Mitochondrial Function Differences between Tumor Tissue of Human Metastatic and Premetastatic CRC
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Patient Selection and Tissue Samples
2.2. Tissue Sample Preparation
2.3. Western Blot Analysis
2.4. IDH Activity
2.5. COX Activity
2.6. ATPase Activity
2.7. mtDNA Quantification
2.8. Statistics
3. Results
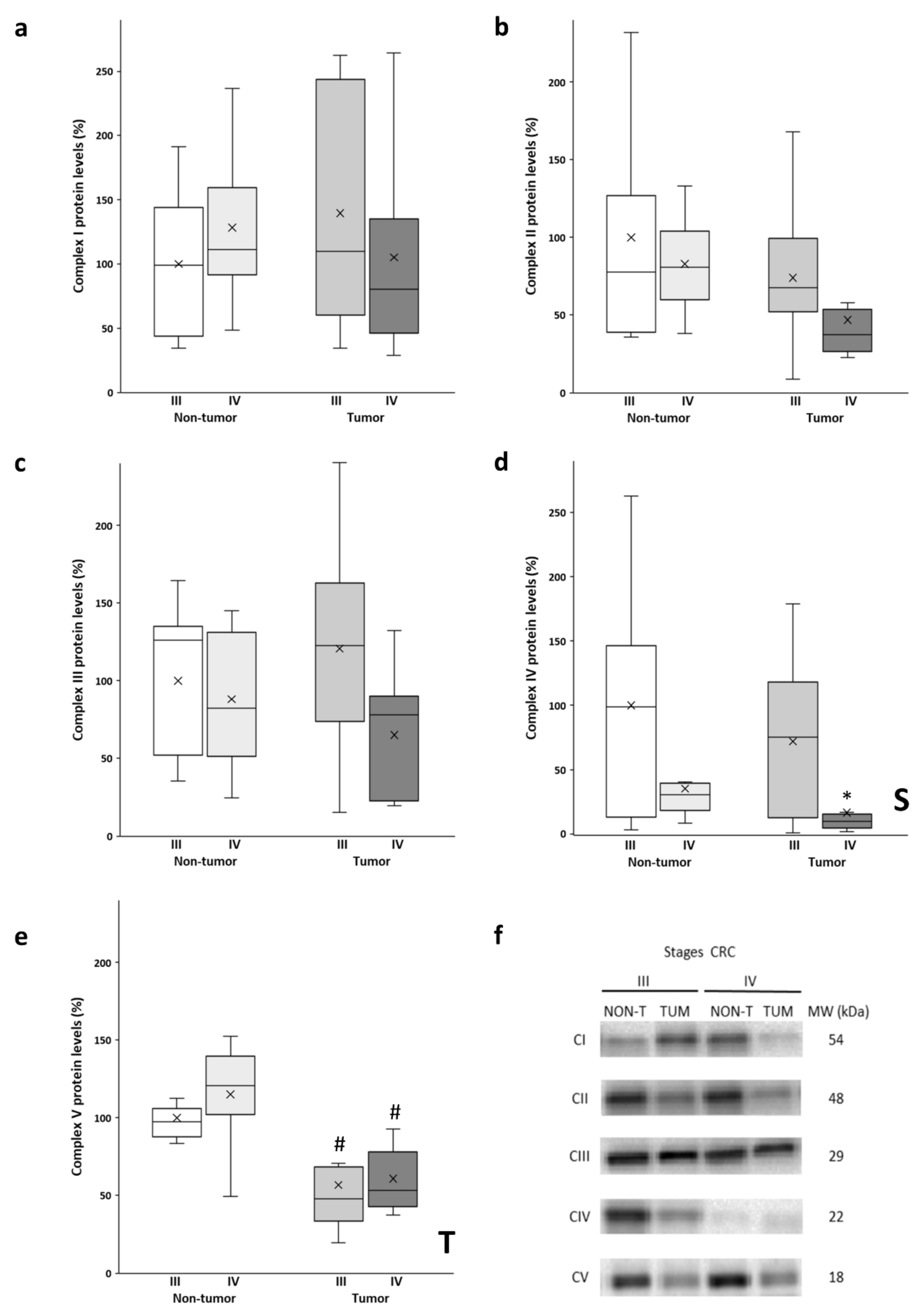
3.1. OXPHOS Complex Protein Expression Levels
3.2. Metabolic Enzyme Levels
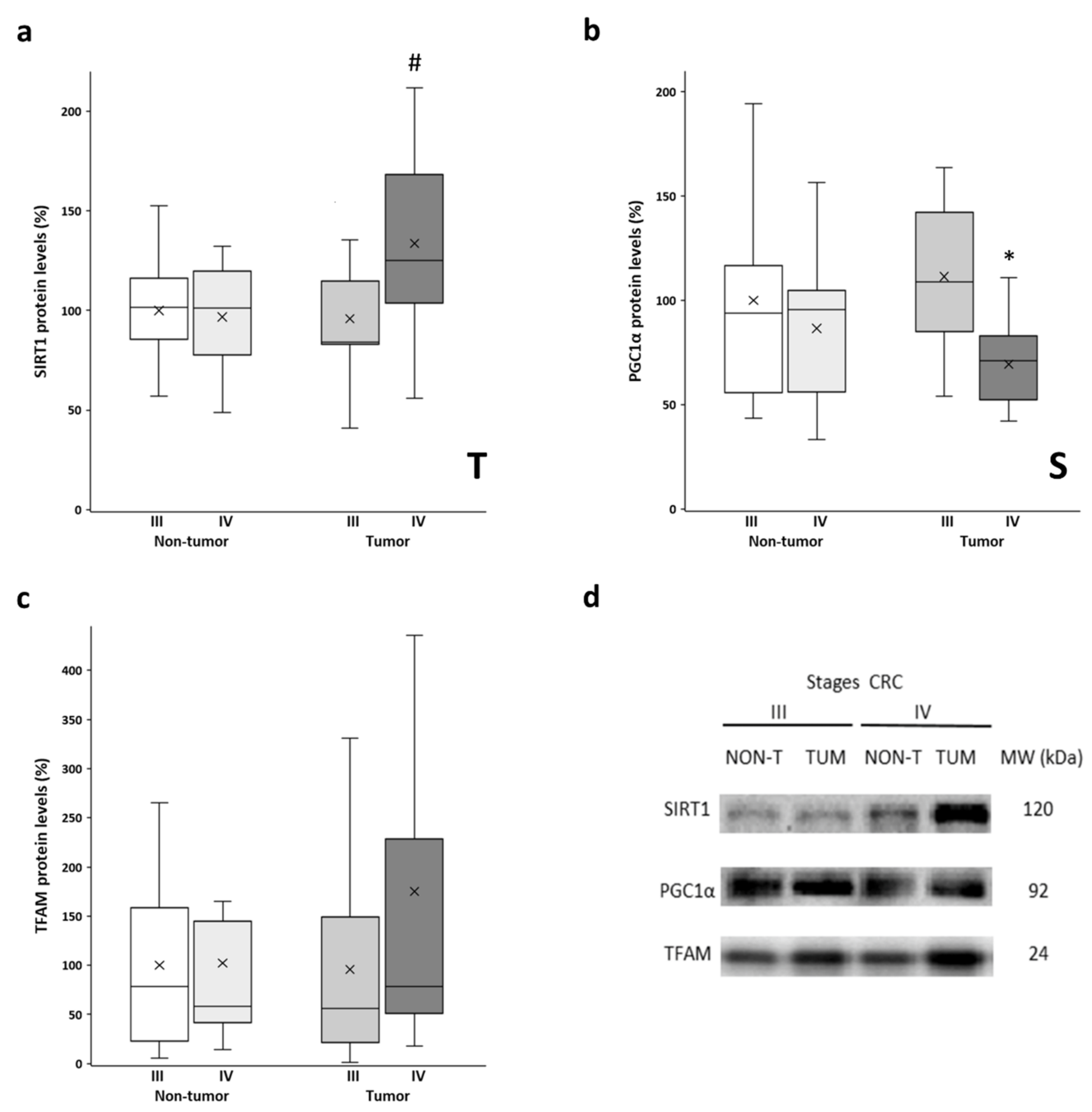
3.3. SIRT1, PGC1α, and TFAM Levels
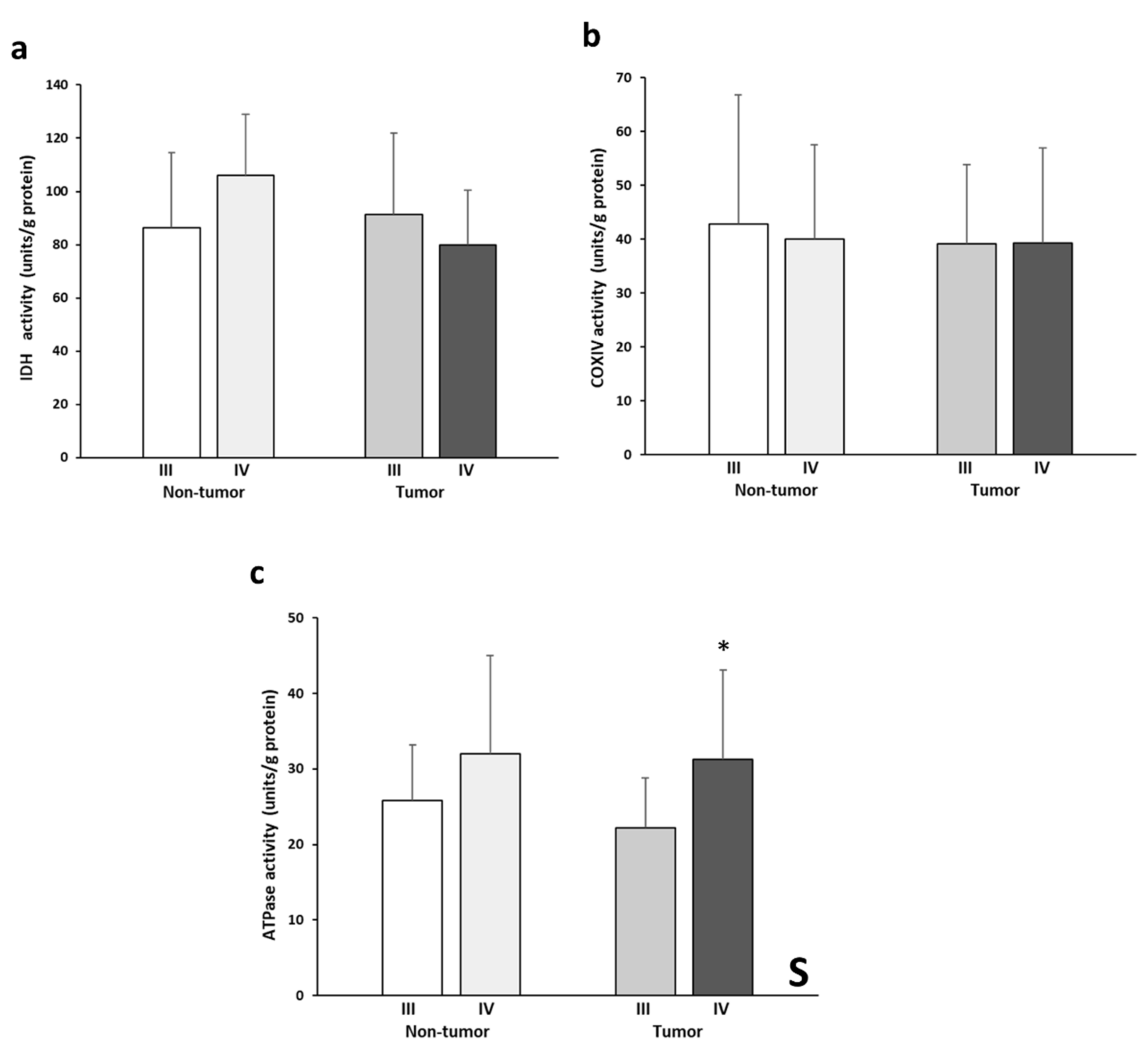
3.4. IDH2, COX, and ATPase Enzymatic Activities
3.5. mtDNA Quantification
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Benson, A.B., 3rd. Epidemiology, disease progression, and economic burden of colorectal cancer. J. Manag. Care Pharm. 2007, 13, S5–S18. [Google Scholar] [CrossRef] [PubMed]
- Lucas, A.S.; O’Neil, B.H.; Goldberg, R.M. A Decade of Advances in Cytotoxic Chemotherapy for Metastatic Colorectal Cancer. Clin. Colorectal Cancer 2011, 10, 238–244. [Google Scholar] [CrossRef] [PubMed]
- Sanoff, H.K.; Sargent, D.J.; Campbell, M.E.; Morton, R.F.; Fuchs, C.S.; Ramanathan, R.K.; Williamson, S.K.; Findlay, B.P.; Pitot, H.C.; Goldberg, R.M. Five-Year Data and Prognostic Factor Analysis of Oxaliplatin and Irinotecan Combinations for Advanced Colorectal Cancer: N9741. J. Clin. Oncol. 2008, 26, 5721–5727. [Google Scholar] [CrossRef] [PubMed]
- Guaragnella, N.; Giannattasio, S.; Moro, L. Mitochondrial dysfunction in cancer chemoresistance. Biochem. Pharmacol. 2014, 92, 62–72. [Google Scholar] [CrossRef]
- Porporato, P.E.; Ry, V.; Payen, L.; Pé Rez-Escuredo, J.; De Saedeleer, C.J.; Danhier, P.; Copetti, T.; Dhup, S.; Tardy, M.; Vazeille, T.; et al. A Mitochondrial Switch Promotes Tumor Metastasis. Cell Rep. 2014, 8, 754–766. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Heiden, M.G.V.; Cantley, L.C.; Thompson, C.B.; Mammalian, P.; Exhibit, C.; Metabolism, A. Understanding the Warburg Effect: The Metabolic Requirements of Cell Proliferation. Science 2009, 324, 1029–1034. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Warburg, O. On the origin of cancer cells. Science 1956, 123, 309–314. [Google Scholar] [CrossRef]
- Moreno-Sánchez, R.; Rodríguez-Enríquez, S.; Marín-Hernández, A.; Saavedra, E. Energy metabolism in tumor cells. FEBS J. 2007, 274, 1393–1418. [Google Scholar] [CrossRef]
- Hanahan, D.; Weinberg, R.A.; Pan, K.H.; Shay, J.W.; Cohen, S.N.; Taylor, M.B.; Clarke, N.W.; Jayson, G.C.; Eshleman, J.R.; Nowak, M.A.; et al. Hallmarks of Cancer: The Next Generation. Cell 2011, 144, 646–674. [Google Scholar] [CrossRef] [Green Version]
- Jose, C.; Bellance, N. Choosing between glycolysis and oxidative phosphorylation: A tumor’s dilemma? Biochim. Biophys. Acta Bioenerg. 2011, 1807, 552–561. [Google Scholar] [CrossRef] [Green Version]
- Potter, V.R. The biochemical approach to the cancer problem. Fed. Proc. 1958, 17, 691–697. [Google Scholar] [PubMed]
- Hernández-López, R.; Torrens-Mas, M.; Pons, D.G.; Company, M.M.; Falcó, E.; Fernández, T.; Ibarra de la Rosa, J.M.; Sastre-Serra, J.; Oliver, J.; Roca, P. Non-tumor adjacent tissue of advanced stage from CRC shows activated antioxidant response. Free Radic. Biol. Med. 2018, 126, 249–258. [Google Scholar] [CrossRef] [PubMed]
- Bergmeyer, H.U.; Bergmeyer, J.; Grassl, M. Methods of Enzymatic Analysis. Vol. 3: Enzymes 1. Oxidoreductases, Transferases; Verlag Chemie: Weinheim, Germany, 1983; Volume 3. [Google Scholar]
- Chrzanowska-Lightowlers, Z.M.A.; Turnbull, D.M.; Lightowlers, R.N. A Microtiter Plate Assay for Cytochrome c Oxidase in Permeabilized Whole Cells. Anal. Biochem. 1993, 214, 45–49. [Google Scholar] [CrossRef] [PubMed]
- Ragan, C.I.; Wilson, M.T.; Darley-Usmar, V.M.; Lowe, P.N. Subfractionation of mitochondria and isolation of the proteins of oxidative phosphorylation. In Mitochondria A Practical Approach; IRL Press Limited: Oxford, UK, 1987; pp. 79–112. [Google Scholar]
- Feichtinger, R.G.; Neureiter, D.; Skaria, T.; Wessler, S.; Cover, T.L.; Mayr, J.A.; Zimmermann, F.A.; Posselt, G.; Sperl, W.; Kofler, B. Oxidative Phosphorylation System in Gastric Carcinomas and Gastritis. Oxidative Med. Cell. Longev. 2017, 2017, 1–14. [Google Scholar] [CrossRef] [Green Version]
- Kumari, S.; Badana, A.K.; Murali Mohan, G.; Shailender, G.; Malla, R.R. Reactive Oxygen Species: A Key Constituent in Cancer Survival. Biomark. Insights 2018, 13, 1177271918755391. [Google Scholar] [CrossRef] [Green Version]
- Nesci, S.; Trombetti, F.; Pagliarani, A.; Ventrella, V.; Algieri, C.; Tioli, G.; Lenaz, G. Molecular and supramolecular structure of the mitochondrial oxidative phosphorylation system: Implications for pathology. Life 2021, 11, 242. [Google Scholar] [CrossRef]
- Galadari, S.; Rahman, A.; Pallichankandy, S.; Thayyullathil, F. Reactive oxygen species and cancer paradox: To promote or to suppress? Free Radic. Biol. Med. 2017, 104, 144–164. [Google Scholar] [CrossRef]
- Valle, A.; Santandreu, F.M.; Garcia-Palmer, F.J.; Roca, P.; Oliver, J. The serum levels of 17beta-estradiol, progesterone and triiodothyronine correlate with brown adipose tissue thermogenic parameters during aging. Cell. Physiol. Biochem. 2008, 22, 337–346. [Google Scholar] [CrossRef]
- Pons, D.G.; Torrens-Mas, M.; Nadal-Serrano, M.; Sastre-Serra, J.; Roca, P.; Oliver, J. The presence of Estrogen Receptor β modulates the response of breast cancer cells to therapeutic agents. Int. J. Biochem. Cell Biol. 2015, 66, 85–94. [Google Scholar] [CrossRef]
- Luna, F.; Roca, P.; Oliver, J.; Antenucci, C.D. Maximal thermogenic capacity and non-shivering thermogenesis in the South American subterranean rodent Ctenomys talarum. J. Comp. Physiol. B 2012, 182, 971–983. [Google Scholar] [CrossRef]
- Blanquer-Rosselló, M.d.M.; Hernández-López, R.; Roca, P.; Oliver, J.; Valle, A. Resveratrol induces mitochondrial respiration and apoptosis in SW620 colon cancer cells. Biochim. Biophys. Acta Gen. Subj. 2017, 1891, 431–440. [Google Scholar] [CrossRef] [PubMed]
- Guevara, R.; Gianotti, M.; Roca, P.; Oliver, J. Age and Sex-Related Changes in Rat Brain Mitochondrial Function. Cell. Physiol. Biochem. 2011, 27, 201–206. [Google Scholar] [CrossRef] [PubMed]
- Torrens-Mas, M.; González-Hedström, D.; Abrisqueta, M.; Roca, P.; Oliver, J.; Sastre-Serra, J. PGC-1α in melanoma: A key factor for antioxidant response and mitochondrial function. J. Cell. Biochem. 2017, 118, 4404–4413. [Google Scholar] [CrossRef] [PubMed]
- Pons, D.G.; Nadal-Serrano, M.; Torrens-Mas, M.; Oliver, J.; Roca, P. The Phytoestrogen Genistein Affects Breast Cancer Cells Treatment Depending on the ERα/ERβ Ratio. J. Cell. Biochem. 2016, 117, 218–229. [Google Scholar] [CrossRef]
- Santandreu, F.M.; Valle, A.; Fernandez de Mattos, S.; Roca, P.; Oliver, J. Hydrogen peroxide regulates the mitochondrial content of uncoupling protein 5 in colon cancer cells. Cell. Physiol. Biochem. 2009, 24, 379–390. [Google Scholar] [CrossRef]
- Acin-Perez, R.; Gatti, D.L.; Bai, Y.; Manfredi, G. Protein phosphorylation and prevention of cytochrome oxidase inhibition by ATP: Coupled mechanisms of energy metabolism regulation. Cell Metab. 2011, 13, 712–719. [Google Scholar] [CrossRef] [Green Version]
- Helling, S.; Vogt, S.; Rhiel, A.; Ramzan, R.; Wen, L.; Marcus, K.; Kadenbach, B. Phosphorylation and kinetics of mammalian cytochrome c oxidase. Mol. Cell. Proteom. 2008, 7, 1714–1724. [Google Scholar] [CrossRef] [Green Version]
- Li, Y.; Park, J.S.; Deng, J.H.; Bai, Y. Cytochrome c oxidase subunit IV is essential for assembly and respiratory function of the enzyme complex. J. Bioenerg. Biomembr. 2006, 38, 283–291. [Google Scholar] [CrossRef] [Green Version]
- Chekulayev, V.; Mado, K.; Shevchuk, I.; Koit, A.; Kaldma, A.; Klepinin, A.; Timohhina, N.; Tepp, K.; Kandashvili, M.; Ounpuu, L.; et al. Metabolic remodeling in human colorectal cancer and surrounding tissues: Alterations in regulation of mitochondrial respiration and metabolic fluxes. Biochem. Biophys. Rep. 2015, 4, 111–125. [Google Scholar] [CrossRef] [Green Version]
- Graziano, F.; Ruzzo, A.; Giacomini, E.; Ricciardi, T.; Aprile, G.; Loupakis, F.; Lorenzini, P.; Ongaro, E.; Zoratto, F.; Catalano, V.; et al. Glycolysis gene expression analysis and selective metabolic advantage in the clinical progression of colorectal cancer. Pharm. J. 2017, 17, 258–264. [Google Scholar] [CrossRef]
- Sjoblom, T.; Jones, S.; Wood, L.D.; Parsons, D.W.; Lin, J.; Barber, T.D.; Mandelker, D.; Leary, R.J.; Ptak, J.; Silliman, N.; et al. The Consensus Coding Sequences of Human Breast and Colorectal Cancers. Science 2006, 314, 268–274. [Google Scholar] [CrossRef] [PubMed]
- Xu, W.; Yang, H.; Liu, Y.; Yang, Y.; Wang, P.; Kim, S.-H.; Ito, S.; Yang, C.; Wang, P.; Xiao, M.-T.; et al. Oncometabolite 2-Hydroxyglutarate Is a Competitive Inhibitor of α-Ketoglutarate-Dependent Dioxygenases. Cancer Cell 2011, 19, 17–30. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jones, A.W.E.; Yao, Z.; Vicencio, J.M.; Karkucinska-Wieckowska, A.; Szabadkai, G. PGC-1 family coactivators and cell fate: Roles in cancer, neurodegeneration, cardiovascular disease and retrograde mitochondria–nucleus signalling. Mitochondrion 2012, 12, 86–99. [Google Scholar] [CrossRef] [PubMed]
- Lagouge, M.; Argmann, C.; Gerhart-Hines, Z.; Meziane, H.; Lerin, C.; Daussin, F.; Messadeq, N.; Milne, J.; Lambert, P.; Elliott, P.; et al. Resveratrol Improves Mitochondrial Function and Protects against Metabolic Disease by Activating SIRT1 and PGC-1α. Cell 2006, 127, 1109–1122. [Google Scholar] [CrossRef]
- Wu, Z.; Puigserver, P.; Andersson, U.; Zhang, C.; Adelmant, G.; Mootha, V.; Troy, A.; Cinti, S.; Lowell, B.; Scarpulla, R.C.; et al. Mechanisms controlling mitochondrial biogenesis and respiration through the thermogenic coactivator PGC-1. Cell 1999, 98, 115–124. [Google Scholar] [CrossRef] [Green Version]
- Puigserver, P.; Wu, Z.; Park, C.W.; Graves, R.; Wright, M.; Spiegelman, B.M. A cold-inducible coactivator of nuclear receptors linked to adaptive thermogenesis. Cell 1998, 92, 829–839. [Google Scholar] [CrossRef] [Green Version]
- Feilchenfeldt, J.; Bründler, M.A.; Soravia, C.; Tötsch, M.; Meier, C.A. Peroxisome proliferator-activated receptors (PPARs) and associated transcription factors in colon cancer: Reduced expression of PPARgamma-coactivator 1 (PGC-1). Cancer Lett. 2004, 203, 25–33. [Google Scholar] [CrossRef]
- Vellinga, T.T.; De Boer, V.C.J.; Fatrai, S.; Van Schelven, S.; Trumpi, K.; Verheem, A.; Snoeren, N.; Emmink, B.L.; Koster, J.; Rinkes, I.H.M.B.; et al. SIRT1/PGC1a-Dependent increase in oxidative phosphorylation supports chemotherapy resistance of colon cancer. Clin. Cancer Res. 2015, 21, 2870–2879. [Google Scholar] [CrossRef] [Green Version]
- Cannino, G.; Di Liegro, C.M.; Rinaldi, A.M. Nuclear–mitochondrial interaction. Mitochondrion 2007, 7, 359–366. [Google Scholar] [CrossRef]
- Kanki, T.; Ohgaki, K.; Gaspari, M.; Gustafsson, C.M.; Fukuoh, A.; Sasaki, N.; Hamasaki, N.; Kang, D. Architectural Role of Mitochondrial Transcription Factor A in Maintenance of Human Mitochondrial DNA. Mol. Cell. Biol. 2004, 24, 9823–9834. [Google Scholar] [CrossRef] [Green Version]
- Kaufman, B.A.; Durisic, N.; Mativetsky, J.M.; Costantino, S.; Hancock, M.A.; Grutter, P.; Shoubridge, E.A. The mitochondrial transcription factor TFAM coordinates the assembly of multiple DNA molecules into nucleoid-like structures. Mol. Biol. Cell 2007, 18, 3225–3236. [Google Scholar] [CrossRef] [PubMed]
- Gabrielson, M.; Björklund, M.; Carlson, J.; Shoshan, M. Expression of mitochondrial regulators PGC1α and TFAM as putative markers of subtype and chemoresistance in epithelial ovarian carcinoma. PLoS ONE 2014, 9, e107109. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Piantadosi, C.A.; Suliman, H.B. Mitochondrial transcription factor A induction by redox activation of nuclear respiratory factor 1. J. Biol. Chem. 2006, 281, 324–333. [Google Scholar] [CrossRef] [Green Version]
- Lu, B.; Lee, J.; Nie, X.; Li, M.; Morozov, Y.I.; Venkatesh, S.; Bogenhagen, D.F.; Temiakov, D.; Suzuki, C.K. Phosphorylation of Human TFAM in Mitochondria Impairs DNA Binding and Promotes Degradation by the AAA+ Lon Protease. Mol. Cell 2013, 49, 121–132. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yoshida, Y.; Izumi, H.; Torigoe, T.; Ishiguchi, H.; Itoh, H.; Kang, D.; Kohno, K. p53 physically interacts with mitochondrial transcription factor A and differentially regulates binding to damaged DNA. Cancer Res. 2003, 63, 3729–3734. [Google Scholar] [PubMed]
- Jiang, X.; Wang, J. Down-regulation of TFAM increases the sensitivity of tumour cells to radiation via p53/TIGAR signalling pathway. J. Cell. Mol. Med. 2019, 23, 4545–4558. [Google Scholar] [CrossRef] [Green Version]
- Li, H.Y.; Lin, X.W.; Geng, S.L.; Xu, W.H. TGF-β and BMP signals regulate insect diapause through Smad1-POU-TFAM pathway. Biochim. Biophys. Acta Mol. Cell Res. 2018, 1865, 1239–1249. [Google Scholar] [CrossRef] [PubMed]
- Wu, K.; Ma, J.; Zhan, Y.; Liu, K.; Ye, Z.; Chen, J.; Xu, K.; Huang, H.; He, Y. Down-Regulation of MicroRNA-214 Contributed to the Enhanced Mitochondrial Transcription Factor A and Inhibited Proliferation of Colorectal Cancer Cells. Cell. Physiol. Biochem. 2018, 49, 545–554. [Google Scholar] [CrossRef]
- Channakkar, A.S.; Singh, T.; Pattnaik, B.; Gupta, K.; Seth, P.; Adlakha, Y.K. MiRNA-137-mediated modulation of mitochondrial dynamics regulates human neural stem cell fate. Stem Cells 2020, 38, 683–697. [Google Scholar] [CrossRef] [Green Version]
- Jiang, J.; Yang, J.; Wang, Z.; Wu, G.; Liu, F. TFAM is directly regulated by miR-23b in glioma. Oncol. Rep. 2013, 30, 2105–2110. [Google Scholar] [CrossRef]
- Gaya-Bover, A.; Hernández-López, R.; Alorda-Clara, M.; Ibarra de la Rosa, J.M.; Falcó, E.; Fernández, T.; Company, M.M.; Torrens-Mas, M.; Roca, P.; Oliver, J.; et al. Antioxidant enzymes change in different non-metastatic stages in tumoral and peritumoral tissues of colorectal cancer. Int. J. Biochem. Cell Biol. 2020, 120, 105698. [Google Scholar] [CrossRef] [PubMed]
- Feng, S.; Xiong, L.; Ji, Z.; Cheng, W.; Yang, H. Correlation between increased copy number of mitochondrial DNA and clinicopathological stage in colorectal cancer. Oncol. Lett. 2011, 2, 899. [Google Scholar] [CrossRef] [PubMed]
- Lee, H.-C.; Yin, P.-H.; Lin, J.-C.; Wu, C.-C.; Chen, C.-Y.; Wu, C.-W.; Chi, C.-W.; Tam, T.-N.; Wei, Y.-H. Mitochondrial Genome Instability and mtDNA Depletion in Human Cancers. Ann. N. Y. Acad. Sci. 2005, 1042, 109–122. [Google Scholar] [CrossRef] [PubMed]
- Lee, H.C.; Wei, Y.H. Mitochondrial DNA instability and metabolic shift in human cancers. Int. J. Mol. Sci. 2009, 10, 674–701. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shuwen, H.; Xi, Y.; Yuefen, P. Can Mitochondria DNA Provide a Novel Biomarker for Evaluating the Risk and Prognosis of Colorectal Cancer? Dis. Markers 2017, 2017, 5189803. [Google Scholar] [CrossRef]
- Wang, Y.; He, S.; Zhu, X.; Qiao, W.; Zhang, J. High Copy Number of Mitochondrial DNA Predicts Poor Prognosis in Patients with Advanced Stage Colon Cancer. Int. J. Biol. Markers 2016, 31, 382–388. [Google Scholar] [CrossRef]

| CRC | Stage III | Stage IV | |||
|---|---|---|---|---|---|
| Non-Tumor Adjacent Tissue | Tumor Tissue | Non-Tumor Adjacent Tissue | Tumor Tissue | ||
| Content of mtDNA | 100 ± 7.66 | 121 ± 2.73 * | 93 ± 2.82 | 118 ± 1.84 * | T |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hernández-López, R.; Torrens-Mas, M.; Pons, D.G.; Company, M.M.; Falcó, E.; Fernández, T.; Ibarra de la Rosa, J.M.; Roca, P.; Oliver, J.; Sastre-Serra, J. Mitochondrial Function Differences between Tumor Tissue of Human Metastatic and Premetastatic CRC. Biology 2022, 11, 293. https://doi.org/10.3390/biology11020293
Hernández-López R, Torrens-Mas M, Pons DG, Company MM, Falcó E, Fernández T, Ibarra de la Rosa JM, Roca P, Oliver J, Sastre-Serra J. Mitochondrial Function Differences between Tumor Tissue of Human Metastatic and Premetastatic CRC. Biology. 2022; 11(2):293. https://doi.org/10.3390/biology11020293
Chicago/Turabian StyleHernández-López, Reyniel, Margalida Torrens-Mas, Daniel G. Pons, Maria M. Company, Esther Falcó, Teresa Fernández, Javier M. Ibarra de la Rosa, Pilar Roca, Jordi Oliver, and Jorge Sastre-Serra. 2022. "Mitochondrial Function Differences between Tumor Tissue of Human Metastatic and Premetastatic CRC" Biology 11, no. 2: 293. https://doi.org/10.3390/biology11020293
APA StyleHernández-López, R., Torrens-Mas, M., Pons, D. G., Company, M. M., Falcó, E., Fernández, T., Ibarra de la Rosa, J. M., Roca, P., Oliver, J., & Sastre-Serra, J. (2022). Mitochondrial Function Differences between Tumor Tissue of Human Metastatic and Premetastatic CRC. Biology, 11(2), 293. https://doi.org/10.3390/biology11020293









