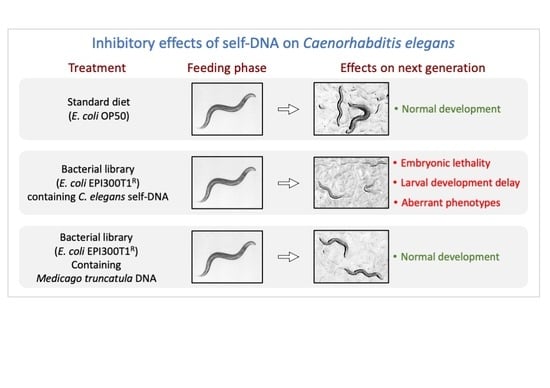
Self-DNA Exposure Induces Developmental Defects and Germline DNA Damage Response in Caenorhabditis elegans
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Caenorhabditis elegans and Escherichia coli Strains
2.2. Fosmid Libraries
2.3. Culture Conditions
2.4. Determination of Average Fosmid Copy Number
2.5. Phenotype Screening in C. elegans
2.6. Time Course of Larval Development (from Hatching to Adulthood)
2.7. Immunostaining in C. elegans Germline
2.8. Quantitative Analysis of Germline Apoptosis
2.9. Analysis of DAPI-Stained Bodies in the Germline Nuclei
2.10. Image Collection and Processing
2.11. Statistical Tools
3. Results
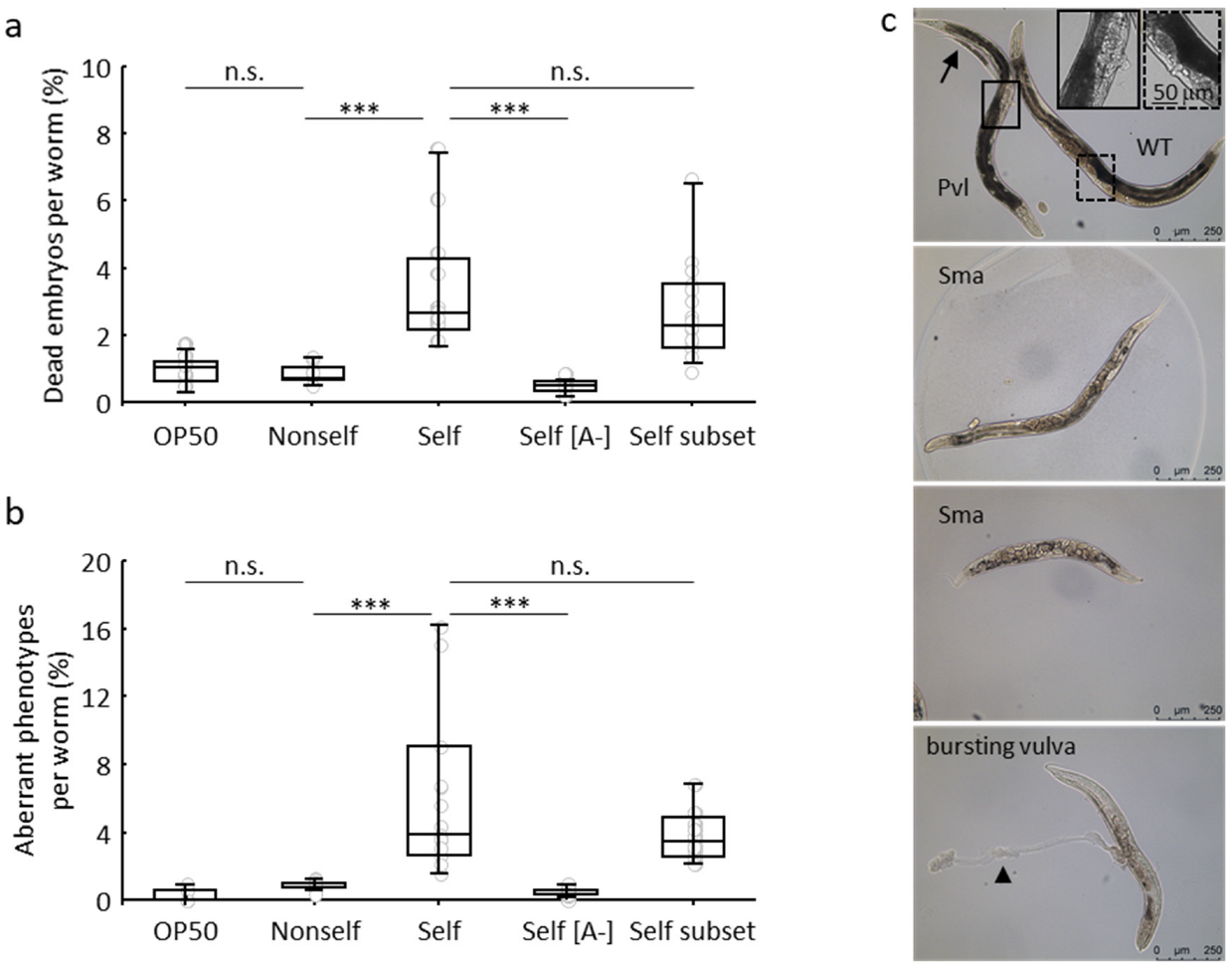
3.1. Negative Effects in C. elegans Fed on a self-DNA Bacterial Genomic Library
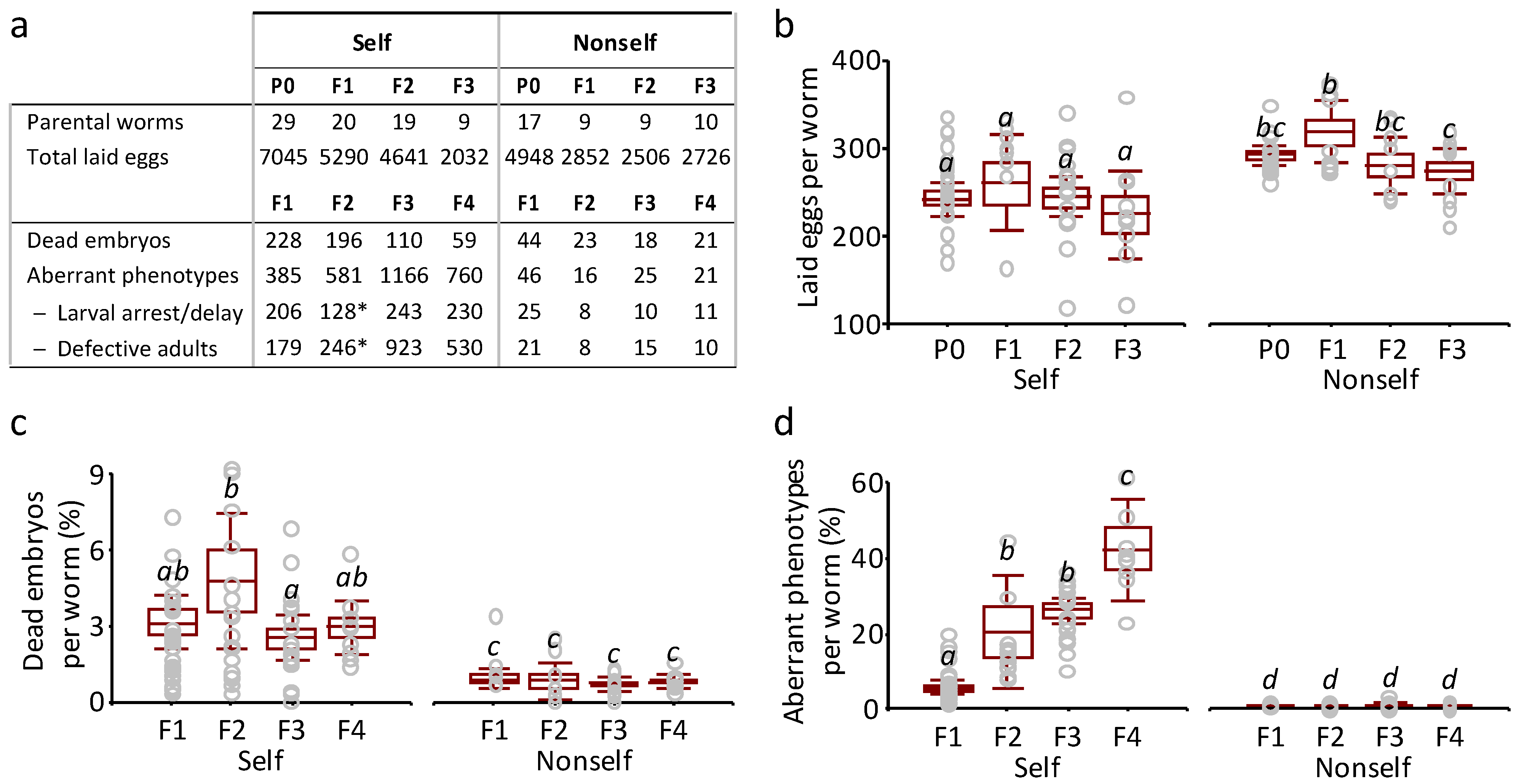
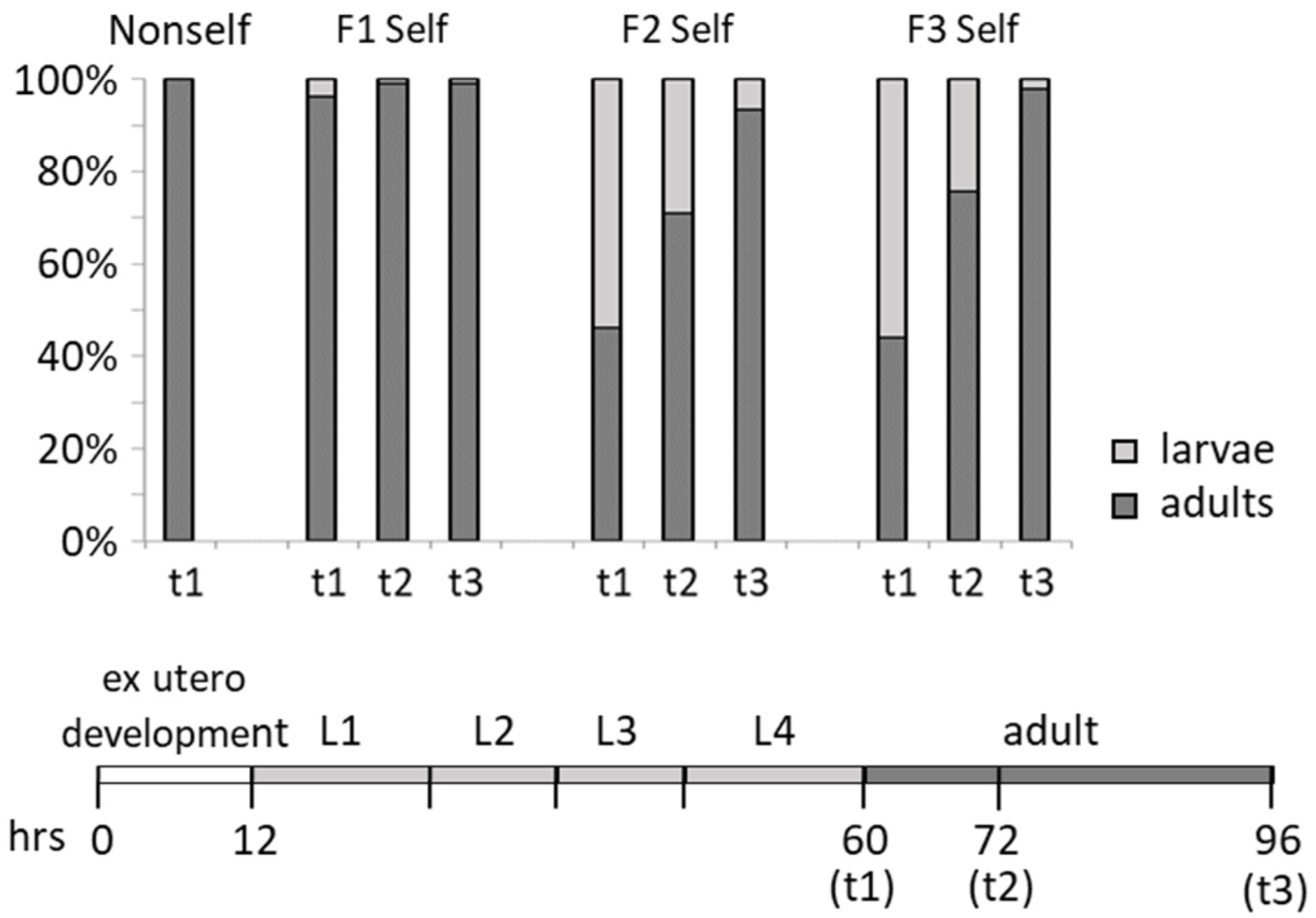
3.2. Detrimental Effects of the Self-DNA Diet across Generations
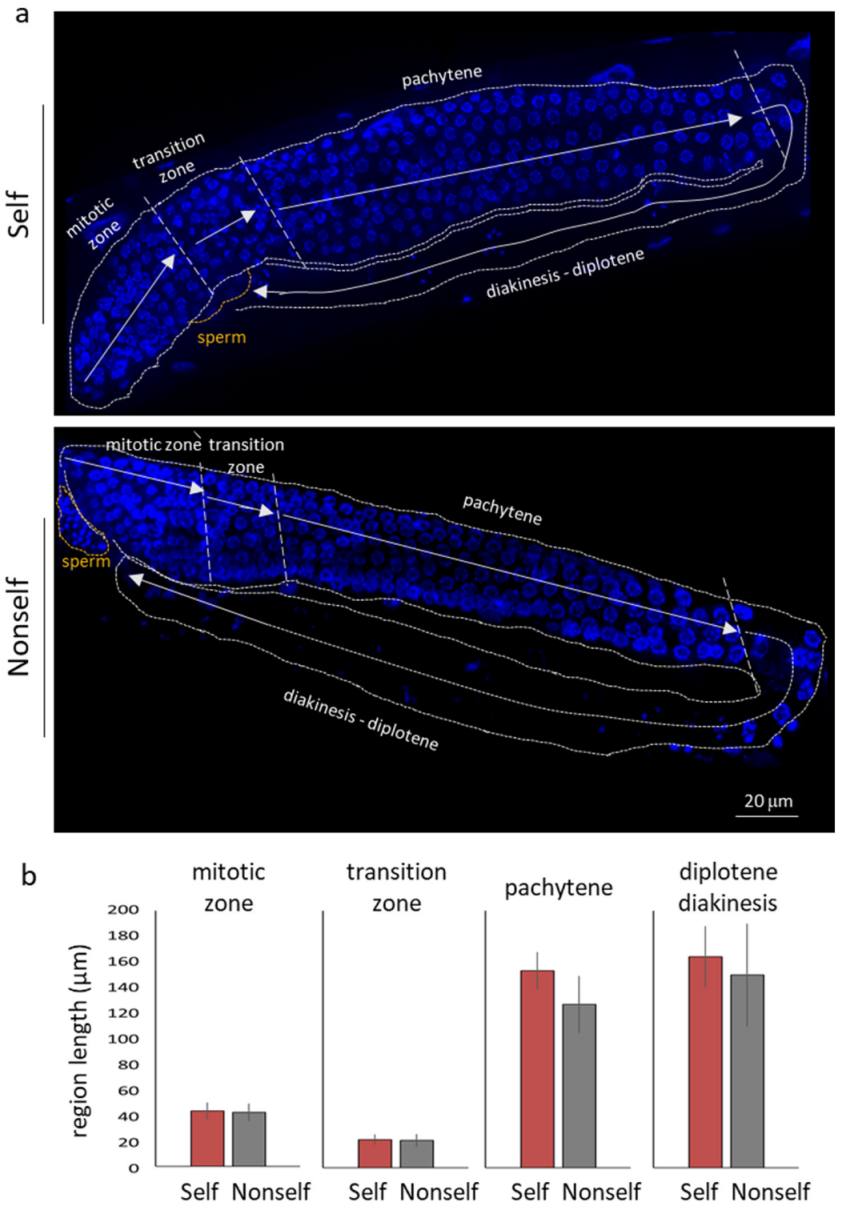
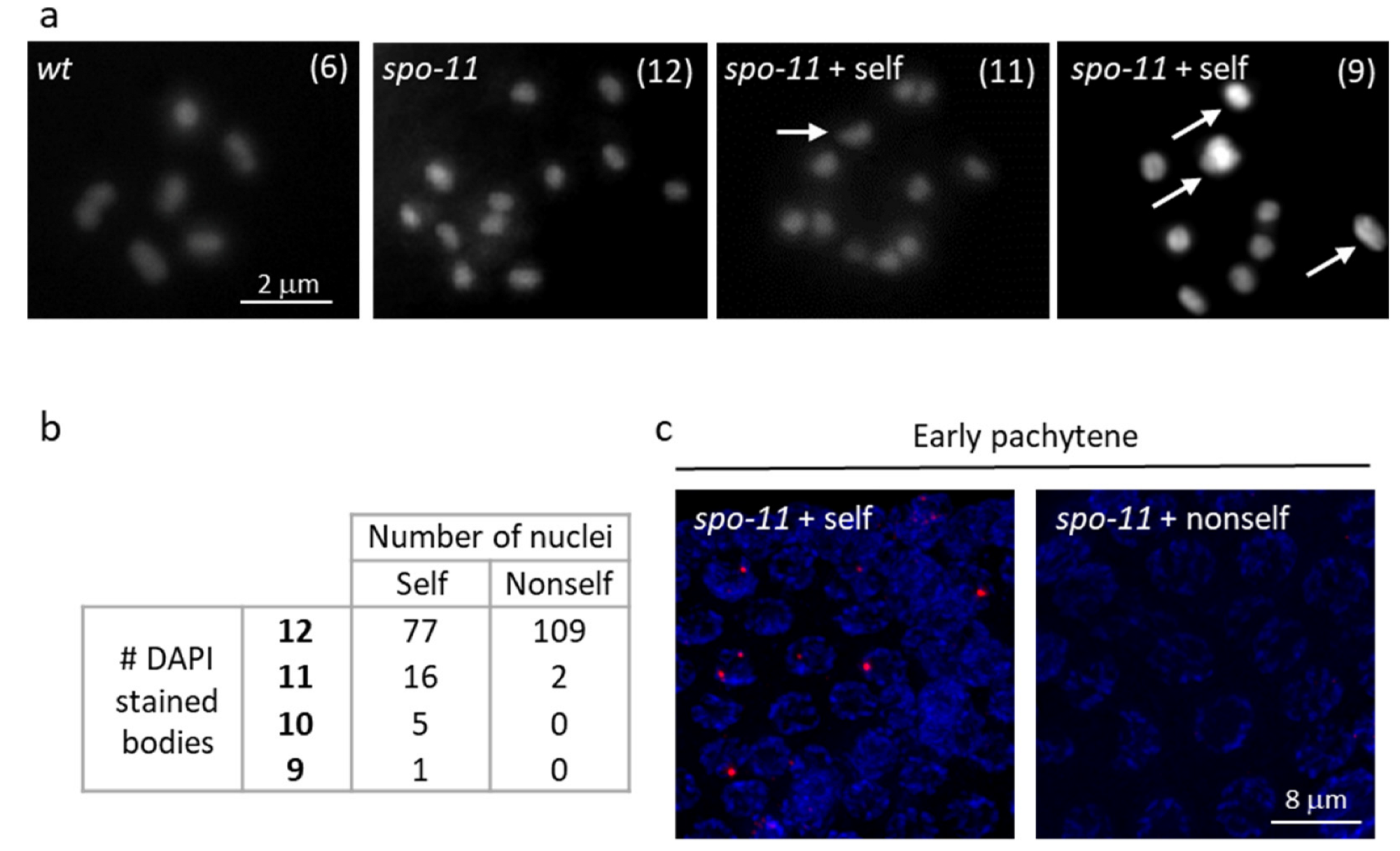
3.3. Self-DNA Feeding Induces DNA Damage and Apoptosis in the Germline
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Hu, M.M.; Shu, H.B. Innate Immune Response to Cytoplasmic DNA: Mechanisms and Diseases. Annu. Rev. Immunol. 2020, 38, 79–98. [Google Scholar] [CrossRef] [PubMed]
- Kawasaki, T.; Kawai, T. Discrimination Between Self and Non-Self-Nucleic Acids by the Innate Immune System. Int. Rev. Cell Mol. Biol. 2019, 344, 1–30. [Google Scholar] [PubMed]
- Mazzoleni, S.; Bonanomi, G.; Incerti, G.; Chiusano, M.L.; Termolino, P.; Mingo, A.; Senatore, M.; Giannino, F.; Cartenì, F.; Rietkerk, M.; et al. Inhibitory and toxic effects of extracellular self-DNA in litter: A mechanism for negative plant-soil feedbacks? New Phytol. 2015, 205, 1195–1210. [Google Scholar] [CrossRef] [PubMed]
- Mazzoleni, S.; Cartenì, F.; Bonanomi, G.; Senatore, M.; Termolino, P.; Giannino, F.; Incerti, G.; Rietkerk, M.; Lanzotti, V.; Chiusano, M.L. Inhibitory effects of extracellular self-DNA: A general biological process? New Phytol. 2015, 206, 127–132. [Google Scholar] [CrossRef]
- Duran-Flores, D.; Heil, M. Extracellular self-DNA as a damage-associated molecular pattern (DAMP) that triggers self-specific immunity induction in plants. Brain Behav. Immun. 2018, 72, 78–88. [Google Scholar] [CrossRef] [PubMed]
- Chiusano, M.L.; Incerti, G.; Colantuono, C.; Termolino, P.; Palomba, E.; Monticolo, F.; Benvenuto, G.; Foscari, A.; Esposito, A.; Marti, L.; et al. Arabidopsis thaliana Response to Extracellular DNA: Self Versus Nonself Exposure. Plants 2021, 10, 1744. [Google Scholar] [CrossRef]
- Matzinger, P. The danger model: A renewed sense of self. Science 2002, 296, 301–305. [Google Scholar] [CrossRef]
- Cohen, L.B.; Troemel, E.R. Microbial pathogenesis and host defense in the nematode C. elegans. Curr. Opin. Microbiol. 2015, 23, 94–101. [Google Scholar] [CrossRef]
- Pujol, N.; Zugasti, O.; Wong, D.; Couillault, C.; Kurz, C.L.; Schulenburg, H.; Ewbank, J.J. Anti-fungal innate immunity in C. elegans is enhanced by evolutionary diversification of antimicrobial peptides. PLoS Pathog. 2008, 4, e1000105. [Google Scholar] [CrossRef]
- Nakad, R.; Schumacher, B. DNA Damage Response and Immune Defense: Links and Mechanisms. Front. Genet. 2016, 7, 147. [Google Scholar] [CrossRef]
- Ermolaeva, M.A.; Segref, A.; Dakhovnik, A.; Ou, H.L.; Schneider, J.I.; Utermöhlen, O.; Hoppe, T.; Schumacher, B. DNA damage in germ cells induces an innate immune response that triggers systemic stress resistance. Nature 2013, 501, 416–420. [Google Scholar] [CrossRef]
- Aballay, A.; Ausubel, F.M. Programmed cell death mediated by ced-3 and ced-4 protects Caenorhabditis elegans from Salmonella typhimurium-mediated killing. Proc. Natl. Acad. Sci. USA 2001, 98, 2735–2739. [Google Scholar] [CrossRef]
- Brenner, S. The genetics of Caenorhabditis elegans. Genetics 1974, 77, 71–94. [Google Scholar] [CrossRef]
- Ichida, H.; Sun, X.; Imanaga, S.; Ito, Y.; Yoneyama, K.; Kuwata, S.; Ohsato, S. Construction and characterization of a copy number-inducible fosmid library of Xanthomonas oryzae pathovar oryzae MAFF311018. Gene 2014, 546, 68–72. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔCT method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Serpe, M.; Forenza, C.; Adamo, A.; Russo, N.; Perugino, G.; Ciaramella, M.; Valenti, A. The DNA alkylguanine DNA alkyltransferase-2 (AGT-2) Of Caenorhabditis elegans is involved in meiosis and early development under physiological conditions. Sci. Rep. 2019, 9, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Germoglio, M.; Valenti, A.; Gallo, I.; Forenza, C.; Santonicola, P.; Silva, N.; Adamo, A. In vivo analysis of FANCD2 recruitment at meiotic DNA breaks in Caenorhabditis elegans. Sci. Rep. 2020, 10, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Germoglio, M.; Adamo, A. A Role in Apoptosis Regulation for the rad-51 Gene of Caenorhabditis elegans. Genetics 2018, 209, 1017–1028. [Google Scholar] [CrossRef] [PubMed]
- Zhang, F.; Berg, M.; Dierking, K.; Félix, M.A.; Shapira, M.; Samuel, B.S.; Schulenburg, H. Caenorhabditis elegans as a Model for Microbiome Research. Front. Microbiol. 2017, 8, 485. [Google Scholar] [CrossRef]
- Timmons, L.; Fire, A. Specific interference by ingested dsRNA. Nature 1998, 395, 854. [Google Scholar] [CrossRef]
- Timmons, L.; Court, D.L.; Fore, A. Ingestion of bacterially expressed dsRNAs can produce specific and potent genetic interference in Caenorhabditis elegans. Gene 2001, 24, 103–112. [Google Scholar] [CrossRef]
- Ballestriero, F.; Thomas, T.; Burke, C.; Egan, S.; Kjelleberg, S. Identification of compounds with bioactivity against the nematode Caenorhabditis elegans by a screen based on the functional genomics of the marine bacterium Pseudoalteromonas tunicata D2. Appl. Environ. Microbiol. 2010, 76, 5710–5717. [Google Scholar] [CrossRef]
- Lam, K.N.; Charles, T.C. Strong spurious transcription likely contributes to DNA insert bias in typical metagenomic clone libraries. Microbiome 2015, 3, 22. [Google Scholar] [CrossRef]
- Yigit, E.; Batista, P.J.; Bei, Y.; Pang, K.M.; Chen, C.C.; Tolia, N.H.; Joshua-Tor, L.; Mitani, S.; Simard, M.J.; Mello, C.C. Analysis of the C. elegans Argonaute family reveals that distinct Argonautes act sequentially during RNAi. Cell 2006, 17, 747–757. [Google Scholar] [CrossRef] [PubMed]
- Sulston, J.E. Post-embryonic development in the ventral cord of Caenorhabditis elegans. Philos. Trans. R. Soc. Lond. B Biol. Sci. 1976, 10, 287–297. [Google Scholar]
- Hevelone, J.; Hartman, P.S. An Endonuclease from Caenorhabditis elegans: Partial Purification and Characterization. Biochem. Genet. 1988, 26, 447–461. [Google Scholar] [CrossRef]
- Wu, Y.C.; Stanfield, G.M.; Horvitz, H.R. NUC-1, a Caenorhabditis elegans DNase II homolog, functions in an intermediate step of DNA degradation during apoptosis. Genes Dev. 2000, 14, 536–548. [Google Scholar] [CrossRef] [PubMed]
- Dernburg, A.F.; McDonald, K.; Moulder, G.; Barstead, R.; Dresser, M.; Villeneuve, A.M. Meiotic recombination in C. elegans initiates by a conserved mechanism and is dispensable for homologous chromosome synapsis. Cell 1998, 94, 387–398. [Google Scholar] [CrossRef]
- Rinaldo, C.; Bazzicalupo, P.; Ederle, S.; Hilliard, M.; La Volpe, A. Roles for Caenorhabditis elegans rad-51 in meiosis and in resistance to ionizing radiation during development. Genetics 2002, 160, 471–479. [Google Scholar] [CrossRef]
- Colaiácovo, M.P.; MacQueen, A.J.; Martinez-Perez, E.; McDonald, K.; Adamo, A.; La Volpe, A.; Villeneuve, A.M. Synaptonemal complex assembly in C. elegans is dispensable for loading strand-exchange proteins but critical for proper completion of recombination. Dev. Cell 2003, 5, 463–474. [Google Scholar] [CrossRef]
- Couteau, F.; Zetka, M. HTP-1 coordinates synaptonemal complex assembly with homolog alignment during meiosis in C. elegans. Genes Dev. 2005, 19, 2744–2756. [Google Scholar] [CrossRef] [PubMed]
- Gumienny, T.L.; Lambie, E.; Hartwieg, E.; Horvitz, H.R.; Hengartner, M.O. Genetic control of programmed cell death in the Caenorhabditis elegans hermaphrodite germline. Development 1999, 126, 1011–1022. [Google Scholar] [CrossRef] [PubMed]
- Adamo, A.; Montemauri, P.; Silva, N.; Ward, J.D.; Boulton, S.J.; La Volpe, A. BRC-1 acts in the inter-sister pathway of meiotic double-strand break repair. EMBO Rep. 2008, 9, 7–12. [Google Scholar] [CrossRef]
- Adamo, A.; Collis, S.J.; Adelman, C.A.; Silva, N.; Horejsi, Z.; Ward, J.D.; Martinez-Perez, E.; Boulton, S.J.; La Volpe, A. Preventing nonhomologous end joining suppresses DNA repair defects of Fanconi anemia. Mol. Cell 2010, 39, 25–35. [Google Scholar] [CrossRef]
- Schumacher, B.; Hofmann, K.; Boulton, S.; Gartner, A. The C. elegans homolog of the p53 tumor suppressor is required for DNA damage-induced apoptosis. Curr. Biol. 2001, 11, 1722–1727. [Google Scholar] [CrossRef]
- Woglar, A.; Jantsch, V. Chromosome movement in meiosis I prophase of Caenorhabditis elegans. Chromosoma 2014, 123, 15–24. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Barbero, F.; Guglielmotto, M.; Capuzzo, A.; Maffei, M.E. Extracellular Self-DNA (esDNA), but Not Heterologous Plant or Insect DNA (etDNA), Induces Plasma Membrane Depolarization and Calcium Signaling in Lima Bean (Phaseolus lunatus) and Maize (Zea mays). Int. J. Mol. Sci. 2016, 17, 1659. [Google Scholar] [CrossRef]
- Vega-Muñoz, I.; Feregrino-Pérez, A.A.; Torres-Pacheco, I.; Guevara-González, R.G. Exogenous fragmented DNA acts as a damage-associated molecular pattern (DAMP) inducing changes in CpG DNA methylation and defence-related responses in Lactuca sativa. Funct. Plant Biol. 2018, 45, 1065–1072. [Google Scholar] [CrossRef]
- Yuen, G.J.; Ausubel, F.M. Both live and dead Entero- cocci activate Caenorhabditis elegans host defense via immune and stress pathways. Virulence 2018, 9, 683–699. [Google Scholar] [CrossRef]
- Kim, D.H.; Ewbank, J.J. Signaling in the innate immune response. WormBook Online Rev. C. Elegans Biol. 2018, 1–35. [Google Scholar] [CrossRef]
- Williams, A.B.; Heider, F.; Messling, J.E.; Rieckher, M.; Bloch, W.; Schumacher, B. Restoration of Proteostasis in the Endoplasmic Reticulum Reverses an Inflammation-LikeResponse to Cytoplasmic DNA in Caenorhabditis elegans. Genetics 2019, 212, 1259–1278. [Google Scholar] [CrossRef]
- Holway, A.H.; Kim, S.H.; La Volpe, A.; Michael, W.M. Checkpoint silencing during the DNA damage response in Caenorhabditis elegans embryos. J. Cell Biol. 2006, 172, 999–1008. [Google Scholar] [CrossRef] [PubMed]
- Roerink, S.F.; Koole, W.; Stapel, L.C.; Romeijn, R.J.; Tijsterman, M. A broad requirement for TLS polymerases η and κ, and interacting sumoylation and nuclear pore proteins, in lesion bypass during C. elegans embryogenesis. PLoS Genet. 2012, 8, e1002800. [Google Scholar] [CrossRef] [PubMed]
- Miller, E.V.; Grandi, L.N.; Giannini, J.A.; Robinson, J.D.; Powell, J.R. The Conserved G-Protein Coupled Receptor FSHR-1 Regulates Protective Host Responses to Infection and Oxidative Stress. PLoS ONE 2015, 10, e0137403. [Google Scholar] [CrossRef] [PubMed]
- Yu, H.; Lai, H.; Lin, T.; Chen, C.; Lo, S.J. Loss of DNase II function in the gonad is associated with a higher expression of antimicrobial genes in Caenorhabditis elegans. Biochem. J. 2015, 470, 145–154. [Google Scholar] [CrossRef] [PubMed]
- Chatzinikolaou, G.; Karakasilioti, I.; Garinis, G.A. DNA damage and innate immunity: Links and trade-offs. Trends Immunol. 2014, 35, 429–435. [Google Scholar] [CrossRef] [PubMed]
- Williams, A.B.; Schumacher, B. DNA damage responses and stress resistance: Concepts from bacterial SOS to metazoan immunity. Mech. Ageing Dev. 2017, 165, 27–32. [Google Scholar] [CrossRef]
- Wallis, D.C.; Nguyen, D.; Uebel, C.J.; Phillips, C.M. Visualization and Quantification of Transposon Activity in Caenorhabditis elegans RNAi Pathway Mutants. G3 2019, 9, 3825–3832. [Google Scholar] [CrossRef]
- Kurhanewicz, N.A.; Dinwiddie, D.; Bush, Z.D.; Libuda, D.E. Elevated Temperatures Cause Transposon-Associated DNA Damage in C. elegans Spermatocytes. Curr. Biol. 2020, 30, 5007–5017. [Google Scholar] [CrossRef]
- Dennis, S.; Sheth, U.; Feldman, J.L.; English, K.A.; Priess, J.R. C. elegans germ cells show temperature and age-dependent expression of Cer1, a Gypsy/Ty3-related retrotransposon. PLoS Pathog. 2012, 8, e1002591. [Google Scholar] [CrossRef]
- Ryan, C.P.; Brownlie, J.C.; Whyard, S. Hsp90 and Physiological Stress Are Linked to Autonomous Transposon Mobility and Heritable Genetic Change in Nematodes. Genome Biol. Evol. 2016, 8, 3794–3805. [Google Scholar] [CrossRef]
- Deniz, Ö.; Frost, J.M.; Branco, M.R. Regulation of transposable elements by DNA modifications. Nat. Rev. Genet. 2019, 20, 417–431. [Google Scholar] [CrossRef] [PubMed]
- Greer, E.L.; Blanco, M.A.; Gu, L.; Sendinc, E.; Liu, J.; Aristizábal-Corrales, D.; Hsu, C.H.; Aravind, L.; He, C.; Shi, Y. DNA Methylation on N6-Adenine in C. elegans. Cell 2015, 161, 868–878. [Google Scholar] [CrossRef]
- Kumar, A. Jump around: Transposons in and out of the laboratory. F1000Research 2020, 9, F1000 Faculty Rev-135. [Google Scholar] [CrossRef]
- Hoeven, R.V.; McCallum, K.C.; Cruz, M.R.; Garsin, D.A. Ce-Duox1/BLI-3 generated reactive oxygen species trigger protective SKN-1 activity via p38 MAPK signaling during infection in C. elegans. PLoS Pathog. 2011, 7, e1002453. [Google Scholar]
- Carbajal-Valenzuela, I.A.; Medina-Ramos, G.; Caicedo-Lopez, L.H.; Jiménez-Hernández, A.; Ortega-Torres, A.E.; Contreras-Medina, L.M.; Torres-Pacheco, I.; Guevara-González, R.G. Extracellular DNA: Insight of a Signal Molecule in Crop Protection. Biology 2021, 10, 1022. [Google Scholar] [CrossRef] [PubMed]
- Collins, R.A.; Wangensteen, O.S.; O’Gorman, E.J.; Mariani, S.; Sims, D.W.; Genner, M.J. Persistence of environmental DNA in marine systems. Commun. Biol. 2018, 1, 185. [Google Scholar] [CrossRef]
- Nagler, M.; Insam, H.; Pietramellara, G.; Ascher-Jenull, J. Extracellular DNA in natural environments: Features, relevance and applications. Appl. Microbiol. Biotechnol. 2018, 102, 6343–6356. [Google Scholar] [CrossRef] [PubMed]
- Pathan, S.I.; Arfaioli, P.; Ceccherini, M.T.; Ascher-Jenull, J.; Pietramellara, G. Preliminary evidences of the presence of extracellular DNA single stranded forms in soil. PLoS ONE 2020, 15, e227296. [Google Scholar] [CrossRef]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Germoglio, M.; Adamo, A.; Incerti, G.; Cartenì, F.; Gigliotti, S.; Storlazzi, A.; Mazzoleni, S. Self-DNA Exposure Induces Developmental Defects and Germline DNA Damage Response in Caenorhabditis elegans. Biology 2022, 11, 262. https://doi.org/10.3390/biology11020262
Germoglio M, Adamo A, Incerti G, Cartenì F, Gigliotti S, Storlazzi A, Mazzoleni S. Self-DNA Exposure Induces Developmental Defects and Germline DNA Damage Response in Caenorhabditis elegans. Biology. 2022; 11(2):262. https://doi.org/10.3390/biology11020262
Chicago/Turabian StyleGermoglio, Marcello, Adele Adamo, Guido Incerti, Fabrizio Cartenì, Silvia Gigliotti, Aurora Storlazzi, and Stefano Mazzoleni. 2022. "Self-DNA Exposure Induces Developmental Defects and Germline DNA Damage Response in Caenorhabditis elegans" Biology 11, no. 2: 262. https://doi.org/10.3390/biology11020262
APA StyleGermoglio, M., Adamo, A., Incerti, G., Cartenì, F., Gigliotti, S., Storlazzi, A., & Mazzoleni, S. (2022). Self-DNA Exposure Induces Developmental Defects and Germline DNA Damage Response in Caenorhabditis elegans. Biology, 11(2), 262. https://doi.org/10.3390/biology11020262