C. tagal (Figure S1). Interestingly, one amino acid site at position 75 was unique in only C. decandra and C. zippeliana (Isoleucine) compared with mangrove and non-mangrove species (Valine) in the family Rhizophoraceae. The rps15 gene was also reported to be related to evolution under positive selection in Araliaceae species []. Knockout of the chloroplast rps15 gene in tobacco leads to a specific reduction in small 30S ribosomal subunits []. Thus, these genes, rps7 and rps15, might be undergoing adaptive evolution in response to stress environments in mangrove forests.
5. Conclusions
In this study, the complete chloroplast genome sequences of Ceriops decandra, C. zippeliana, C. tagal, and Avicennia lanata were sequenced and compared. The chloroplast genome of A. lanata (seaward zone) is compact compared with the three Ceriops species (middle zone). The chloroplast genomes are mostly conserved in genome organization, gene order, and gene content; however, gene gain and loss have been found among them. The occurrence of contraction or expansion of IR regions in Avicennia and Ceriops species would be a result of decreasing and increasing gene duplications in the IR regions, respectively. Phylogenetic analysis showed that C. decandra is closer to C. zippeliana than to C. tagal in the family Rhizophoraceae, and A. lanata is clustered with A. marina in the family Acanthaceae, which supports convergent evolution between the two genera. The different chloroplast repeats and SSRs in the four mangrove species can be used as genetic markers, and two species-specific primer sets have been developed for species identification among the three Ceriops species in this work. The rps7 gene was identified under positive selection among mangrove species and might correlate with adaptive selection under coastal environments. Hence, these results could not only provide valuable genetic information of mangrove Ceriops and Avicennia species but also offer molecular markers for species identification and a candidate gene in response to climatic stress conditions of coastal environments.
Supplementary Materials
The following supporting information can be downloaded at https://www.mdpi.com/article/10.3390/biology11030383/s1: Figure S1: Multiple protein alignment of rps15 genes among 19 species; Table S1: Sample locations and Illumina raw reads of four mangrove species; Table S2: List of chloroplast accession numbers and genes for phylogenetic analysis; Table S3: List of chloroplast accession numbers and genes for gene selective pressure analysis; Table S4: Repeats in the chloroplast genomes of Ceriops and Avicennia species; Table S5: SSRs in the chloroplast genomes of Ceriops and Avicennia species; Table S6: List of specific sequences in a IR region in three Ceriops species; Table S7: Ka/Ks values between Ceriops species and assumed ancestors as well as between Avicennia species and assumed ancestors.
Author Contributions
P.R.-a., W.P., and S.T. designed the experiment and revised the manuscript; P.R.-a. prepared the original draft; P.R.-a. and W.K. analyzed the data; T.Y. and P.P. prepared the material and performed the formal analysis; C.M., W.M., and D.J. validated the data. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by the National Science and Technology Development Agency (P1952261), Thailand.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
The chloroplast genome sequences of Ceriops decandra, Ceriops zippeliana, Ceriops tagal, and Avicennia lanata were submitted to the National Center for Biotechnology Information (NCBI), with the accession numbers OK272497, OK272496, OK258322, and OK258321, respectively.
Acknowledgments
We thank the research team from Mangrove Forest Research Center for sample collection.
Conflicts of Interest
The authors declare no conflict of interest.
References
- FAO. The world’s mangroves 1980–2005. In FAO Forestry Paper; FAO: Rome, Italy, 2007. [Google Scholar]
- Lee, S.Y.; Primavera, J.H.; Dahdouh-Guebas, F.; Mckee, K.; Bosire, J.O.; Cannicci, S.; Diele, K.; Fromard, F.; Koedam, N.; Marchand, C.; et al. Ecological role and services of tropical mangrove ecosystems: A reassessment. Glob. Ecol. Biogeogr. 2014, 23, 726–743. [Google Scholar] [CrossRef]
- Spalding, M.; Blasco, F.; Field, C. World Mangrove Atlas; The International Society for Mangrove Ecosystems: Okinawa, Japan, 2010. [Google Scholar]
- Ellison, A.M.; Farnsworth, E.J. Mangrove Communities. In Marine Community Ecology; Bertness, M.D., Gaines, S.D., Hay, M.E., Eds.; Sinauer Associates: Sunderland, MA, USA, 2001. [Google Scholar]
- Kathiresan, K.; Bingham, B.L. Biology of mangroves and mangrove ecosystems. Adv. Mar. Biol. 2001, 40, 81–251. [Google Scholar] [CrossRef]
- Vannucci, M. What is so special about mangroves? Braz. J. Biol. 2001, 61, 599–603. [Google Scholar] [CrossRef] [PubMed]
- Tomlinson, P.B. The Botany of Mangroves, 2nd ed.; Cambridge University Press: Cambridge, UK, 1986. [Google Scholar]
- Duke, N.C.; Maynecke, J.-O.; Dittmann, S.; Ellison, A.M.; Anger, K.; Berger, U.; Cannicci, S.; Diele, K.; Ewel, K.C.; Field, C.D.; et al. A world without mangroves? Science 2007, 317, 41–43. [Google Scholar] [CrossRef]
- Polidoro, B.A.; Carpenter, K.E.; Collins, L.; Duke, N.C.; Ellison, A.M.; Ellison, J.C.; Farnsworth, E.J.; Fernando, E.S.; Kathiresan, K.; Koedam, N.E.; et al. The loss of species: Mangrove extinction risk and geographic areas of global concern. PLoS ONE 2010, 5, e10095. [Google Scholar] [CrossRef]
- Ward, R.D.; Friess, D.A.; Day, R.H.; Mackenzie, R.A. Impacts of climate change on mangrove ecosystems: A region by region overview. Ecosyst. Health Sustain. 2016, 2, e01211. [Google Scholar] [CrossRef]
- Saddhe, A.A.; Jamdade, R.A.; Kumar, K. Assessment of mangroves from Goa, west coast India using DNA barcode. Springerplus 2016, 5, 1554. [Google Scholar] [CrossRef] [PubMed]
- Sandilyan, S.; Kathiresan, K. Mangrove conservation: A global perspective. Biodivers. Conserv. 2012, 21, 3523–3542. [Google Scholar] [CrossRef]
- Shi, S.; Huang, Y.; Zeng, K.; Tan, F.; He, H.; Huang, J.; Fu, Y. Molecular phylogenetic analysis of mangroves: Independent evolutionary origins of vivipary and salt secretion. Mol. Phylogenet. Evol. 2005, 34, 159–166. [Google Scholar] [CrossRef]
- Wee, A.K.S.; Mori, G.M.; Lira, C.F.; Núñez-Farfán, J.; Takayama, K.; Faulks, L.; Shi, S.; Tsuda, Y.; Suyama, Y.; Yamamoto, T.; et al. The integration and application of genomic information in mangrove conservation. Conserv. Biol. 2019, 33, 206–209. [Google Scholar] [CrossRef]
- Shearman, J.R.; Naktang, C.; Sontirod, C.; Kongkachana, W.; U-thoomporn, S.; Jomchai, N.; Maknual, C.; Yamprasai, S.; Promchoo, W.; Ruang-areerate, P.; et al. Assembly of a hybrid mangrove, Bruguiera hainesii, and its two ancestral contributors, Bruguiera cylindrica and Bruguiera gymnorhiza. Genomics, 2022; in press. [Google Scholar]
- Hu, M.J.; Sun, W.H.; Tsai, W.C.; Xiang, S.; Lai, X.K.; Chen, D.Q.; Liu, X.D.; Wang, Y.F.; Le, Y.X.; Chen, S.M.; et al. Chromosome-scale assembly of the Kandelia obovata genome. Hortic. Res. 2020, 7, 75. [Google Scholar] [CrossRef] [PubMed]
- Xu, S.; He, Z.; Zhang, Z.; Guo, Z.; Guo, W.; Lyu, H.; Li, J.; Yang, M.; Du, Z.; Huang, Y.; et al. The origin, diversification and adatation of a major mangrove clade (Rhizophoraceae) revealed by whole-genome sequencing. Natl. Sci Rev. 2017, 4, 721–734. [Google Scholar] [CrossRef]
- Natarajan, P.; Murugesan, A.K.; Govindan, G.; Gopalakrishnan, A.; Kumar, R.; Duraisamy, P.; Balaji, R.; Tanuja; Shyamli, P.S.; Parida, A.K.; et al. A reference-grade genome identifies salt-tolerance genes from the salt-secreting mangrove species Avicennia marina. Commun. Biol. 2021, 4, 851. [Google Scholar] [CrossRef]
- Pootakham, W.; Sonthirod, C.; Naktang, C.; Kongkachana, W.; U-thoomporn, S.; Phetchawang, P.; Maknual, C.; Jiumjamrassil, D.; Pravinvongvuthi, T.; Tangphatsornruang, S. A de novo reference assembly of the yellow mangrove Ceriops zippeliana genome. G3 Genes Genomes Genet. 2022; in press. [Google Scholar]
- Pootakham, W.; Naktang, C.; Sonthirod, C.; Kongkachana, W.; Yoocha, T.; Jomchai, N.; Maknual, C.; Chumriang, P.; Pravinvongvuthi, T.; Tangphatsornruang, S. De novo reference assembly of the upriver orange mangrove (Bruguiera sexangula) genome. Genome Biol. Evol. 2022; submitted. [Google Scholar]
- Pootakham, W.; Naktang, C.; Sonthirod, C.; Kongkachana, W.; Narong, N.; Sangsrakru, D.; Maknual, C.; Jiumjamrassil, D.; Chumriang, P.; Tangphatsornruang, S. Chromosome-level genome assembly and population genetic analysis of the Indian mangrove (Ceriops tagal). Hortic. Res. 2022; submitted. [Google Scholar]
- Pootakham, W.; Sonthirod, C.; Naktang, C.; Kongkachana, W.; Sangsrakru, D.; U-thoomporn, S.; Maknual, C.; Meepol, W.; Promchoo, W.; Maprasop, P.; et al. A chromosome-scale reference genome assembly of yellow mangrove (Bruguiera parviflora) reveals a whole genome duplication event associated with the Rhizophoraceae lineage. Mol. Ecol. Resour. 2022, 2022, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Pootakham, W.; Kongkachana, W.; Naktang, C.; Sonthirod, C.; Yoocha, T.; Jomchai, N.; Maknual, C.; Meepol, W.; Yamprasai, S.; Tangphatsornruang, S. A chromosome-scale genome assembly of the Ceriops decandra mangrove. BMC Genom. Data, 2022; submitted. [Google Scholar]
- Giesen, W.; Wulffraat, S.; Zieen, M.; Scholten, L. Mangrove Guidebook for Southeast Asia; FAO and Wetlands International: Bangkok, Thailand, 2006. [Google Scholar]
- Raju, A.J.S.; Karyamsetty, H.J. Reproductive ecology of mangrove trees Ceriops tagal (Perr.) C. B. Robinson (Rhizophoraceae). Acta Bot. Croat. 2008, 67, 201–208, ISSN 0365-0588. [Google Scholar]
- Thatoi, H.; Samantaray, D.; Das, S.K. The genus Avicennia, a pioneer group of dominant mangrove plant species with potential medicinal values: A review. Front. Life Sci. 2016, 9, 267–291. [Google Scholar] [CrossRef]
- Shimony, C.; Fahn, A.; Reinhold, L. Ultrastructure and ion gradients in the salt glands of Avicennia marina (Forssk.) Vierh. New Phytol. 1973, 72, 27–36. [Google Scholar] [CrossRef]
- Duke, N. Australia’s Mangroves: The Authoritative Guide to Australia’s Mangrove Plants; University of Queensland: Brisbane, Australia, 2006; ISBN 9780646461960. [Google Scholar]
- Ding, H. Rhizophoraceae. Flora Males. Ser. 1 Spermatophyta 1958, 5, 429–493. [Google Scholar]
- Sheue, C.R.; Rashid, S.M.A.; Yong, J.W.H.; Yang, Y.P. Ceriops zippeliana Blume (Rhizophoraceae), a new record of a mangrove species in Singapore. Taiwania 2010, 55, 72–77. [Google Scholar] [CrossRef]
- Sheue, C.R.; Liu, H.Y.; Tsai, C.C.; Rashid, S.M.A.; Yong, J.W.H.; Yang, Y.P. On the morphology and molecular basis of segregation of Ceriops zippeliana and C. decandra (Rhizophoraceae) from Asia. Blumea J. Plant Taxon. Plant Geogr. 2009, 54, 220–227. [Google Scholar] [CrossRef][Green Version]
- Huang, Y.; Tan, F.; Su, G.; Deng, S.; He, H.; Shi, S. Population genetic structure of three tree species in the mangrove genus Ceriops (Rhizophoraceae) from the Indo West Pacific. Genetica 2008, 133, 47–56. [Google Scholar] [CrossRef] [PubMed]
- Duke, N.; Kathiresan, K.; Salmo, S.G., III; Fernando, E.S.; Peras, J.R.; Sukardjo, S.; Miyagi, T. Ceriops decandra. IUCN Red List Threat. Species 2010, 2010, e.T178853A7627935. [Google Scholar]
- Duke, N.C. A systematic revision of the mangrove genus avicennia (Avicenniaceae) in Australasia. Aust. Syst. Bot. 1991, 4, 299–324. [Google Scholar] [CrossRef]
- Mori, G.M.; Zucchi, M.I.; Sampaio, I.; Souza, A.P. Species distribution and introgressive hybridization of two Avicennia species from the Western Hemisphere unveiled by phylogeographic patterns Phylogenetics and phylogeography. BMC Evol. Biol. 2015, 15, 61. [Google Scholar] [CrossRef] [PubMed]
- Duke, N.C. Mangrove floristics and biogeography. Trop. Mangrove Ecosyst. 1992, 41, 63–100. [Google Scholar] [CrossRef]
- Chua, L.S. Avicennia lanata. IUCN Red List Threat. Species 1998, 1998, e.T31819A9662485. [Google Scholar]
- Birky, C.W. Uniparental inheritance of mitochondrial and chloroplast genes: Mechanisms and evolution. Proc. Natl. Acad. Sci. USA 1995, 92, 11331–11338. [Google Scholar] [CrossRef]
- Asaf, S.; Khan, A.L.; Numan, M.; Al-Harrasi, A. Mangrove tree (Avicennia marina): Insight into chloroplast genome evolutionary divergence and its comparison with related species from family Acanthaceae. Sci. Rep. 2021, 11, 3586. [Google Scholar] [CrossRef]
- Yang, Y.; Zhang, Y.; Chen, Y.; Gul, J.; Zhang, J.; Liu, Q.; Chen, Q. Complete chloroplast genome sequence of the mangrove species Kandelia obovata and comparative analyses with related species. PeerJ 2019, 7, e7713. [Google Scholar] [CrossRef]
- Shi, C.; Han, K.; Li, L.; Seim, I.; Lee, S.M.Y.; Xu, X.; Yang, H.; Fan, G.; Liu, X. Complete chloroplast genomes of 14 mangroves: Phylogenetic and comparative genomic analyses. Biomed Res. Int. 2020, 2020, 8731857. [Google Scholar] [CrossRef] [PubMed]
- Ruang-areerate, P.; Kongkachana, W.; Naktang, C.; Sonthirod, C.; Narong, N.; Jomchai, N.; Maprasop, P.; Maknual, C.; Phormsin, N.; Shearman, J.R.; et al. Complete chloroplast genome sequences of five Bruguiera species (Rhizophoraceae): Comparative analysis and phylogenetic relationships. PeerJ 2021, 9, 1–22. [Google Scholar] [CrossRef] [PubMed]
- Tangphatsornruang, S.; Sangsrakru, D.; Chanprasert, J.; Uthaipaisanwong, P.; Yoocha, T.; Jomchai, N.; Tragoonrung, S. The chloroplast genome sequence of mungbean (Vigna radiata) determined by high-throughput pyrosequencing: Structural organization and phylogenetic relationships. DNA Res. 2010, 17, 11–22. [Google Scholar] [CrossRef]
- Tangphatsornruang, S.; Uthaipaisanwong, P.; Sangsrakru, D.; Chanprasert, J.; Yoocha, T.; Jomchai, N.; Tragoonrung, S. Characterization of the complete chloroplast genome of Hevea brasiliensis reveals genome rearrangement, RNA editing sites and phylogenetic relationships. Gene 2011, 475, 104–112. [Google Scholar] [CrossRef] [PubMed]
- Uthaipaisanwong, P.; Chanprasert, J.; Shearman, J.R.; Sangsrakru, D.; Yoocha, T.; Jomchai, N.; Jantasuriyarat, C.; Tragoonrung, S.; Tangphatsornruang, S. Characterization of the chloroplast genome sequence of oil palm (Elaeis guineensis Jacq.). Gene 2012, 500, 172–180. [Google Scholar] [CrossRef]
- Doyle, J.J.; Doyle, J.L. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem. Bull. 1987, 19, 11–15. [Google Scholar]
- Dierckxsens, N.; Mardulyn, P.; Smits, G. NOVOPlasty: De novo assembly of organelle genomes from whole genome data. Nucleic Acids Res. 2017, 45, e18. [Google Scholar] [CrossRef]
- Jin, J.J.; Yu, W.B.; Yang, J.B.; Song, Y.; DePamphilis, C.W.; Yi, T.S.; Li, D.Z. GetOrganelle: A fast and versatile toolkit for accurate de novo assembly of organelle genomes. Genome Biol. 2020, 21, 241. [Google Scholar] [CrossRef]
- Freudenthal, J.A.; Pfaff, S.; Terhoeven, N.; Korte, A.; Ankenbrand, M.J.; Förster, F. A systematic comparison of chloroplast genome assembly tools. Genome Biol. 2020, 21, 254. [Google Scholar] [CrossRef]
- Tillich, M.; Lehwark, P.; Pellizzer, T.; Ulbricht-Jones, E.S.; Fischer, A.; Bock, R.; Greiner, S. GeSeq—Versatile and accurate annotation of organelle genomes. Nucleic Acids Res. 2017, 45, W6–W11. [Google Scholar] [CrossRef]
- Laslett, D.; Canback, B. ARAGORN, A program to detect tRNA genes and tmRNA genes in nucleotide sequences. Nucleic Acids Res. 2004, 32, 11–16. [Google Scholar] [CrossRef] [PubMed]
- Greiner, S.; Lehwark, P.; Bock, R. OrganellarGenomeDRAW (OGDRAW) version 1.3.1: Expanded toolkit for the graphical visualization of organellar genomes. Nucleic Acids Res. 2019, 47, W59–W64. [Google Scholar] [CrossRef]
- Frazer, K.A.; Pachter, L.; Poliakov, A.; Rubin, E.M.; Dubchak, I. VISTA: Computational tools for comparative genomics. Nucleic Acids Res. 2004, 32, 273–279. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Yang, Y.; Li, J.; Jin, Y.; Liu, Q.; Zhang, Y. The complete chloroplast genome sequence of a medicinal mangrove tree Ceriops tagal and its phylogenetic analysis. Mitochondrial DNA Part B Resour. 2019, 4, 267–268. [Google Scholar] [CrossRef]
- Amiryousefi, A.; Hyvönen, J.; Poczai, P. IRscope: An online program to visualize the junction sites of chloroplast genomes. Bioinformatics 2018, 34, 3030–3031. [Google Scholar] [CrossRef]
- Kurtz, S.; Choudhuri, J.V.; Ohlebusch, E.; Schleiermacher, C.; Stoye, J.; Giegerich, R. REPuter: The manifold applications of repeat analysis on a genomic scale. Nucleic Acids Res. 2001, 29, 4633–4642. [Google Scholar] [CrossRef]
- Thiel, T.; Michalek, W.; Varshney, R.K.; Graner, A. Exploiting EST databases for the development and characterization of gene-derived SSR-markers in barley (Hordeum vulgare L.). Theor. Appl. Genet. 2003, 106, 411–422. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef]
- Stamatakis, A. RAxML version 8: A tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 2014, 30, 1312–1313. [Google Scholar] [CrossRef]
- Edgar, R.C. MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004, 32, 1792–1797. [Google Scholar] [CrossRef]
- Wang, D.; Zhang, Y.; Zhang, Z.; Zhu, J.; Yu, J. KaKs_Calculator 2.0: A toolkit incorporating gamma-series methods and sliding window strategies. Genom. Proteom. Bioinform. 2010, 8, 77–80. [Google Scholar] [CrossRef]
- Mo, Z.; Lou, W.; Chen, Y.; Jia, X.; Zhai, M.; Guo, Z.; Xuan, J. The chloroplast genome of Carya illinoinensis: Genome structure, adaptive evolution, and phylogenetic analysis. Forests 2020, 11, 207. [Google Scholar] [CrossRef]
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2021; Volume 1, ISBN 3900051070. [Google Scholar]
- Untergasser, A.; Cutcutache, I.; Koressaar, T.; Ye, J.; Faircloth, B.C.; Remm, M.; Rozen, S.G. Primer3-new capabilities and interfaces. Nucleic Acids Res. 2012, 40, e115. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Zhong, J.; Yuan, C.C. Complete chloroplast genome of a mangrove natural hybrid, Rhizophora × lamarckii. Mitochondrial DNA Part B Resour. 2019, 4, 1465–1466. [Google Scholar] [CrossRef]
- Li, Y.C.; Li, S.Y.; Zhang, T.H.; Qin, L.L.; An, Y.D.; Pang, Y.K.; Jiang, G.F. Characterization of the complete chloroplast genome of mangrove Rhizophora apiculata Blume (Rhizophoraceae). Mitochondrial DNA Part B Resour. 2021, 6, 2071–2073. [Google Scholar] [CrossRef]
- Li, C.T.; Guo, P.; Huang, H.R.; Pei, N.C.; Shi, M.M.; Yan, H.F. The complete chloroplast genome of Rhizophora stylosa and its phylogenetic implications. Mitochondrial DNA Part B Resour. 2019, 4, 374–375. [Google Scholar] [CrossRef]
- Li, H.; Ma, D.; Li, J.; Wei, M.; Zheng, H.; Zhu, X. Illumina sequencing of complete chloroplast genome of Avicennia marina, a pioneer mangrove species. Mitochondrial DNA Part B Resour. 2020, 5, 2131–2132. [Google Scholar] [CrossRef]
- Li, C.T.; Pei, N.C.; Yin, Y.; Yan, H.F.; Shi, M.-M.; Huang, H.-R. The complete chloroplast genome of a true mangrove Sonneratia apetala and its phylogenetic implications. Mitochondrial DNA Part B Resour. 2019, 4, 160–161. [Google Scholar] [CrossRef]
- Yu, T.; Hinsinger, D.D.; Strijk, J.S.; Wee, A.K.S. The first complete chloroplast genome of a major mangrove species Sonneratia alba Sm. and its implications on conservation efforts. Mitochondrial DNA Part B Resour. 2018, 3, 500–502. [Google Scholar] [CrossRef]
- Hameed, A.; Ahmed, M.Z.; Hussain, T.; Aziz, I.; Ahmad, N.; Gul, B.; Nielsen, B.L. Effects of salinity stress on chloroplast structure and function. Cells 2021, 10, 2023. [Google Scholar] [CrossRef]
- Daniell, H.; Lin, C.S.; Yu, M.; Chang, W.J. Chloroplast genomes: Diversity, evolution, and applications in genetic engineering. Genome Biol. 2016, 17, 134. [Google Scholar] [CrossRef]
- Jansen, R.K.; Cai, Z.; Raubeson, L.A.; Daniell, H.; Depamphilis, C.W.; Leebens-Mack, J.; Müller, K.F.; Guisinger-Bellian, M.; Haberle, R.C.; Hansen, A.K.; et al. Analysis of 81 genes from 64 plastid genomes resolves relationships in angiosperms and identifies genome-scale evolutionary patterns. Proc. Natl. Acad. Sci. USA 2007, 104, 19369–19374. [Google Scholar] [CrossRef] [PubMed]
- Millen, R.S.; Olmstead, R.G.; Adams, K.L.; Palmer, J.D.; Lao, N.T.; Heggie, L.; Kavanagh, T.A.; Hibberd, J.M.; Gray, J.C.; Morden, C.W.; et al. Many parallel losses of infa from chloroplast dna during angiosperm evolution with multiple independent transfers to the nucleus. Plant Cell 2001, 13, 645–658. [Google Scholar] [CrossRef] [PubMed]
- Kersten, B.; Rampant, P.F.; Mader, M.; Le Paslier, M.C.; Bounon, R.; Berard, A.; Vettori, C.; Schroeder, H.; Leplé, J.C.; Fladung, M. Genome sequences of Populus tremula chloroplast and mitochondrion: Implications for holistic poplar breeding. PLoS ONE 2016, 11, e0147209. [Google Scholar] [CrossRef]
- Li, Y.; Zhou, J.G.; Chen, X.L.; Cui, Y.X.; Xu, Z.C.; Li, Y.H.; Song, J.Y.; Duan, B.Z.; Yao, H. Gene losses and partial deletion of small single-copy regions of the chloroplast genomes of two hemiparasitic Taxillus species. Sci. Rep. 2017, 7, 12834. [Google Scholar] [CrossRef]
- Mehmood, F.; Abdullah; Ubaid, Z.; Shahzadi, I.; Ahmed, I.; Waheed, M.T.; Poczai, P.; Mirza, B. Plastid genomics of Nicotiana (Solanaceae): Insights into molecular evolution, positive selection and the origin of the maternal genome of Aztec tobacco (Nicotiana rustica). PeerJ 2020, 8, e9552. [Google Scholar] [CrossRef]
- Alqahtani, A.A.; Jansen, R.K. The evolutionary fate of rpl32 and rps16 losses in the Euphorbia schimperi (Euphorbiaceae) plastome. Sci. Rep. 2021, 11, 7466. [Google Scholar] [CrossRef]
- Guo, X.; Castillo-Ramírez, S.; González, V.; Bustos, P.; Fernández-Vázquez, J.L.; Santamaría, R.; Arellano, J.; Cevallos, M.A.; Dávila, G. Rapid evolutionary change of common bean (Phaseolus vulgaris L.) plastome, and the genomic diversification of legume chloroplasts. BMC Genom. 2007, 8, 228. [Google Scholar] [CrossRef]
- Ueda, M.; Nishikawa, T.; Fujimoto, M.; Takanashi, H.; Arimura, S.I.; Tsutsumi, N.; Kadowaki, K.I. Substitution of the gene for chloroplast RPS16 was assisted by generation of a dual targeting signal. Mol. Biol. Evol. 2008, 25, 1566–1575. [Google Scholar] [CrossRef]
- Raman, G.; Park, V.; Kwak, M.; Lee, B.; Park, S.J. Characterization of the complete chloroplast genome of Arabis stellari and comparisons with related species. PLoS ONE 2017, 12, e0183197. [Google Scholar] [CrossRef]
- Raman, G.; Park, S.; Lee, E.M.; Park, S.J. Evidence of mitochondrial DNA in the chloroplast genome of Convallaria keiskei and its subsequent evolution in the Asparagales. Sci. Rep. 2019, 9, 5028. [Google Scholar] [CrossRef] [PubMed]
- Abdullah; Henriquez, C.L.; Mehmood, F.; Hayat, A.; Sammad, A.; Waseem, S.; Waheed, M.T.; Matthews, P.J.; Croat, T.B.; Poczai, P.; et al. Chloroplast genome evolution in the Dracunculus clade (Aroideae, Araceae). Genomics 2021, 113, 183–192. [Google Scholar] [CrossRef] [PubMed]
- Henriquez, C.L.; Abdullah; Ahmed, I.; Carlsen, M.M.; Zuluaga, A.; Croat, T.B.; McKain, M.R. Evolutionary dynamics of chloroplast genomes in subfamily Aroideae (Araceae). Genomics 2020, 112, 2349–2360. [Google Scholar] [CrossRef] [PubMed]
- Khan, A.; Asaf, S.; Khan, A.L.; Shehzad, T.; Al-rawahi, A.; Al-harrasi, A. Comparative chloroplast genomics of endangered euphorbia species: Insights into hotspot divergence, repetitive sequence variation, and phylogeny. Plants 2020, 9, 199. [Google Scholar] [CrossRef]
- Okumura, S.; Sawada, M.; Park, Y.W.; Hayashi, T.; Shimamura, M.; Takase, H.; Tomizawa, K.I. Transformation of poplar (Populus alba) plastids and expression of foreign proteins in tree chloroplasts. Transgenic Res. 2006, 15, 637–646. [Google Scholar] [CrossRef]
- Shrestha, B.; Gilbert, L.E.; Ruhlman, T.A.; Jansen, R.K. Rampant nuclear transfer and substitutions of plastid genes in Passiflora. Genome Biol. Evol. 2020, 12, 1313–1329. [Google Scholar] [CrossRef]
- Ueda, M.; Fujimoto, M.; Arimura, S.I.; Murata, J.; Tsutsumi, N.; Kadowaki, K.I. Loss of the rpl32 gene from the chloroplast genome and subsequent acquisition of a preexisting transit peptide within the nuclear gene in Populus. Gene 2007, 402, 51–56. [Google Scholar] [CrossRef]
- Yaradua, S.S.; Alzahrani, D.A.; Albokhary, E.J.; Abba, A.; Bello, A. Complete chloroplast genome sequence of Justicia flava: Genome comparative analysis and phylogenetic relationships among Acanthaceae. Biomed Res. Int. 2019, 2019, 4370258. [Google Scholar] [CrossRef]
- Alzahrani, D.A.; Yaradua, S.S.; Yaradua, S.S.; Albokhari, E.J.; Albokhari, E.J.; Abba, A. Complete chloroplast genome sequence of Barleria prionitis, comparative chloroplast genomics and phylogenetic relationships among Acanthoideae. BMC Genom. 2020, 21, 393. [Google Scholar] [CrossRef]
- Zhu, A.; Guo, W.; Gupta, S.; Fan, W.; Mower, J.P. Evolutionary dynamics of the plastid inverted repeat: The effects of expansion, contraction, and loss on substitution rates. New Phytol. 2016, 209, 1747–1756. [Google Scholar] [CrossRef]
- Li, Y.C.; Korol, A.B.; Fahima, T.; Nevo, E. Microsatellites within genes: Structure, function, and evolution. Mol. Biol. Evol. 2004, 21, 991–1007. [Google Scholar] [CrossRef] [PubMed]
- Roth, C.; Liberles, D.A. A systematic search for positive selection in higher plants (Embryophytes). BMC Plant Biol. 2006, 6, 12. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Fargo, D.C.; Boynton, J.E.; Gillham, N.W. Chloroplast ribosomal protein S7 of chlamydomonas binds to chloroplast mRNA leader sequences and may be involved in translation initiation. Plant Cell 2001, 13, 207–218. [Google Scholar] [CrossRef]
- Redwan, R.M.; Saidin, A.; Kumar, S.V. Complete chloroplast genome sequence of MD-2 pineapple and its comparative analysis among nine other plants from the subclass Commelinidae. BMC Plant Biol. 2015, 15, 196. [Google Scholar] [CrossRef] [PubMed]
- Han, K.; Shi, C.; Li, L.; Seim, I.; Lee, S.M.Y.; Xu, X.; Yang, H.; Fan, G.; Liu, X. Lineage-specific evolution of mangrove plastid genomes. Plant Genome 2020, 13, e20019. [Google Scholar] [CrossRef] [PubMed]
- Kim, K.; Nguyen, V.B.; Dong, J.; Wang, Y.; Park, J.Y.; Lee, S.C.; Yang, T.J. Evolution of the Araliaceae family inferred from complete chloroplast genomes and 45S nrDNAs of 10 Panax-related species. Sci. Rep. 2017, 7, 4917. [Google Scholar] [CrossRef] [PubMed]
- Fleischmann, T.T.; Scharff, L.B.; Alkatib, S.; Hasdorf, S.; Schöttler, M.A.; Bock, R. Nonessential plastid-encoded ribosomal proteins in tobacco: A developmental role for plastid translation and implications for reductive genome evolution. Plant Cell 2011, 23, 3137–3155. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).