Probing Genome-Scale Model Reveals Metabolic Capability and Essential Nutrients for Growth of Probiotic Limosilactobacillus reuteri KUB-AC5
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Metabolic Network Construction of L. reuteri KUB-AC5
2.2. Model Development of L. reuteri KUB-AC5
2.3. Cultivation of L. reuteri KUB-AC5
2.4. Model Validation for L. reuteri KUB-AC5 Growth Using Flux Balance Analysis
2.5. Using GSMM of L. reuteri KUB-AC5 as a Scaffold for Growth Simulation and Integrative Transcriptomics Analysis
2.5.1. Identifying the Essential/Preferable Nutrients for L. reuteri KUB-AC5 Growth
2.5.2. Identifying Key Metabolic Routes for Enhancing L. reuteri KUB-AC5 Growth
3. Results and Discussion
3.1. Characteristics of the L. reuteri KUB-AC5 Model (iTN656) and Comparative Analysis with other Related Models
3.2. Phenotypic Characteristics of L. reuteri KUB-AC5 for in Silico Model Validation
3.3. Identifying the Essential Nutrients for L. reuteri KUB-AC5 Growth through Single and Double Omission Analysis
3.4. Alternation of Carbon Sources for Optimizing L. reuteri KUB-AC5 Growth and Biomass Production
3.5. Putative Metabolic Routes for Enhancing Growth of L. reuteri KUB-AC5
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Zheng, J.; Wittouck, S.; Salvetti, E.; Franz, C.; Harris, H.M.B.; Mattarelli, P.; O’Toole, P.W.; Pot, B.; Vandamme, P.; Walter, J.; et al. A taxonomic note on the genus Lactobacillus: Description of 23 novel genera, emended description of the genus Lactobacillus Beijerinck 1901, and union of Lactobacillaceae and Leuconostocaceae. Int. J. Syst. Evol. Microbiol. 2020, 70, 2782–2858. [Google Scholar] [CrossRef] [PubMed]
- Gourbeyre, P.; Denery, S.; Bodinier, M. Probiotics, prebiotics, and synbiotics: Impact on the gut immune system and allergic reactions. J. Leukoc. Biol. 2011, 89, 685–695. [Google Scholar] [CrossRef] [PubMed]
- Nitisinprasert, S.; Nilphai, V.; Bunyun, P.; Sukyai, P.; Doi, K.; Sonomoto, K. Screening and identification of effective thermotolerant lactic acid bacteria producing antimicrobial activity against Escherichia coli and Salmonella sp. resistant to antibiotics. Agric. Nat. Resour. 2000, 34, 387–400. [Google Scholar]
- Nakphaichit, M.; Thanomwongwattana, S.; Phraephaisarn, C.; Sakamoto, N.; Keawsompong, S.; Nakayama, J.; Nitisinprasert, S. The effect of including Lactobacillus reuteri KUB-AC5 during post-hatch feeding on the growth and ileum microbiota of broiler chickens. Poult. Sci. 2011, 90, 2753–2765. [Google Scholar] [CrossRef]
- Nakphaichit, M.; Sobanbua, S.; Siemuang, S.; Vongsangnak, W.; Nakayama, J.; Nitisinprasert, S. Protective effect of Lactobacillus reuteri KUB-AC5 against Salmonella Enteritidis challenge in chickens. Benef. Microbes 2019, 10, 43–54. [Google Scholar] [CrossRef] [PubMed]
- Buddhasiri, S.; Sukjoi, C.; Kaewsakhorn, T.; Nambunmee, K.; Nakphaichit, M.; Nitisinprasert, S.; Thiennimitr, P. Anti-inflammatory effect of probiotic Limosilactobacillus reuteri KUB-AC5 against Salmonella infection in a mouse colitis model. Front. Microbiol. 2021, 12, 2463. [Google Scholar] [CrossRef]
- Shah, N.P. Functional cultures and health benefits. Int. Dairy J. 2007, 17, 1262–1277. [Google Scholar] [CrossRef]
- Martins, E.M.F.; Ramos, A.M.; Vanzela, E.S.L.; Stringheta, P.C.; de Oliveira Pinto, C.L.; Martins, J.M. Products of vegetable origin: A new alternative for the consumption of probiotic bacteria. Food Res. Int. 2013, 51, 764–770. [Google Scholar] [CrossRef]
- Hébert, E.M.; Raya, R.R.; de Giori, G.S. Nutritional requirements of Lactobacillus delbrueckii subsp. lactis in a chemically defined medium. Curr. Microbiol. 2004, 49, 341–345. [Google Scholar] [CrossRef]
- Partanen, L.; Marttinen, N.; Alatossava, T. Fats and fatty acids as growth factors for Lactobacillus delbrueckii. Syst. Appl. Microbiol. 2001, 24, 500–506. [Google Scholar] [CrossRef]
- Hayek, S.A.; Gyawali, R.; Aljaloud, S.O.; Krastanov, A.; Ibrahim, S.A. Cultivation media for lactic acid bacteria used in dairy products. J. Dairy Res. 2019, 86, 490–502. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mardis, E.R. Next-generation DNA sequencing methods. Annu. Rev. Genom. Hum. Genet. 2008, 9, 387–402. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jatuponwiphat, T.; Namrak, T.; Supataragul, A.; Nitisinprasert, S.; Nakphaichit, M.; Vongsangnak, W. Comparative genome analysis reveals metabolic traits associated with probiotics properties in Lactobacillus reuteri KUB-AC5. Gene Rep. 2019, 17, 100536. [Google Scholar] [CrossRef]
- Jatuponwiphat, T.; Namrak, T.; Nitisinprasert, S.; Nakphaichit, M.; Vongsangnak, W. Integrative growth physiology and transcriptome profiling of probiotic Limosilactobacillus reuteri KUB-AC5. PeerJ 2021, 9, e12226. [Google Scholar] [CrossRef]
- Teusink, B.; Wiersma, A.; Molenaar, D.; Francke, C.; de Vos, W.M.; Siezen, R.J.; Smid, E.J. Analysis of growth of Lactobacillus plantarum WCFS1 on a complex medium using a genome-scale metabolic model. J. Biol. Chem. 2006, 281, 40041–40048. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Flahaut, N.A.; Wiersma, A.; van de Bunt, B.; Martens, D.E.; Schaap, P.J.; Sijtsma, L.; Dos Santos, V.A.; de Vos, W.M. Genome-scale metabolic model for Lactococcus lactis MG1363 and its application to the analysis of flavor formation. Appl. Microbiol. Biotechnol. 2013, 97, 8729–8739. [Google Scholar] [CrossRef] [PubMed]
- Kristjansdottir, T.; Bosma, E.F.; Branco Dos Santos, F.; Ozdemir, E.; Herrgard, M.J.; Franca, L.; Ferreira, B.; Nielsen, A.T.; Gudmundsson, S. A metabolic reconstruction of Lactobacillus reuteri JCM 1112 and analysis of its potential as a cell factory. Microb Cell Fact. 2019, 18, 186. [Google Scholar] [CrossRef] [Green Version]
- Luo, H.; Li, P.; Wang, H.; Roos, S.; Ji, B.; Nielsen, J. Genome-scale insights into the metabolic versatility of Limosilactobacillus reuteri. BMC Biotechnol. 2021, 21, 46. [Google Scholar] [CrossRef]
- Henikoff, S.; Henikoff, J.G. Amino acid substitution matrices from protein blocks. Proc. Natl. Acad. Sci. USA 1992, 89, 10915–10919. [Google Scholar] [CrossRef] [Green Version]
- Nguyen, N.N.; Vongsangnak, W.; Shen, B.; Nguyen, P.V.; Leong, H. Megafiller: A retrofitted protein function predictor for filling gaps in metabolic networks. J. Proteom. Bioinf. 2014, S9, 1–11. [Google Scholar] [CrossRef] [Green Version]
- Wang, H.; Marcisauskas, S.; Sanchez, B.J.; Domenzain, I.; Hermansson, D.; Agren, R.; Nielsen, J.; Kerkhoven, E.J. RAVEN 2.0: A versatile toolbox for metabolic network reconstruction and a case study on Streptomyces coelicolor. PLoS Comput. Biol. 2018, 14, e1006541. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kanehisa, M.; Furumichi, M.; Tanabe, M.; Sato, Y.; Morishima, K. KEGG: New perspectives on genomes, pathways, diseases and drugs. Nucleic Acids Res. 2017, 45, D353–D361. [Google Scholar] [CrossRef] [Green Version]
- Caspi, R.; Billington, R.; Fulcher, C.A.; Keseler, I.M.; Kothari, A.; Krummenacker, M.; Latendresse, M.; Midford, P.E.; Ong, Q.; Ong, W.K.; et al. The MetaCyc database of metabolic pathways and enzymes. Nucleic Acids Res. 2018, 46, D633–D639. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Grego, T.; Pesquita, C.; Bastos, H.P.; Couto, F.M. Chemical entity recognition and resolution to ChEBI. ISRN Bioinform. 2012, 2012, 619427. [Google Scholar] [CrossRef] [PubMed]
- Moretti, S.; Tran, V.D.T.; Mehl, F.; Ibberson, M.; Pagni, M. MetaNetX/MNXref: Unified namespace for metabolites and biochemical reactions in the context of metabolic models. Nucleic Acids Res. 2020, 49, D570–D574. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.T.; Hou, C.L.; Zhang, J.; Zeng, X.F.; Qiao, S.Y. Fermentation conditions influence the fatty acid composition of the membranes of Lactobacillus reuteri I5007 and its survival following freeze-drying. Lett. Appl. Microbiol. 2014, 59, 398–403. [Google Scholar] [CrossRef]
- Hamsupo, K.; Sukyai, P.; Nitisinprasert, S.; Wanchaitanawong, P. Different growth media and growth phases affecting on spray drying and freeze drying of Lactobacillus reuteri KUB-AC5. Agric. Nat. Resour. 2005, 39, 718–724. [Google Scholar]
- Orth, J.D.; Thiele, I.; Palsson, B.O. What is flux balance analysis? Nat. Biotechnol. 2010, 28, 245–248. [Google Scholar] [CrossRef]
- Väremo, L.; Nielsen, J.; Nookaew, I. Enriching the gene set analysis of genome-wide data by incorporating directionality of gene expression and combining statistical hypotheses and methods. Nucleic Acids Res. 2013, 41, 4378–4391. [Google Scholar] [CrossRef]
- Abriouel, H.; Pérez Montoro, B.; Casimiro-Soriguer, C.S.; Pérez Pulido, A.J.; Knapp, C.W.; Caballero Gómez, N.; Castillo-Gutiérrez, S.; Estudillo-Martínez, M.D.; Gálvez, A.; Benomar, N. Insight into potential probiotic markers predicted in Lactobacillus pentosus MP-10 genome sequence. Front. Microbiol. 2017, 8, 891. [Google Scholar] [CrossRef] [Green Version]
- Byun, B.Y.; Lee, S.J.; Mah, J.H. Antipathogenic activity and preservative effect of levan (β-2,6-fructan), a multifunctional polysaccharide. Int. J. Food Sci. 2014, 49, 238–245. [Google Scholar] [CrossRef]
- Franceus, J.; Desmet, T. Sucrose phosphorylase and related enzymes in glycoside hydrolase family 13: Discovery, application and engineering. Int. J. Mol. Sci. 2020, 21, 2526. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Reid, S.J.; Abratt, V.R. Sucrose utilisation in bacteria: Genetic organisation and regulation. Appl. Microbiol. Biotechnol. 2005, 67, 312–321. [Google Scholar] [CrossRef] [PubMed]
- Schwab, C.; Walter, J.; Tannock, G.W.; Vogel, R.F.; Ganzle, M.G. Sucrose utilization and impact of sucrose on glycosyltransferase expression in Lactobacillus reuteri. Syst. Appl. Microbiol. 2007, 30, 433–443. [Google Scholar] [CrossRef] [PubMed]
- Ayad, A.A.; Gad El-Rab, D.A.; Ibrahim, S.A.; Williams, L.L. Nitrogen sources effect on Lactobacillus reuteri growth and performance cultivated in date palm (Phoenix dactylifera L.) by-products. Fermentation 2020, 6, 64. [Google Scholar] [CrossRef]
- Zhang, G.; Mills, D.A.; Block, D.E. Development of chemically defined media supporting high-cell-density growth of lactococci, enterococci, and streptococci. Appl. Environ. Microbiol. 2009, 75, 1080–1087. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Santos, F.; Teusink, B.; Molenaar, D.; van Heck, M.; Wels, M.; Sieuwerts, S.; de Vos, W.M.; Hugenholtz, J. Effect of amino acid availability on vitamin B12 production in Lactobacillus reuteri. Appl. Environ. Microbiol. 2009, 75, 3930–3936. [Google Scholar] [CrossRef] [Green Version]
- Saulnier, D.M.; Santos, F.; Roos, S.; Mistretta, T.A.; Spinler, J.K.; Molenaar, D.; Teusink, B.; Versalovic, J. Exploring metabolic pathway reconstruction and genome-wide expression profiling in Lactobacillus reuteri to define functional probiotic features. PLoS ONE 2011, 6, e18783. [Google Scholar] [CrossRef] [Green Version]
- Silvestroni, A.; Connes, C.; Sesma, F.; De Giori, G.S.; Piard, J.C. Characterization of the melA locus for alpha-galactosidase in Lactobacillus plantarum. Appl. Environ. Microbiol. 2002, 68, 5464–5471. [Google Scholar] [CrossRef] [Green Version]
- Wisselink, H.W.; Weusthuis, R.A.; Eggink, G.; Hugenholtz, J.; Grobben, G.J. Mannitol production by lactic acid bacteria: A review. Int. Dairy J. 2002, 12, 151–161. [Google Scholar] [CrossRef]
- Moffatt, B.A.; Ashihara, H. Purine and pyrimidine nucleotide synthesis and metabolism. Arab. Book/Am. Soc. Plant Biol. 2002, 1, e0018. [Google Scholar] [CrossRef] [PubMed] [Green Version]
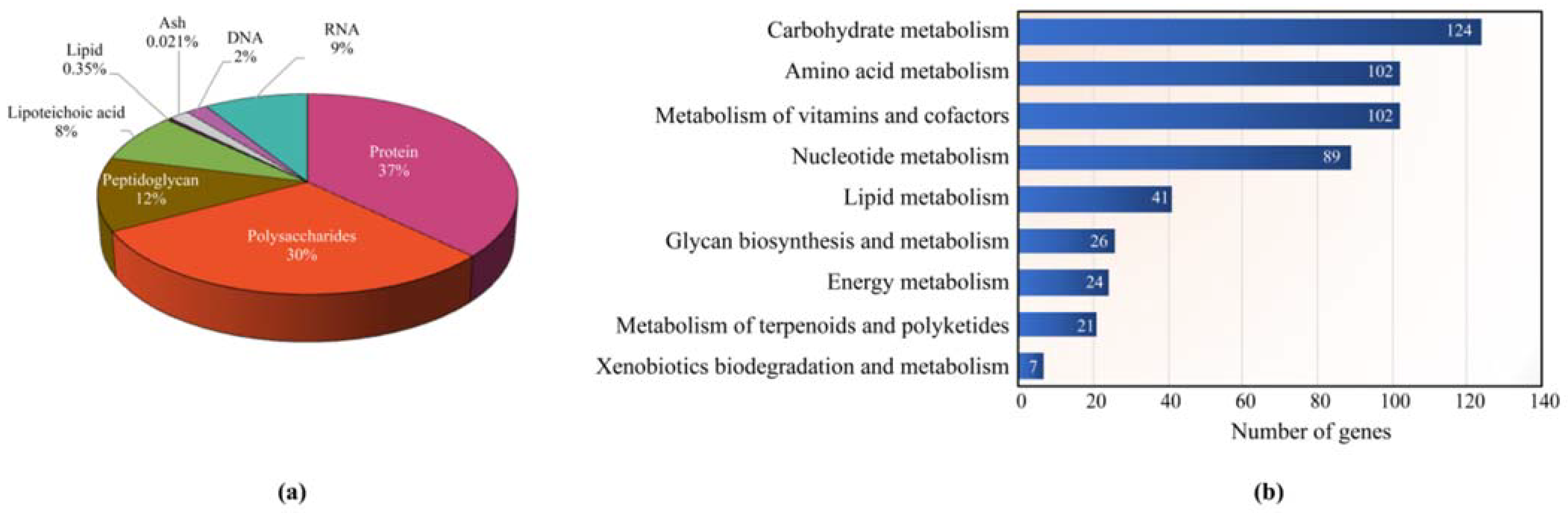
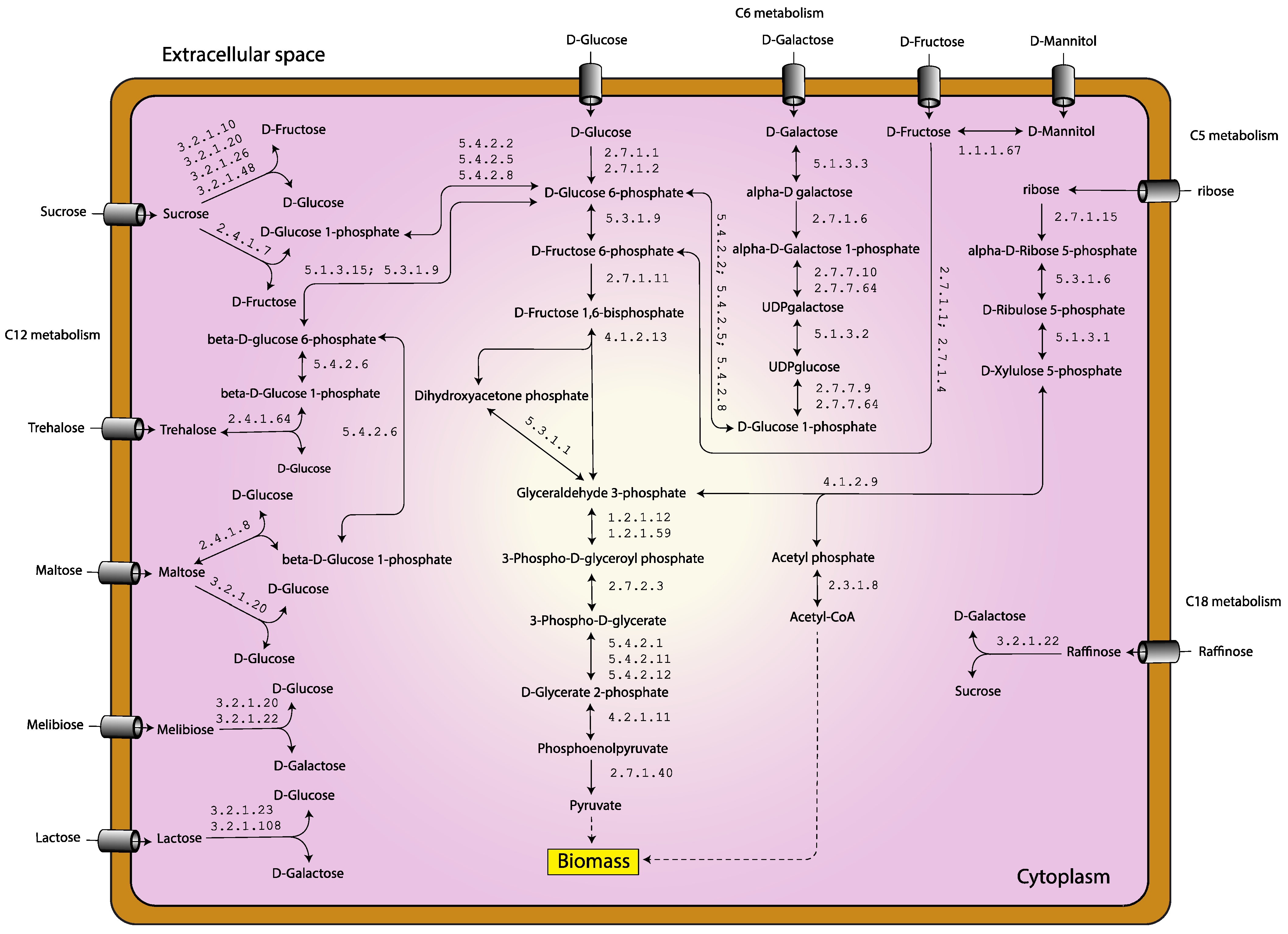
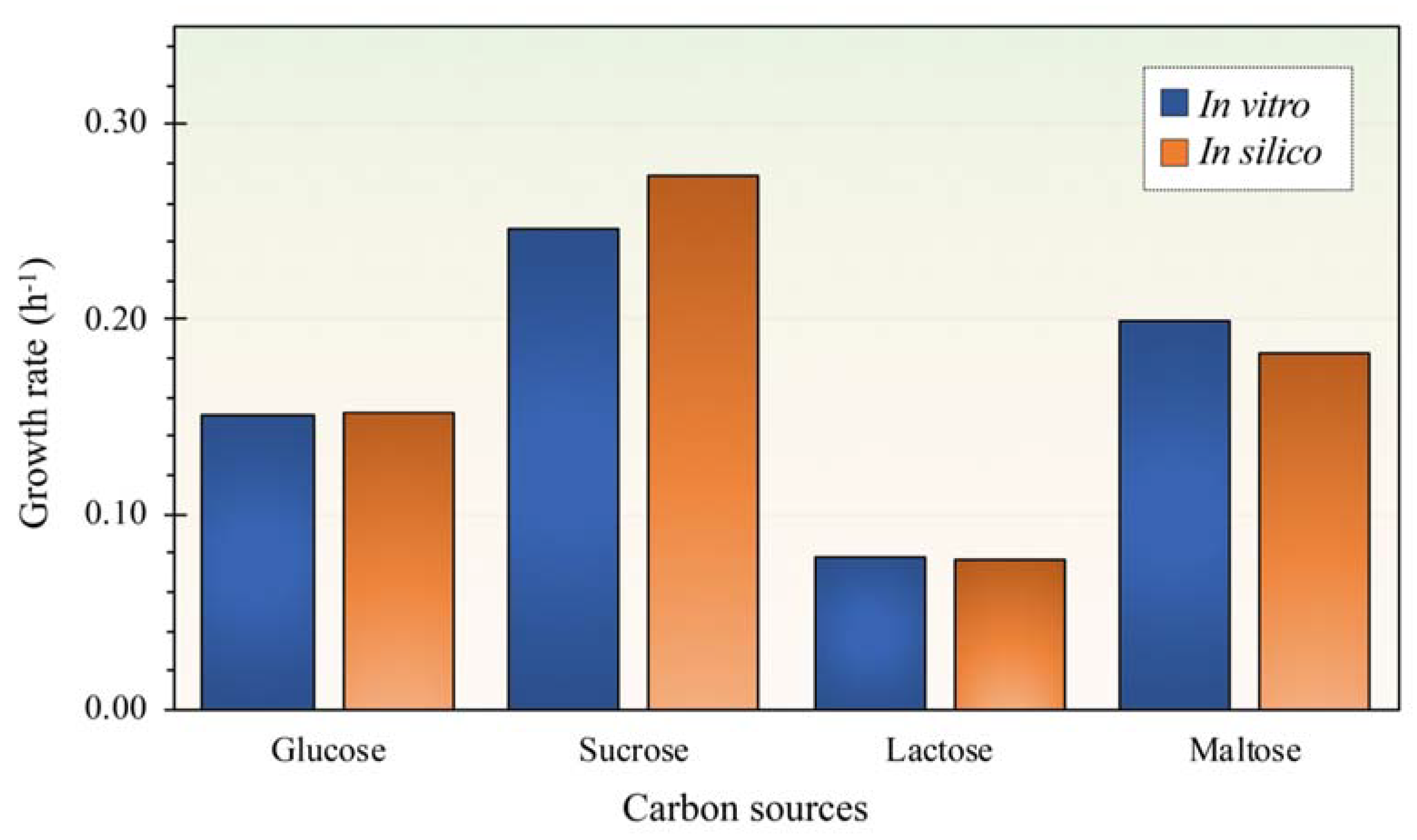
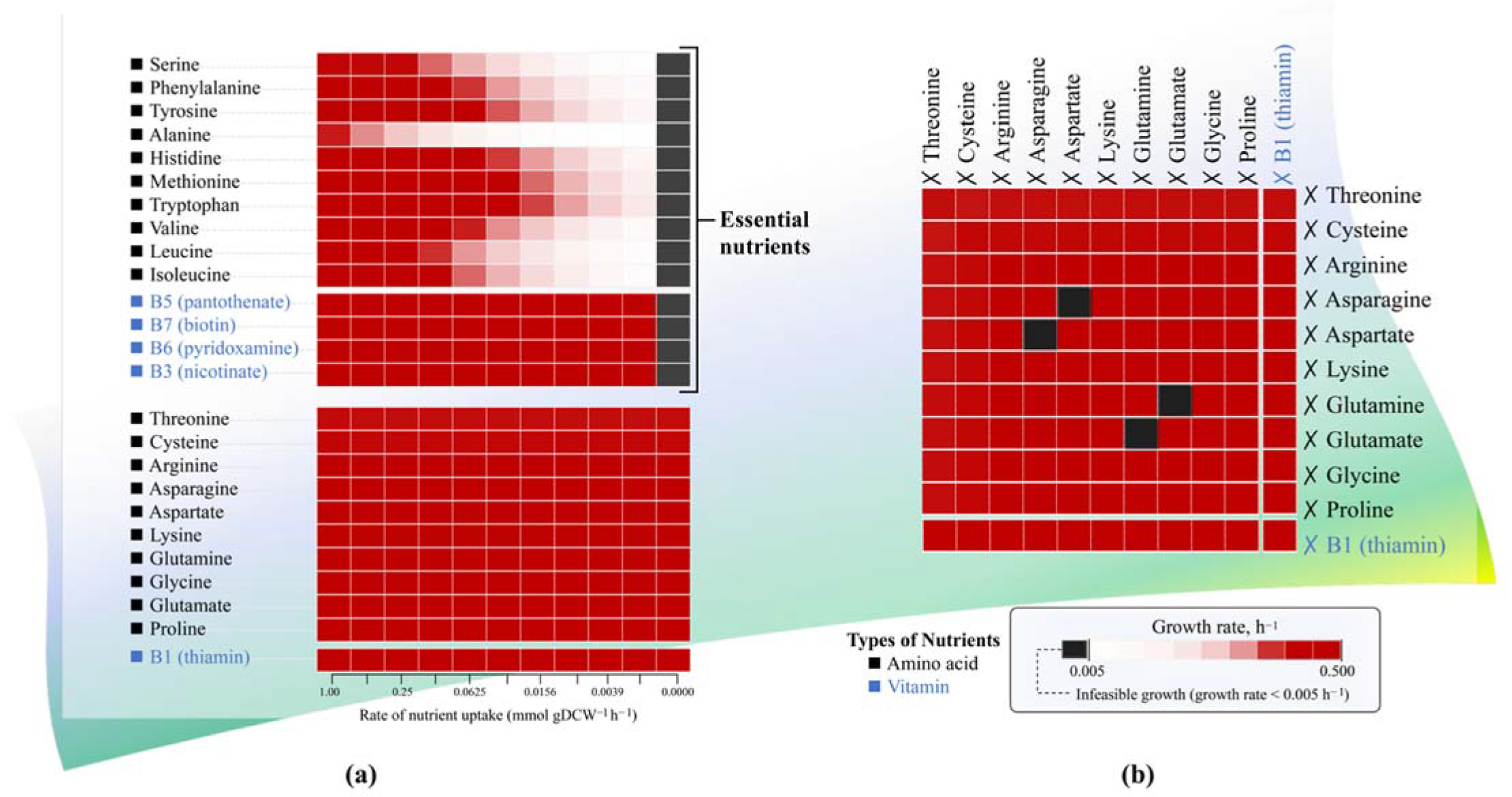
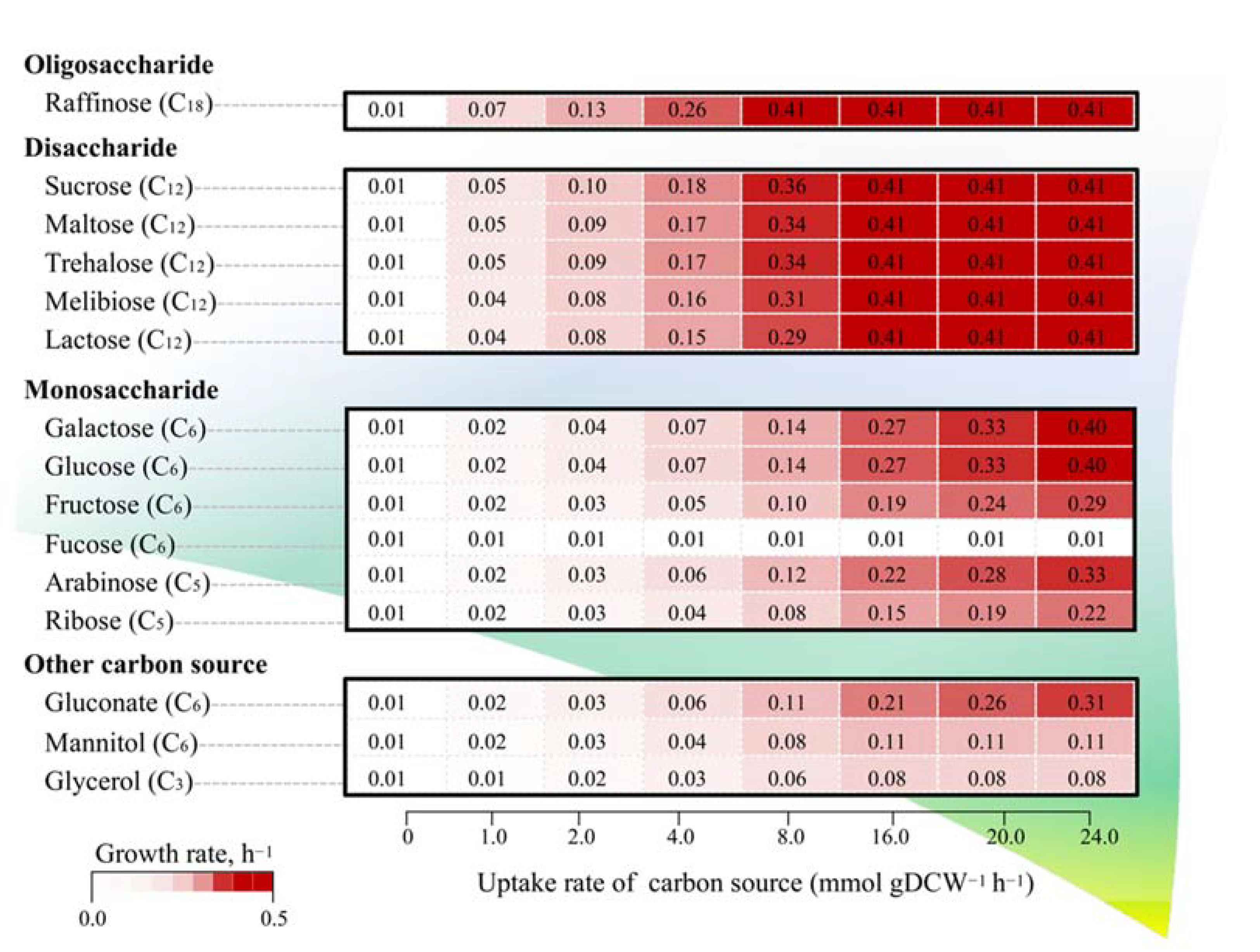
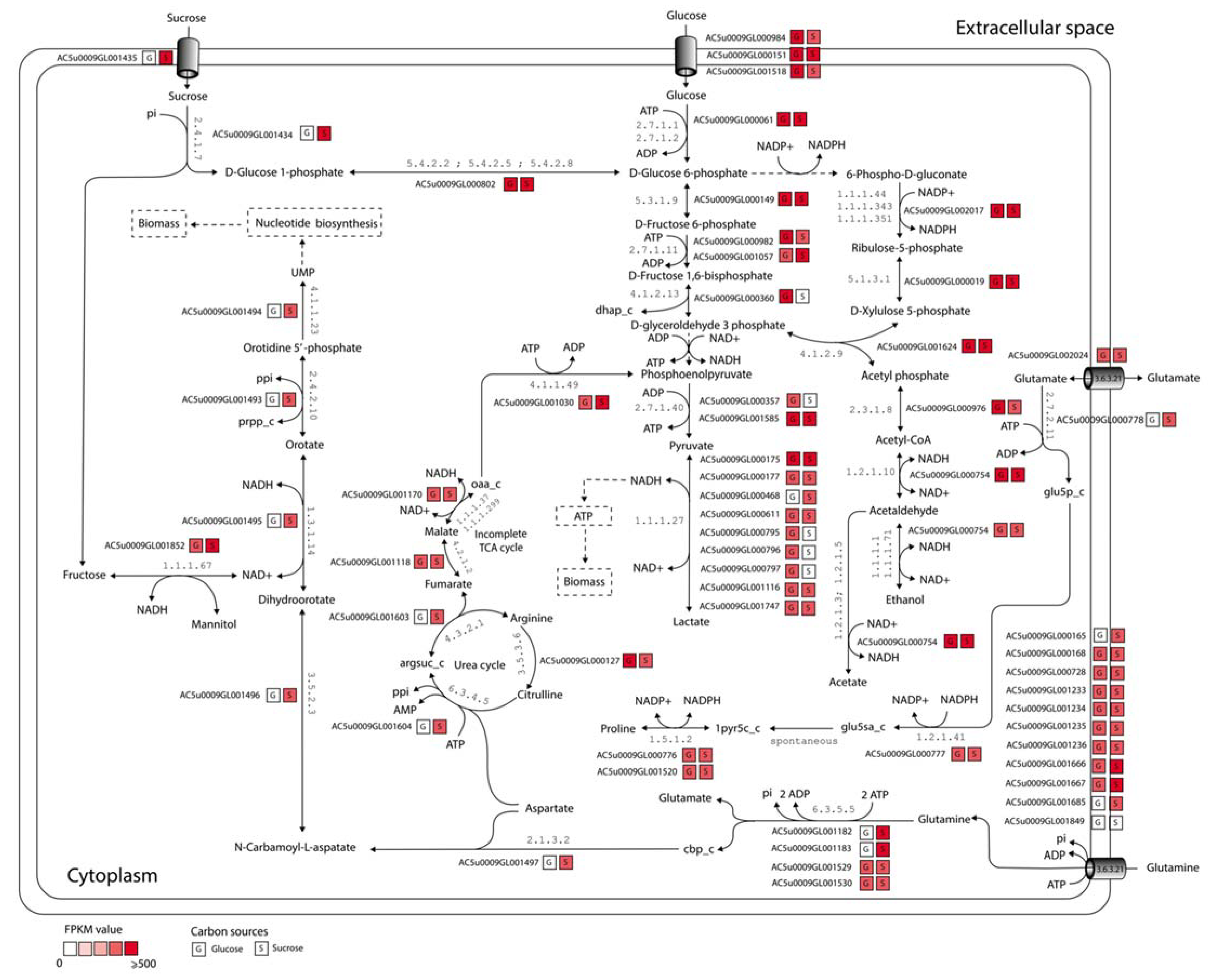
| Genomic Characteristics | KUB-AC5 (This Study) | JCM 1112 1 | ATCC PTA 6475 2 |
|---|---|---|---|
| Genome size (Mb) | 2.19 3 | 2.04 | 2.04 |
| Total number of protein-coding genes | 2196 3 | 1943 | 2019 |
| Metabolic Network Characteristics | iTN656 (This study) | LbReuteri | iHL622 |
| Total genes | 656 | 530 | 622 |
| Total metabolites | 831 | 660 | 713 |
| Total metabolic reactions | 953 | 714 | 869 |
| Gene-protein-reaction (GPR) associations | 810 | 606 | 709 |
| Compartments | 2 | 2 | 2 |
| Characteristic | Glucose | Maltose | Sucrose | Lactose |
|---|---|---|---|---|
| Growth rate (μmax, h−1) * | 0.151 ± 0.004 c | 0.199 ± 0.009 b | 0.247 ± 0.003 a | 0.078 ± 0.005 d |
| Biomass production (gDCW L−1) * | 1.010 ± 0.026 c | 1.240 ± 0.053 b | 1.650 ± 0.030 a | 0.753 ± 0.025 d |
| Substrate uptake rate (mmol gDCW−1 h−1) | 6.634 ± 0.684 a | 3.310 ± 0.764 c | 5.033 ± 0.310 b | 0.940 ± 0.322 d |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Namrak, T.; Raethong, N.; Jatuponwiphat, T.; Nitisinprasert, S.; Vongsangnak, W.; Nakphaichit, M. Probing Genome-Scale Model Reveals Metabolic Capability and Essential Nutrients for Growth of Probiotic Limosilactobacillus reuteri KUB-AC5. Biology 2022, 11, 294. https://doi.org/10.3390/biology11020294
Namrak T, Raethong N, Jatuponwiphat T, Nitisinprasert S, Vongsangnak W, Nakphaichit M. Probing Genome-Scale Model Reveals Metabolic Capability and Essential Nutrients for Growth of Probiotic Limosilactobacillus reuteri KUB-AC5. Biology. 2022; 11(2):294. https://doi.org/10.3390/biology11020294
Chicago/Turabian StyleNamrak, Thanawat, Nachon Raethong, Theeraphol Jatuponwiphat, Sunee Nitisinprasert, Wanwipa Vongsangnak, and Massalin Nakphaichit. 2022. "Probing Genome-Scale Model Reveals Metabolic Capability and Essential Nutrients for Growth of Probiotic Limosilactobacillus reuteri KUB-AC5" Biology 11, no. 2: 294. https://doi.org/10.3390/biology11020294
APA StyleNamrak, T., Raethong, N., Jatuponwiphat, T., Nitisinprasert, S., Vongsangnak, W., & Nakphaichit, M. (2022). Probing Genome-Scale Model Reveals Metabolic Capability and Essential Nutrients for Growth of Probiotic Limosilactobacillus reuteri KUB-AC5. Biology, 11(2), 294. https://doi.org/10.3390/biology11020294








