Invasion of African Clarias gariepinus Drives Genetic Erosion of the Indigenous C. batrachus in Bangladesh
Abstract
Simple Summary
Abstract
1. Introduction
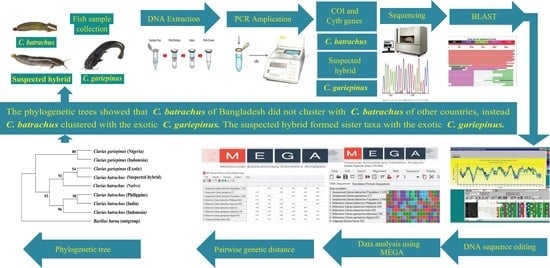
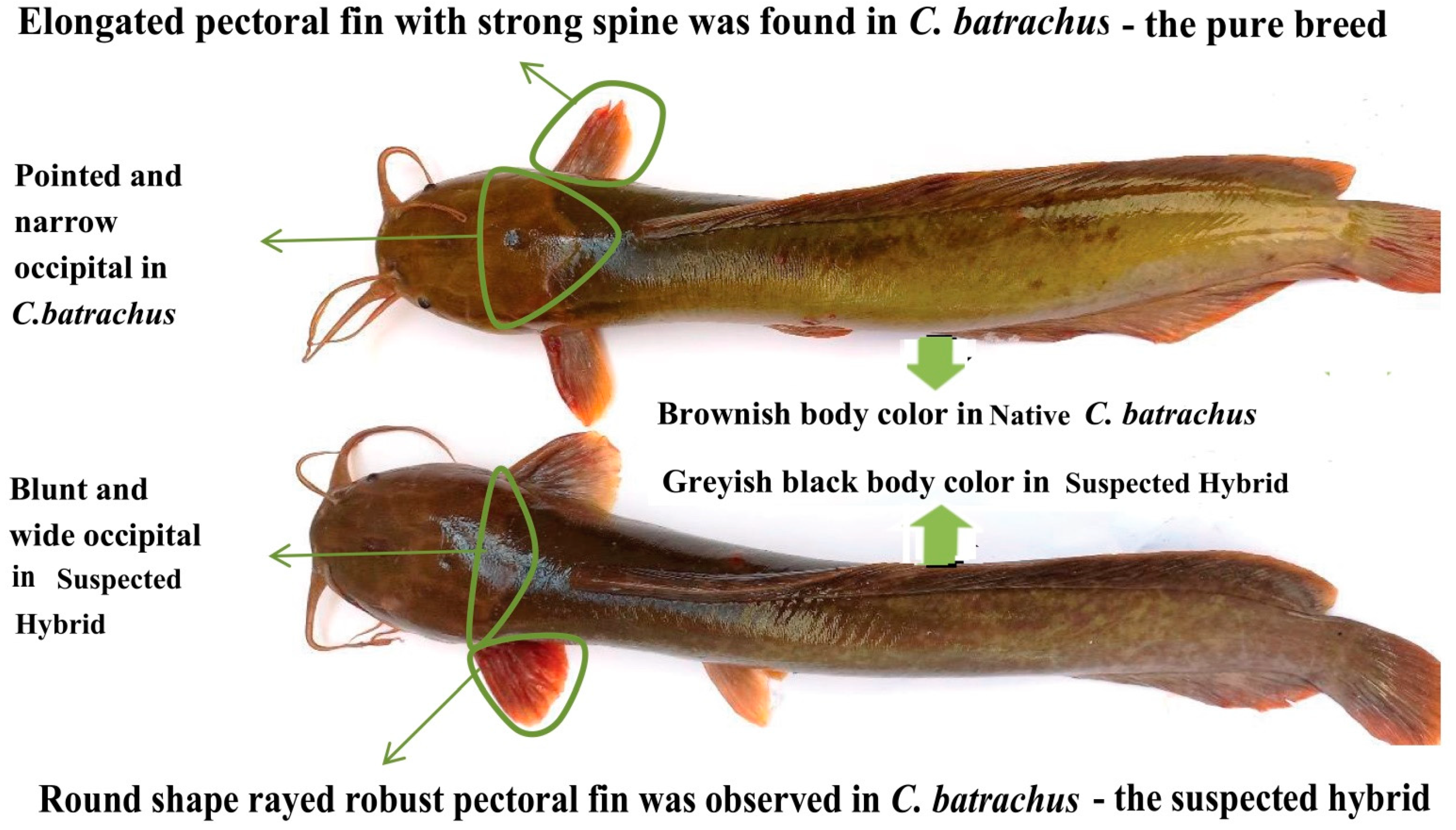
2. Materials and Methods
3. Results
3.1. Sequence Analysis of COI and Cytb Genes
3.2. Sequence Analysis of COI and Cytb Genes
3.3. Transition/Transversion Bias
3.4. Homogeneity of Substitution Patterns of COI Sequences
3.5. Genetic Distance
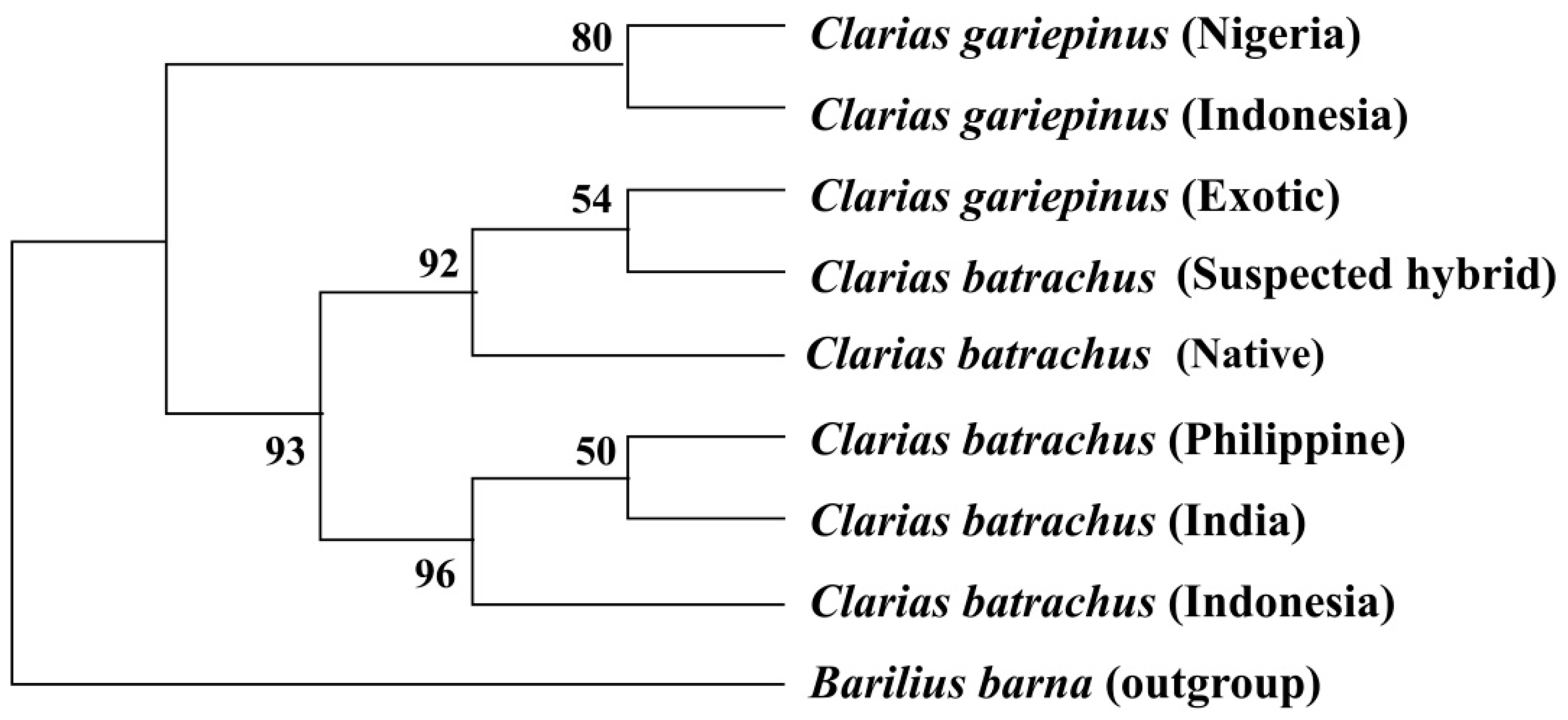
3.6. Phylogenetic Tree Using COI Nucleotide Sequences by Maximum Likelihood Methods
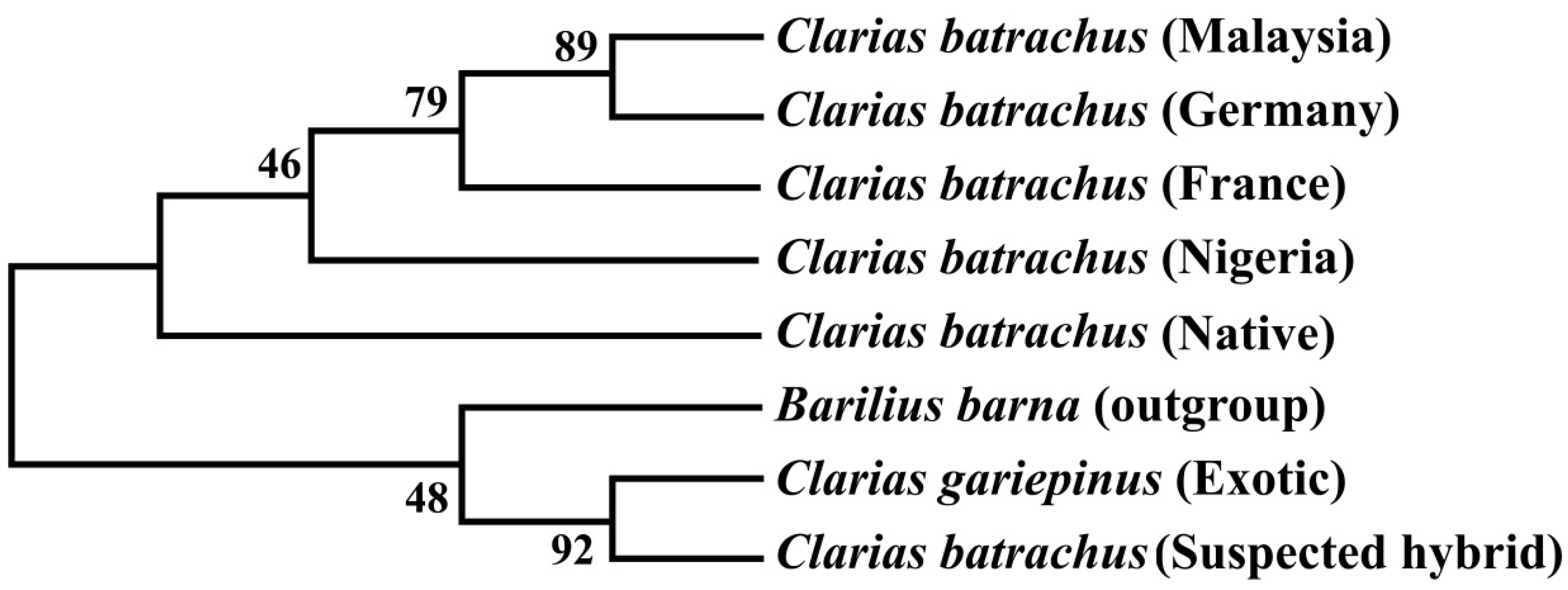
3.7. Phylogenetic Tree Using Cytb Nucleotide Sequences by Maximum Likelihood Method
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Linnaeus, C. Systema Naturae: Per Regna Tria Naturae, Secundum Classes, Ordines, Genera, Species, Cum Characteribus, Differentiis, Synonymis, Locis; Editio decima, reformata [10th revised edition]; Laurentius Salvius: Stockholm, Sweden, 1758; Volume 1, p. 824. [Google Scholar]
- Lakra, W.S.; Goswami, M.; Gopalakrishnan, A. Molecular identification and phylogenetic relationships of seven Indian Sciaenids (Pisces: Perciformes, Sciaenidae) based on 16S rRNA and cytochrome c oxidase subunit I mitochondrial genes. Mol. Biol. Rep. 2008, 36, 831–839. [Google Scholar] [CrossRef] [PubMed]
- Pouyaud, L.; Paradis, E. The phylogenetic structure of habitat shift and morphological convergence in Asian Clarias (Teleostei, Siluriformes: Clariidae). J. Zool. Syst. Evol. Res. 2009, 47, 344–356. [Google Scholar] [CrossRef]
- Rahman, A.K.A. Freshwater Fishes of Bangladesh, 2nd ed.; University of Dhaka: Dhaka, Bangladesh, 2005. [Google Scholar]
- Pradit, S.; Noppradit, P.; Goh, B.P.; Sornplang, K.; Ong, M.C.; Towatana, P. Occurrence of microplastics and trace metals in fish and shrimp from Songkhla Lake, Thailand during the COVID-19 pandemic. Appl. Ecol. Environ. Res. 2021, 19, 1085–1106. [Google Scholar] [CrossRef]
- Sahoo, S.K.; Giri, S.S.; Sahu, A.K.; Ayyappan, S. Experimental Hybridization between Catfish Clarias batrachus (Linn.) x Clarias gariepinus (Bur.) and Performance of the Offspring in Rearing Operations. Asian Fish. Sci. 2003, 16, 157–166. [Google Scholar] [CrossRef]
- Huang, C.F.; Lin, Y.H.; Chen, J.D. The use of RAPD markers to assess catfish hybridization. Biodivers. Conserv. 2005, 14, 3003–3014. [Google Scholar] [CrossRef]
- Szyper, J.P.; Tamaru, C.S.; Howerton, R.D.; Hopkins, K.D.; Fast, A.W.; Weidenbach, R.P. Maturation, Hatchery and Nursery Tech-Niques for Chinese Catfish, Clarias fuscus, in Hawaii. Aquaculture Extension Bulletin Summer 2001, UNIHJ-SEAGRANT-AB-01-01. Available online: http://citeseerx.ist.psu.edu/viewdoc/download?doi=10.1.1.548.9712&rep=rep1&type=pdf (accessed on 11 October 2021).
- Na-Nakorn, U.; Kamonrat, W.; Ngamsiri, T. Genetic diversity of walking catfish, Clarias macrocephalus, in Thailand and evidence of genetic introgression from introduced farmed C. gariepinus. Aquaculture 2004, 240, 145–163. [Google Scholar] [CrossRef]
- Froese, R.; Pauly, D. (Eds.) Clarias batrachus. Available online: https://www.fishbase.org/summary/Clarias-batrachus.html (accessed on 11 October 2021).
- Adan, R.I.Y. Catfish culture in Southeast Asia. SEAFDEC Asian Aquac. 2000, 22, 16–17. [Google Scholar]
- Thakur, N.K. A biological profile of African catfish Clarias gariepinus and impact of its introduction in Asia. In Fish Genetics and Biodiversity Conservation; Ponniah, A.G., Das, P., Verma, S.R., Eds.; Natcon Publications: Muzzafarnagar, India, 1998; pp. 275–292. [Google Scholar]
- Rahman, M.A.; Bhadra, A.; Begum, N.; Islam, M.S.; Hussain, M.G. Production of hybrid vigour through crossbreeding between Clarias batrachus Lin. and Clarias gariepinus Bur. Aquaculture 1995, 138, 125–130. [Google Scholar] [CrossRef]
- Welcomme, R.L.; Vidthayanon, C. The Impacts of Introduction and Stocking of Exotic Species in the Mekong Basin and Policies for Their Control; MRC Technical Paper; Mekong River Commission: Phnom Penh, Cambodia, 2003; p. 65. [Google Scholar]
- DOF (Department of Fisheries). Matshya Pakkhya Saranika; Department of Fisheries, Ministry of Fisheries and Livestock: Dhaka, Bangladesh, 2014; p. 208. [Google Scholar]
- Khedkar, G.D.; Anita, D.; Rushidkumar, N.; Shinde, D.; Haymer, D. High rates of substitution of the native catfish Clarias batrachus by Clarias gariepinus in India. Mitochondrial DNA Part A 2016, 27, 569–574. [Google Scholar] [CrossRef]
- Hebert, P.D.; Ratnasingham, S.; de Waard, J.R. Barcoding animal life: Cytochrome c oxidase subunit 1 divergences among closely related species. Proc. Biol. Sci. 2003, 270, S96–S99. [Google Scholar] [CrossRef]
- Parvez, I.; Mahajebin, T.; Clarke, M.; Chhanda, M.S.; Sultana, S. Genetic variation of native and introduced climbing perch, Anabas testudineus (Bloch, 1792) derived from mitochondrial DNA analyse. Ecol. Genet. Genom. 2020, 17, 100067. [Google Scholar] [CrossRef]
- Abbas, E.M.; Soliman, T.; El-Magd, M.A.; Farrag, M.M.S.; Ismail, R.F.; Kato, M. Phylogeny and DNA Barcoding of the Family Sparidae Inferred from Mitochondrial DNA of the Egyptian Waters. J. Fish. Aquat. Sci. 2017, 12, 73–81. [Google Scholar] [CrossRef][Green Version]
- Basheer, V.S.; Vineesh, N.; Bineesh, K.K.; Kumar, R.G.; Mohitha, C.; Venu, S.; Kathirvelpandian, A.; Gopalakrishnan, A.; Jena, J.K. Mitochondrial signatures for identification of grouper species from Indian waters. Mitochondrial DNA Part A 2017, 28, 451–457. [Google Scholar] [CrossRef] [PubMed]
- Chandra, S.; Barat, A.; Singh, M.; Singh, B.K.; Matura, R. DNA barcoding of Indian coldwater fishes of genus Schizothorax (Family: Cyprinidae) from western Himalaya. J. Fish. Mar. Sci. 2012, 4, 430–435. [Google Scholar]
- Malakar, A.K.; Lakra, W.S.; Goswami, M.S.; Minhra, R.M. Molecular identification of three Ompok species using mitochondrial COI gene. Mitochondrial DNA 2012, 23, 20–24. [Google Scholar] [CrossRef] [PubMed]
- Wong, L.L.; Peatman, E.; Lu, J.; Kucuktas, H.; He, S.; Zhou, C.; Na-nakorn, U.; Liu, Z. DNA barcoding of catfish: Species authentication and phylogenetic assessment. PLoS ONE 2011, 6, e17812. [Google Scholar] [CrossRef]
- Kochzius, M.; Seidel, C.; Antoniou, A.; Botla, S.K.; Campo, D.; Cariani, A.; Vazquez, E.G.; Hauschild, J.; Hervet, C.; Hjorleifsdottir, S.; et al. Identifying fishes through DNA barcodes and microarrays. PLoS ONE 2010, 5, e12620. [Google Scholar] [CrossRef]
- Zhang, J.; Hanner, R. Molecular approach to the identification of fish in the South China Sea. PLoS ONE 2012, 7, e30621. [Google Scholar] [CrossRef]
- An, H.S.; Kim, M.J.; Lee, J.W.; Lee, W.O. Molecular identification of Korean catfish (Siluriformes) based on two genetic markers. Genes Genet. 2012, 34, 695–702. [Google Scholar] [CrossRef]
- Pallavi; Goswami, M.; Nautiyal, P.; Malakar, A.K.; Nagpure, N.S. Genetic divergence and molecular phylogenetics of Puntius spp. based on the mitochondrial cytochrome b gene. Mitochondrial DNA 2012, 23, 477–483. [Google Scholar] [CrossRef]
- Talwar, P.K.; Jhingran, A.G. Inland Fisheries of India and Adjacent Countries; Oxford-IBH Publishing Co. Pvt. Ltd.: New Delhi, India, 1991; Volume 2. [Google Scholar]
- Safi, N.; Rauf, A.; Akhtar, T.; Minhas, R.A.; Ulfat, S.B. Comparative isolation and amplification of cytochrome oxidase 1 DNA from Oncorhynchus mykiss (rainbow trout) of azad jammu & kasmir. Int. J. Fish. Aquat. Stud. 2016, 4, 196–199. [Google Scholar]
- Hall, T.A. BioEdit: A user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp. Ser. 1999, 41, 95–98. [Google Scholar]
- Tamura, K.; Stecher, G.; Peterson, D.; Filipski, A.; Kumar, S. Molecular Evolutionary Genetics Analysis version 6.0. Molecular Biology and Evolution. Mol. Biol. Evol. 2012, 30, 2725–2729. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Gadagkar, S.R. Disparity Index: A simple statistic to measure and test the homogeneity of substitution patterns between molecular sequences. Genetics 2001, 158, 1321–1327. [Google Scholar] [CrossRef]
- Kimura, M. A simple method for estimating evolutionary rate of base substitutions through comparative studies of nucleotide sequences. J. Mol. Evol. 1980, 15, 111–120. [Google Scholar] [CrossRef]
- Falade, M.O.; Opene, A.J.; Benson, O. DNA barcoding of Clarias gariepinus, Coptodon zillii and Sarotherodon melanotheron from Southwestern Nigeria. F1000Research 2016, 5, 1268. [Google Scholar] [CrossRef]
- Devassy, A.; Kumar, R.; Shajitha, P.P.; John, R.; Padmakumar, K.G.; Basheer, V.S.; Gopalakrishnan, A.; Mathew, L. Genetic identification and phylogenetic relationships of Indian clariids based on mitochondrial COI sequences. DNA Mapp. Seq. 2016, 27, 3777–3780. [Google Scholar] [CrossRef]
- Sobita, N.; Basudha, C.; Sharma, S.K. Molecular characterization of an indigenous barb Pethia manipurensis (Cyprinidae: Barbinae) from Manipur, India and its relationship with other cyprinid fishes inferring from mitochondrial cox1 gene sequences. Indian J. Anim. Res. 2019, 53, 181–186. [Google Scholar] [CrossRef]
- Jaafar, T.N. DNA Barcoding and Population Genetic Structure of Malaysian Marine Fishes (Doctoral Dissertation, Prifysgol Bangor University). 2014. Available online: http://ethos.bl.uk/OrderDetails.do?uin=uk.bl.ethos.612627 (accessed on 29 October 2021).
- Yu, S.C.S.; Quilang, J.P. Molecular phylogeny of catfishes (Teleostei: Siluriformes) in the Philippines using the mitochondrial genes COI, Cyt b, 16S rRNA and the nuclear genes Rag1 and Rag2. Philipp. J. Sci. 2014, 143, 2015. [Google Scholar]

| Gene Name | Accession No | Species Name | Status | Country |
|---|---|---|---|---|
| COI | MG988399 | C. batrachus | Sequenced | Bangladesh |
| MG988400 | C. gariepinus | Sequenced | Bangladesh | |
| MG988401 | Suspected hybrid | Sequenced | Bangladesh | |
| HQ682681.1 | C. batrachus | Reference | Philippine | |
| KU692438.1 | C. batrachus | Reference | Indonesia | |
| KF214296 | C. batrachus | Reference | India | |
| KU69244.1 | C. gariepinus | Reference | Indonesia | |
| HM882828.1 | C. gariepinus | Reference | Nigeria | |
| Cytochrome b | MG988402 | C. batrachus | Sequenced | Bangladesh |
| MG988403 | C. gariepinus | Sequenced | Bangladesh | |
| MG988404 | Suspected hybrid | Sequenced | Bangladesh | |
| KR007705.1 | C. batrachus | Reference | Germany | |
| JF280859.1 | C. batrachus | Reference | Malaysia | |
| GU906881.1 | C. gariepinus | Reference | Nigeria | |
| AF235924.1 | C. gariepinus | Reference | France |
| No | Species Name | 1 | 2 | 3 | 4 | 5 | 6 | 7 |
|---|---|---|---|---|---|---|---|---|
| 1 | C. batrachus 1 (MG988399) | |||||||
| 2 | C. batrachus (Philiphine) | 0.189 | ||||||
| 3 | C. batrachus (Indonesia) | 0.691 | 0.000 | |||||
| 4 | C. batrachus (India) | 0.000 | 0.000 | 0.000 | ||||
| 5 | C. gariepinus (MG988400) | 0.000 | 1.328 | 1.961 | 0.900 | |||
| 6 | C. gariepinus (Indonesia) | 0.318 | 0.000 | 0.000 | 0.000 | 1.370 | ||
| 7 | C. gariepinus (Nigeria) | 0.146 | 0.000 | 0.000 | 0.000 | 1.068 | 0.000 | |
| 8 | Suspected hybrid (MG988401) | 0.000 | 1.258 | 1.909 | 0.923 | 0.000 | 1.440 | 1.235 |
| A | T | C | G | |
|---|---|---|---|---|
| A | - | 6.9605% | 6.2622% | 8.7613% |
| T | 5.9107% | - | 15.3969% | 4.5926% |
| C | 5.9107% | 17.1140% | - | 4.5926% |
| G | 11.2759% | 6.9605% | 6.2622% | - |
| Sl.No | Species Name | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 |
|---|---|---|---|---|---|---|---|---|---|
| 1 | C. batrachus 1 (MG988399) | 0.189 | 0.691 | 0.000 * | 0.000 * | 0.318 | 0.146 | 0.000 * | |
| 2 | C. batrachus (Philiphine) | 0.194 | 0.000 * | 0.000 * | 1.328 | 0.000 * | 0.000 * | 1.258 | |
| 3 | C. batrachus (Indonesia) | 0.108 | 1.000 | 0.000 * | 1.961 | 0.000 * | 0.000 * | 1.909 | |
| 4 | C. batrachus (India) | 1.000 | 1.000 | 1.000 | 0.900 | 0.000 * | 0.000 * | 0.923 | |
| 5 | C. gariepinus (MG988400) | 1.000 | 0.044 | 0.012 | 0.100 | 1.370 | 1.068 | 0.000 * | |
| 6 | C. gariepinus (Indonesia) | 0.234 | 1.000 | 1.000 | 1.000 | 0.032 | 0.000 * | 1.440 | |
| 7 | C. gariepinus (Nigeria) | 0.332 | 1.000 | 1.000 | 1.000 | 0.050 | 1.000 | 1.235 | |
| 8 | Suspected hybrid (MG988401) | 1.000 | 0.024 | 0.022 | 0.082 | 1.000 | 0.026 | 0.046 |
| No | Species Name | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 |
|---|---|---|---|---|---|---|---|---|---|
| 1 | C. batrachus 1 (MG988399) | ||||||||
| 2 | C. batrachus (Philiphine) | 4.931 | |||||||
| 3 | C. batrachus (Indonesia) | 3.283 | 3.110 | ||||||
| 4 | C. batrachus (India) | 4.122 | 2.137 | 2.302 | |||||
| 5 | C. gariepinus((MG988400) | 0.339 | 5.005 | 4.400 | 4.635 | ||||
| 6 | C. gariepinus (Indonesia) | 2.837 | 4.987 | 4.638 | 6.238 | 2.594 | |||
| 7 | C. gariepinus (Nigeria) | 4.055 | 4.636 | 5.095 | 3.147 | 4.025 | 2.315 | ||
| 8 | Suspectred hybrid (MG988401)) | 0.311 | 4.706 | 4.297 | 4.763 | 0.295 | 2.631 | 4.629 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Parvez, I.; Rumi, R.A.; Ray, P.R.; Hassan, M.M.; Sultana, S.; Pervin, R.; Suwanno, S.; Pradit, S. Invasion of African Clarias gariepinus Drives Genetic Erosion of the Indigenous C. batrachus in Bangladesh. Biology 2022, 11, 252. https://doi.org/10.3390/biology11020252
Parvez I, Rumi RA, Ray PR, Hassan MM, Sultana S, Pervin R, Suwanno S, Pradit S. Invasion of African Clarias gariepinus Drives Genetic Erosion of the Indigenous C. batrachus in Bangladesh. Biology. 2022; 11(2):252. https://doi.org/10.3390/biology11020252
Chicago/Turabian StyleParvez, Imran, Rukaya Akter Rumi, Purnima Rani Ray, Mohammad Mahbubul Hassan, Shirin Sultana, Rubaiya Pervin, Suvit Suwanno, and Siriporn Pradit. 2022. "Invasion of African Clarias gariepinus Drives Genetic Erosion of the Indigenous C. batrachus in Bangladesh" Biology 11, no. 2: 252. https://doi.org/10.3390/biology11020252
APA StyleParvez, I., Rumi, R. A., Ray, P. R., Hassan, M. M., Sultana, S., Pervin, R., Suwanno, S., & Pradit, S. (2022). Invasion of African Clarias gariepinus Drives Genetic Erosion of the Indigenous C. batrachus in Bangladesh. Biology, 11(2), 252. https://doi.org/10.3390/biology11020252