First Descriptive Analysis of the Faecal Microbiota of Wild and Anthropized Barbary Macaques (Macaca sylvanus) in the Region of Bejaia, Northeast Algeria
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Study Sites and Groups
2.2. Behavioural Data Collection
2.3. Faecal Sample
2.4. Extraction of Bacterial DNA
2.5. PCR Amplification and Product Quantification
2.6. Bioinformatics Analyses
2.7. Data Analysis
3. Results
3.1. Diet Composition of the Tourist-Provisioned Group
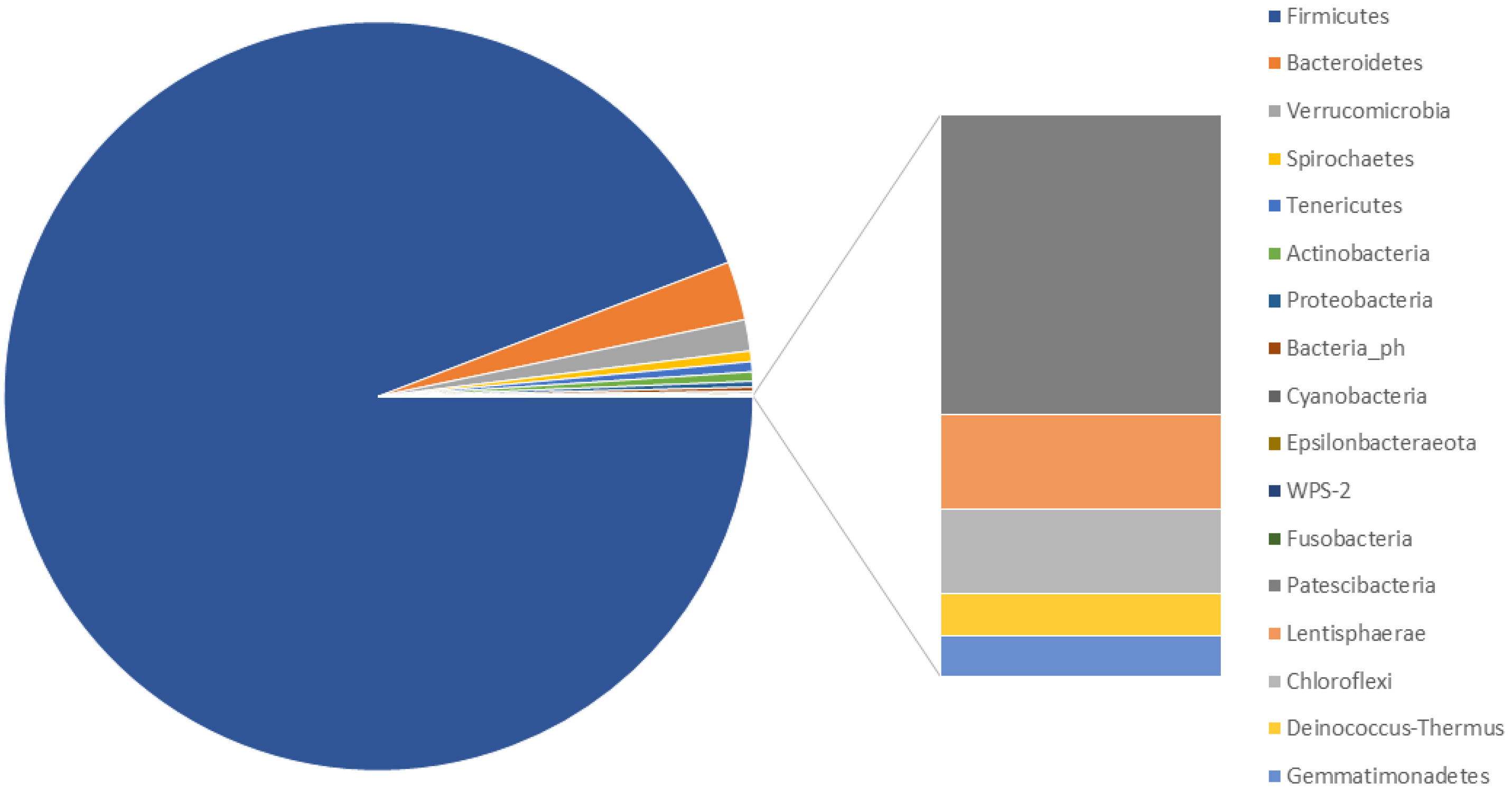
3.2. Characterisation of the Faecal Microbiota of M. sylvanus
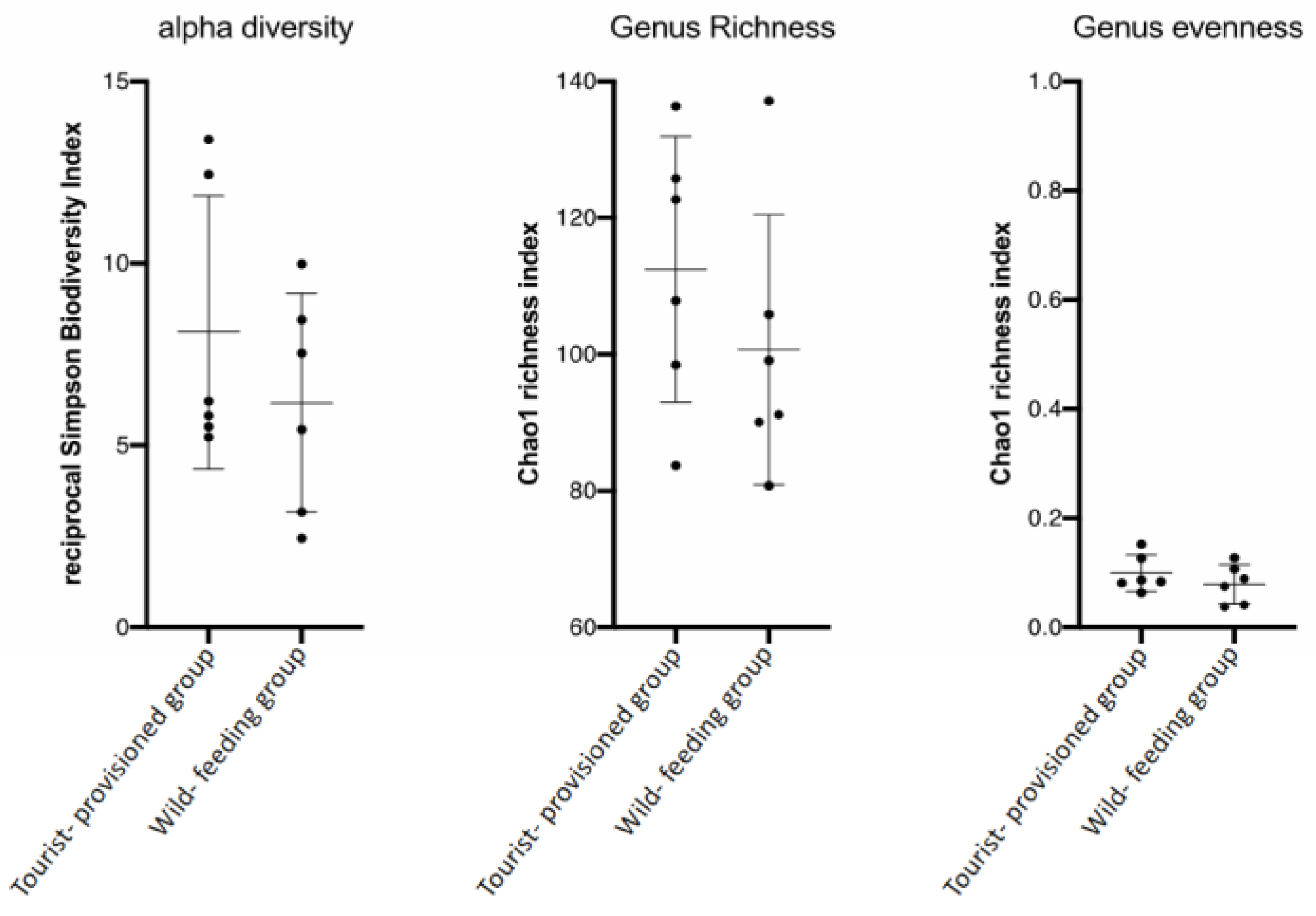
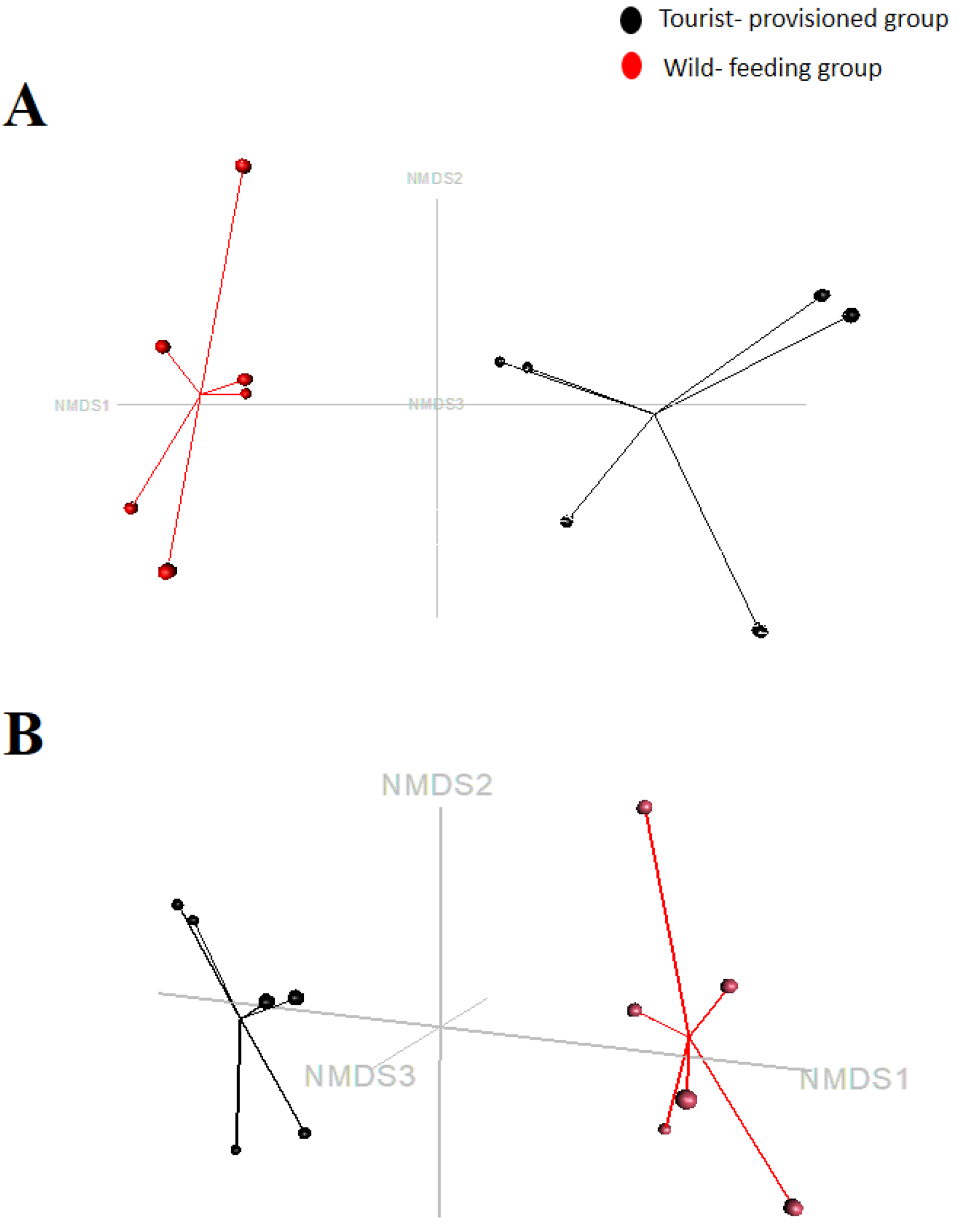
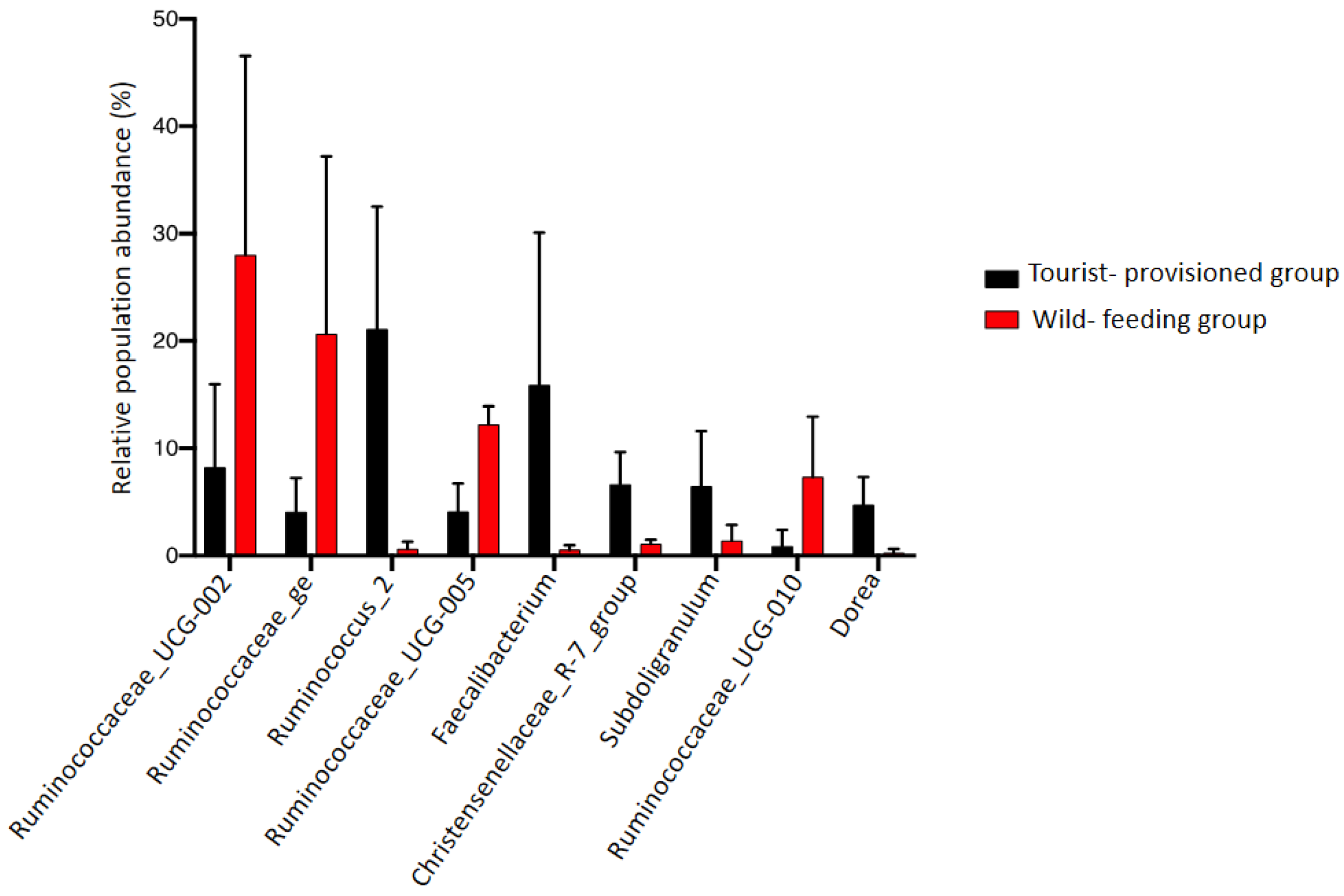
3.3. Evaluation of the Impact of Food Provisioning on the Faecal Microbiota of Barbary Macaques
4. Discussion
4.1. Diet Composition of the Tourist-Provisioned Group
4.2. Characterisation of M. sylvanus Faecal Microbiota
4.3. Impact of Food Provisioning on the Gut Microbiota of the Barbary Macaque
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Clayton, J.B.; Gomez, A.; Amato, K.; Knights, D.; Travis, D.A.; Blekhman, R.; Knight, R.; Leigh, S.; Stumpf, R.; Wolf, T.; et al. The gut microbiome of nonhuman primates: Lessons in ecology and evolution. Am. J. Primatol. 2018, 80, e22867. [Google Scholar] [CrossRef] [PubMed]
- Bayer, E.A.; Lamed, R.; White, B.A.; Flint, H.J. From cellulosomes to cellulosomics. Chem. Rec. 2008, 8, 364–377. [Google Scholar] [CrossRef] [PubMed]
- Round, J.L.; Mazmanian, S.K. The gut microbiota shapes intestinal immune responses during health and disease. Nat. Rev. Immunol. 2009, 9, 313–323. [Google Scholar] [CrossRef] [PubMed]
- Hume, I.D. Fermentation in the Hindgut of Mammals; Springer: Boston, MA, USA, 1997; pp. 84–115. [Google Scholar] [CrossRef]
- Milton, K.; McBee, R.H. Rates of fermentative digestion in the howler monkey, Alouatta palliata (primates: Ceboidea). Comp. Biochem. Physiol. Part A Physiol. 1983, 74, 29–31. [Google Scholar] [CrossRef]
- Popovich, D.; Jenkins, D.J.A.; Kendall, C.W.C.; Dierenfeld, E.S.; Carroll, R.W.; Tariq, N.; Vidgen, E. The Western Lowland Gorilla Diet Has Implications for the Health of Humans and Other Hominoids. J. Nutr. 1997, 127, 2000–2005. [Google Scholar] [CrossRef] [Green Version]
- Turnbaugh, P.J.; Ley, R.E.; Mahowald, M.A.; Magrini, V.; Mardis, E.R.; Gordon, J.I. An obesity-associated gut microbiome with increased capacity for energy harvest. Nature 2006, 444, 1027–1031. [Google Scholar] [CrossRef]
- Menni, C.; Jackson, A.M.; Pallister, T.; Steves, C.J.; Spector, T.D.; Valdes, A.M. Gut microbiome diversity and high-fibre intake are related to lower long-term weight gain. Int. J. Obes. 2017, 41, 1099–1105. [Google Scholar] [CrossRef] [Green Version]
- Brown, C.T.; Davis-Richardson, A.G.; Giongo, A.; Gano, K.A.; Crabb, D.B.; Mukherjee, N.; Casella, G.; Drew, J.C.; Ilonen, J.; Knip, M.; et al. Gut Microbiome Metagenomics Analysis Suggests a Functional Model for the Development of Autoimmunity for Type 1 Diabetes. PLoS ONE 2011, 6, e25792. [Google Scholar] [CrossRef] [Green Version]
- Bhattacharjee, S.; Lukiw, W.J. Alzheimer’s disease and the microbiome. Front. Cell. Neurosci. 2013, 7, 153. [Google Scholar] [CrossRef] [Green Version]
- Gevers, D.; Kugathasan, S.; Denson, L.A.; Vázquez-Baeza, Y.; Van Treuren, W.; Ren, B.; Schwager, E.; Knights, D.; Song, S.J.; Yassour, M.; et al. The Treatment-Naive Microbiome in New-Onset Crohn’s Disease. Cell Host Microbe 2014, 15, 382–392. [Google Scholar] [CrossRef] [Green Version]
- Hooper, L.V.; Midtvedt, T.; Gordon, J.I. How Host-Microbial Interactions Shape the Nutrient Environment of the Mammalian Intestine. Annu. Rev. Nutr. 2002, 22, 283–307. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Barelli, C.; Albanese, D.; Donati, C.; Pindo, M.; Dallago, C.; Rovero, F.; Cavalieri, D.; Tuohy, K.; Hauffe, H.C.; De Filippo, C. Habitat fragmentation is associated to gut microbiota diversity of an endangered primate: Implications for conservation. Sci. Rep. 2015, 5, 14862. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ley, R.E.; Hamady, M.; Lozupone, C.; Turnbaugh, P.J.; Ramey, R.R.; Bircher, J.S.; Schlegel, M.L.; Tucker, T.A.; Schrenzel, M.D.; Knight, R.; et al. Evolution of Mammals and Their Gut Microbes. Science 2008, 320, 1647–1651. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Amato, K.R.; Leigh, S.; Kent, A.; Mackie, R.I.; Yeoman, C.J.; Stumpf, R.M.; Wilson, B.A.; Nelson, K.E.; White, B.A.; Garber, P.A. The Gut Microbiota Appears to Compensate for Seasonal Diet Variation in the Wild Black Howler Monkey (Alouatta pigra). Microb. Ecol. 2014, 69, 434–443. [Google Scholar] [CrossRef] [PubMed]
- Hale, V.L.; Tan, C.L.; Niu, K.; Yang, Y.; Zhang, Q.; Knight, R.; Amato, K.R. Gut microbiota in wild and captive Guizhou snub-nosed monkeys, Rhinopithecus brelichi. Am. J. Primatol. 2019, 81, e22989. [Google Scholar] [CrossRef] [PubMed]
- Clayton, J.B.; Al-Ghalith, G.A.; Long, H.T.; Van Tuan, B.; Cabana, F.; Huang, H.; Vangay, P.; Ward, T.; Van Minh, V.; Tam, N.A.; et al. Associations Between Nutrition, Gut Microbiome, and Health in A Novel Nonhuman Primate Model. Sci. Rep. 2018, 8, 11159. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mallott, E.K.; Amato, K.R.; Garber, P.A.; Malhi, R.S. Influence of fruit and invertebrate consumption on the gut microbiota of wild white-faced capuchins (Cebus capucinus ). Am. J. Phys. Anthr. 2018, 165, 576–588. [Google Scholar] [CrossRef] [PubMed]
- Estrada, A.; Garber, P.A.; Rylands, A.B.; Roos, C.; Fernandez-Duque, E.; Di Fiore, A.; Nekaris, K.A.-I.; Nijman, V.; Heymann, E.W.; Lambert, J.E.; et al. Impending extinction crisis of the world’s primates: Why primates matter. Sci. Adv. 2017, 3, e1600946. [Google Scholar] [CrossRef] [Green Version]
- Chen, T.; Li, Y.; Liang, J.; Li, Y.; Huang, Z. Gut microbiota of provisioned and wild rhesus macaques (Macaca mulatta) living in a limestone forest in southwest Guangxi, China. MicrobiologyOpen 2019, 9, e981. [Google Scholar] [CrossRef] [Green Version]
- Wallis, J.; Benrabah, M.E.; Pilot, M.; Majolo, B.; Waters, S. Macaca sylvanus. The IUCN Red List of Threatened Species 2020: e. T12561A50043570. 2020. Available online: https://www.researchgate.net/profile/Sian-Waters/publication/344465217_Macaca_sylvanus_Barbary_Macaque_THE_IUCN_RED_LIST_OF_THREATENED_SPECIES/links/5f799632a6fdcc0086558213/Macaca-sylvanus-Barbary-Macaque-THE-IUCN-RED-LIST-OF-THREATENED-SPECIES.pdf (accessed on 24 October 2021).
- Ciani, A.C.; Palentini, L.; Arahou, M.; Martinoli, L.; Capiluppi, C.; Mouna, M. Population decline of Macaca sylvanus in the middle atlas of Morocco. Biol. Conserv. 2005, 121, 635–641. [Google Scholar] [CrossRef]
- Van Lavieren, E.; Wich, S.A. Decline of the Endangered Barbary macaque Macaca sylvanus in the cedar forest of the Middle Atlas Mountains, Morocco. Oryx 2009, 44, 133–138. [Google Scholar] [CrossRef] [Green Version]
- Bachiri, T.; Lalaoui, R.; Bakour, S.; Allouache, M.; Belkebla, N.; Rolain, J.M.; Touati, A. First Report of the Plasmid-Mediated Colistin Resistance Genemcr-1inEscherichia coliST405 Isolated from Wildlife in Bejaia, Algeria. Microb. Drug Resist. 2018, 24, 890–895. [Google Scholar] [CrossRef] [PubMed]
- Borg, C.; Majolo, B.; Qarro, M.; Semple, S. A Comparison of Body Size, Coat Condition and Endoparasite Diversity of Wild Barbary Macaques Exposed to Different Levels of Tourism. Anthrozoös 2014, 27, 49–63. [Google Scholar] [CrossRef]
- Medkour, H.; Amona, I.; Laidoudi, Y.; Davoust, B.; Bitam, I.; Levasseur, A.; Akiana, J.; Diatta, G.; Pacheco, L.; Gorsane, S.; et al. Parasitic Infections in African Humans and Non-Human Primates. Pathogens 2020, 9, 561. [Google Scholar] [CrossRef]
- Maibeche, Y.; Moali, A.; Yahi, N.; Ménard, N. Is Diet Flexibility an Adaptive Life Trait for Relictual and Peri-Urban Populations of the Endangered Primate Macaca sylvanus? PLoS ONE 2015, 10, e0118596. [Google Scholar] [CrossRef] [Green Version]
- Altmann, J. Observational Study of Behavior: Sampling Methods. Behaviour 1974, 49, 227–267. [Google Scholar] [CrossRef] [Green Version]
- Rodriguez, C.; Taminiau, B.; Korsak, N.; Avesani, V.; Van Broeck, J.; Brach, P.; Delmée, M.; Daube, G. Longitudinal survey of Clostridium difficile presence and gut microbiota composition in a Belgian nursing home. BMC Microbiol. 2016, 16, 229. [Google Scholar] [CrossRef] [Green Version]
- Rognes, T.; Flouri, T.; Nichols, B.; Quince, C.; Mahé, F. VSEARCH: A versatile open source tool for metagenomics. PeerJ 2016, 4, e2584. [Google Scholar] [CrossRef]
- Schloss, P.D.; Westcott, S.L.; Ryabin, T.; Hall, J.R.; Hartmann, M.; Hollister, E.B.; Lesniewski, R.A.; Oakley, B.B.; Parks, D.H.; Robinson, C.J.; et al. Introducing mothur: Open-Source, Platform-Independent, Community-Supported Software for Describing and Comparing Microbial Communities. Appl. Environ. Microbiol. 2009, 75, 7537–7541. [Google Scholar] [CrossRef] [Green Version]
- Kozich, J.J.; Westcott, S.L.; Baxter, N.T.; Highlander, S.K.; Schloss, P.D. Development of a dual-index sequencing strategy and curation pipeline for analyzing amplicon sequence data on the MiSeq Illumina sequencing platform. Appl. Environ. Microbiol. 2013, 79, 5112–5120. [Google Scholar] [CrossRef] [Green Version]
- Excoffier, L.; Smouse, P.E.; Quattro, J.M. Analysis of molecular variance inferred from metric distances among DNA haplotypes: Application to human mitochondrial DNA restriction data. Genetics 1992, 131, 479–491. [Google Scholar] [CrossRef] [PubMed]
- Stewart, C.N.; Excoffier, L. Assessing population genetic structure and variability with RAPD data: Application to Vaccinium macrocarpon (American Cranberry). J. Evol. Biol. 1996, 9, 153–171. [Google Scholar] [CrossRef]
- Bauer, A.S.; Arndt, T.P.; Leslie, E.K.; Pearl, D.L.; Turner, P.V. Obesity in rhesus and cynomolgus macaques: A comparative review of the condition and its implications for research. Comp. Med. 2011, 61, 514–526. [Google Scholar] [PubMed]
- Gower, J.C. Some Distance Properties of Latent Root and Vector Methods Used in Multivariate Analysis. Biometrika 1966, 53, 325. [Google Scholar] [CrossRef]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef] [Green Version]
- Ménard, N.; Qarro, M. Bark stripping and water availability: A comparative study between Moroccan and Algerian Barbary macaques (Macaca sylvanus). Rev. Ecot. 1999, 54, 123–132. [Google Scholar]
- Ménard, N.; Rantier, Y.; Foulquier, A.; Qarro, M.; Chillasse, L.; Vallet, D.; Pierre, J.-S.; Butet, A. Impact of human pressure and forest fragmentation on the Endangered Barbary macaque Macaca sylvanus in the Middle Atlas of Morocco. Oryx 2013, 48, 276–284. [Google Scholar] [CrossRef] [Green Version]
- Derlet, R.W.; Albertson, T.E. Activated charcoal-Past, present and future. West J. Med. 1986, 145, 493–496. [Google Scholar]
- Zellner, T.; Prasa, D.; Färber, E.; Hoffmann-Walbeck, P.; Genser, D.; Eyer, F. The Use of Activated Charcoal to Treat Intoxications. Deutsch. Aerzteblatt Int. 2019, 116, 311–317. [Google Scholar] [CrossRef]
- Orams, M.B. Feeding wildlife as a tourism attraction: A review of issues and impacts. Tour. Manag. 2002, 23, 281–293. [Google Scholar] [CrossRef]
- Jia, T.; Zhao, S.; Knott, K.; Li, X.; Liu, Y.; Li, Y.; Chen, Y.; Yang, M.; Lu, Y.; Wu, J.; et al. The gastrointestinal tract microbiota of northern white-cheeked gibbons (Nomascus leucogenys) varies with age and captive condition. Sci. Rep. 2018, 8, 3214. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, X.; Fan, P.; Che, R.; Li, H.; Yi, L.; Zhao, N.; Garber, P.A.; Li, F.; Jiang, Z. Fecal bacterial diversity of wild Sichuan snub-nosed monkeys (Rhinopithecus roxellana). Am. J. Primatol. 2018, 80, e22753. [Google Scholar] [CrossRef] [PubMed]
- Morotomi, M.; Nagai, F.; Watanabe, Y. Description of Christensenella minuta gen. nov., sp. nov., isolated from human faeces, which forms a distinct branch in the order Clostridiales, and proposal of Christensenellaceae fam. nov. Int. J. Syst. Evol. Microbiol. 2012, 62, 144–149. [Google Scholar] [CrossRef] [PubMed]
- Zhu, H.; Zeng, D.; Wang, Q.; Wang, N.; Zeng, B.; Niu, L.; Ni, X. Diarrhea-Associated Intestinal Microbiota in Captive Sichuan Golden Snub-Nosed Monkeys (Rhinopithecus roxellana). Microbes Environ. 2018, 33, 249–256. [Google Scholar] [CrossRef] [Green Version]
- Amato, K.R.; Martinez-Mota, R.; Righini, N.; Raguet-Schofield, M.; Corcione, F.P.; Marini, E.; Humphrey, G.; Gogul, G.; Gaffney, J.; Lovelace, E.; et al. Phylogenetic and ecological factors impact the gut microbiota of two Neotropical primate species. Oecologia 2015, 180, 717–733. [Google Scholar] [CrossRef]
- Clayton, J.B.; Vangay, P.; Huang, H.; Ward, T.; Hillmann, B.M.; Al-Ghalith, G.A.; Travis, D.A.; Long, H.T.; Van Tuan, B.; Van Minh, V.; et al. Captivity humanizes the primate microbiome. Proc. Natl. Acad. Sci. USA 2016, 113, 10376–10381. [Google Scholar] [CrossRef] [Green Version]
- Orkin, J.D.; Campos, F.A.; Myers, M.S.; Hernandez, S.E.C.; Guadamuz, A.; Melin, A.D. Seasonality of the gut microbiota of free-ranging white-faced capuchins in a tropical dry forest. ISME J. 2018, 13, 183–196. [Google Scholar] [CrossRef] [Green Version]
- Lee, W.; Hayakawa, T.; Kiyono, M.; Yamabata, N.; Hanya, G. Gut microbiota composition of Japanese macaques associates with extent of human encroachment. Am. J. Primatol. 2019, 81, e23072. [Google Scholar] [CrossRef]
- Christopherson, M.R.; Dawson, A.J.; Stevenson, D.M.; Cunningham, A.C.; Bramhacharya, S.; Weimer, P.J.; Kendziorski, C.; Suen, G. Unique aspects of fiber degradation by the ruminal ethanologen Ruminococcus albus 7 revealed by physiological and transcriptomic analysis. BMC Genom. 2014, 15, 1066. [Google Scholar] [CrossRef] [Green Version]
- Koike, S.; Kobayashi, Y. Fibrolytic Rumen Bacteria: Their Ecology and Functions. Asian-Australas. J. Anim. Sci. 2009, 22, 131–138. [Google Scholar] [CrossRef]
- Ferreira-Halder, C.V.; de Sousa Faria, A.V.; Andrade, S.S. Action and function of Faecalibacterium prausnitzii in health and disease. Best Pract. Res. Clin. Gastroenterol. 2017, 31, 643–648. [Google Scholar] [CrossRef] [PubMed]
- Remely, M.; Tesar, I.; Hippe, B.; Gnauer, S.; Rust, P.; Haslberger, A. Gut microbiota composition correlates with changes in body fat content due to weight loss. Benef. Microbes 2015, 6, 431–439. [Google Scholar] [CrossRef] [PubMed]
- Zhou, H.; Zhou, S.-Y.; Gillilland, M.; Li, J.-Y.; Lee, A.; Gao, J.; Zhang, G.; Xu, X.; Owyang, C. Bile acid toxicity in Paneth cells contributes to gut dysbiosis induced by high-fat feeding. JCI Insight 2020, 5, e138881. [Google Scholar] [CrossRef] [PubMed]
- Derrien, M.; Vaughan, E.E.; Plugge, C.M.; De Vos, W.M. Akkermansia muciniphila gen. nov., sp. nov., a human intestinal mucin-degrading bacterium. Int. J. Syst. Evol. Microbiol. 2004, 54, 1469–1476. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Everard, A.; Belzer, C.; Geurts, L.; Ouwerkerk, J.P.; Druart, C.; Bindels, L.B.; Guiot, Y.; Derrien, M.; Muccioli, G.G.; Delzenne, N.M.; et al. Cross-talk between Akkermansia muciniphila and intestinal epithelium controls di-et-induced obesity. Proc Natl. Acad. Sci. USA 2013, 110, 9066–9071. [Google Scholar] [CrossRef] [Green Version]
- Zhang, T.; Ji, X.; Lu, G.; Zhang, F. The potential of Akkermansia muciniphila in inflammatory bowel disease. Appl. Microbiol. Biotechnol. 2021, 105, 5785–5794. [Google Scholar] [CrossRef]
- Hanya, G.; Tackmann, J.; Sawada, A.; Lee, W.; Pokharel, S.S.; Maciel, V.G.D.C.; Toge, A.; Kuroki, K.; Otsuka, R.; Mabuchi, R.; et al. Fermentation Ability of Gut Microbiota of Wild Japanese Macaques in the Highland and Lowland Yakushima: In Vitro Fermentation Assay and Genetic Analyses. Microb. Ecol. 2020, 80, 459–474. [Google Scholar] [CrossRef]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Boumenir, M.; Hornick, J.-L.; Taminiau, B.; Daube, G.; Brotcorne, F.; Iguer-Ouada, M.; Moula, N. First Descriptive Analysis of the Faecal Microbiota of Wild and Anthropized Barbary Macaques (Macaca sylvanus) in the Region of Bejaia, Northeast Algeria. Biology 2022, 11, 187. https://doi.org/10.3390/biology11020187
Boumenir M, Hornick J-L, Taminiau B, Daube G, Brotcorne F, Iguer-Ouada M, Moula N. First Descriptive Analysis of the Faecal Microbiota of Wild and Anthropized Barbary Macaques (Macaca sylvanus) in the Region of Bejaia, Northeast Algeria. Biology. 2022; 11(2):187. https://doi.org/10.3390/biology11020187
Chicago/Turabian StyleBoumenir, Mourad, Jean-Luc Hornick, Bernard Taminiau, Georges Daube, Fany Brotcorne, Mokrane Iguer-Ouada, and Nassim Moula. 2022. "First Descriptive Analysis of the Faecal Microbiota of Wild and Anthropized Barbary Macaques (Macaca sylvanus) in the Region of Bejaia, Northeast Algeria" Biology 11, no. 2: 187. https://doi.org/10.3390/biology11020187
APA StyleBoumenir, M., Hornick, J.-L., Taminiau, B., Daube, G., Brotcorne, F., Iguer-Ouada, M., & Moula, N. (2022). First Descriptive Analysis of the Faecal Microbiota of Wild and Anthropized Barbary Macaques (Macaca sylvanus) in the Region of Bejaia, Northeast Algeria. Biology, 11(2), 187. https://doi.org/10.3390/biology11020187







