Simple Summary
COVID-19 emerged as a new disease with quick transmission and a high mortality rate at the end of 2019, caused by SARS-CoV-2. Common features of the coronavirus family helped resolve structural and entry mechanism characteristics of SARS-CoV-2. Still, rapid mutagenesis leads to the fast evolution of the virus and the emergence of new strains that differ in infectivity, morbidity, and mortality. Besides differences in the viral genome, genetic variability in the host defense and immune systems may also play a role in the outcome of virus–host interactions. Furthermore, epigenetic mechanisms may also influence the outcomes, including miRNA gene silencing and DNA methylation, which may be heavily influenced by SARS-CoV-2. Molecular biomarkers are intensively investigated as potential predictive and prognostic biomarkers of the disease course and treatment response. We reviewed new data regarding the mechanisms behind fast virus mutagenesis, infectivity, and potential human genetic and epigenetic characteristics that may lead to a more severe or lethal outcome of the disease.
Abstract
The rapid spread of COVID-19 outbreak lead to a global pandemic declared in March 2020. The common features of corona virus family helped to resolve structural characteristics and entry mechanism of SARS-CoV-2. However, rapid mutagenesis leads to the emergence of new strains that may have different reproduction rates or infectivity and may impact the course and severity of the disease. Host related factors may also play a role in the susceptibility for infection as well as the severity and outcomes of the COVID-19. We have performed a literature and database search to summarize potential viral and host-related genomic and epigenomic biomarkers, such as genetic variability, miRNA, and DNA methylation in the molecular pathway of SARS-CoV-2 entry into the host cell, that may be related to COVID-19 susceptibility and severity. Bioinformatics tools may help to predict the effect of mutations in the spike protein on the binding to the ACE2 receptor and the infectivity of the strain. SARS-CoV-2 may also target several transcription factors and tumour suppressor genes, thus influencing the expression of different host genes and affecting cell signalling. In addition, the virus may interfere with RNA expression in host cells by exploiting endogenous miRNA and its viral RNA. Our analysis showed that numerous human miRNA may form duplexes with different coding and non-coding regions of viral RNA. Polymorphisms in human genes responsible for viral entry and replication, as well as in molecular damage response and inflammatory pathways may also contribute to disease prognosis and outcome. Gene ontology analysis shows that proteins encoded by such polymorphic genes are highly interconnected in regulation of defense response. Thus, virus and host related genetic and epigenetic biomarkers may help to predict the course of the disease and the response to treatment.
1. Introduction
COVID-19 emerged as a new disease with quick transmission and a high mortality rate at the end of 2019, caused by SARS-CoV-2. In mid-2020, the pandemic was declared. Since then, more than 200,000 scientific papers have been published on COVID-19.
SARS-CoV-2 enters the cell through the binding of S1 proteins with the help of the ACE2 receptor [1,2] (Figure 1). TMPRSS2 and furin are necessary for proteolytic activation of the virus SARS-CoV-2 [3], since furin and TMPRSS2 inhibitors were shown to block SARS-CoV-2. [3]. TMPRSS2 cleaves S1/S2 at the cleavage site in SARS-CoV-2, a process requiring pre-cleavage by furin [4,5]. Once inside the lysosome, the viral envelope gets degraded with lysosome protease. RNA-dependent RNA polymerase (RdRp) is first to be translated. With the help of RdRp, a negative strand of RNA is produced that serves as a template for the multiplication of viral RNA. It was suggested that SARS-CoV-2 RNAs could be reverse transcribed and integrated into the human genome [6]. The large CpG island (18 CpG) in region 151–368 could contribute to the regulation of the expression of the viral template at the appropriate time for virus reactivation (UCSC Genome Browser, CpG islands, http://genome.ucsc.edu/ (accessed 15 December 2021)).
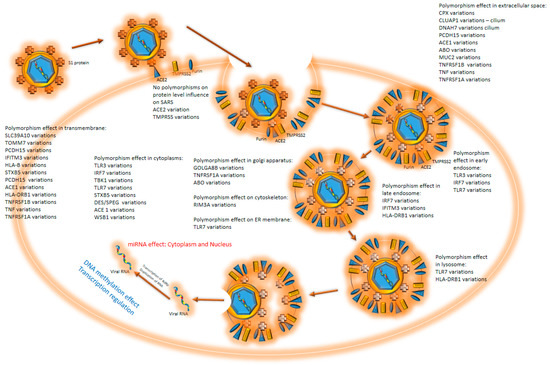
Figure 1.
COVID 19 cell entry: virus enters the cell through interaction with ACE2 protein. Upon binding, the protease cleaves S1 protein. S1 interacts with different cell membrane proteins, which causes the membrane to envelop the virus, accelerating endocytosis. The endosome goes through all phases to the lysosome, where RNA is released from the endosome and enters the cytoplasm. At all stages, polymorphisms may influence the virus entry and duplication. When virus RNA is released into the cytoplasm, miRNA effects step in. Virus offensive mechanisms contra cell defense efforts and continue to duplicate. Human DNA methylation effects should be noticed throughout the process.
From the cytoplasm, where proteins are translated, proteins get transported into the endoplasmic reticulum (ER), where they are post-translationally modified and transported towards the Golgi apparatus (GA) via the intermediate compartment (IC) [7]. IC virion is assembled by wrapping virion proteins around N protein, bound to viral RNA. The assembled viral particles may be exocytosed from the cell [2,8]. SARS-CoV-2 was detected in multiple organs of a COVID-19 patient who had died because of a multiorgan failure. Besides the respiratory system (e.g., lungs and trachea), it also infected the kidneys, small intestines, pancreas, blood vessels, and other tissues, such as sweat glands and vascular endothelial cells in the skin [9].
SARS-CoV-2 replicates more actively and effectively in human lung tissues than SARS-CoV; a higher viral load was found likely due to ongoing immune evasion mechanisms or defective viral clearance [10]. Mutations in the S1 protein or other regions involved in binding and entry into the human cell have been associated with the infectivity of a different strain of SARS-CoV-2.
Patients infected with SARS-CoV-2 may have no symptoms or develop a critical illness. Five different categories of severity exist according to the NIH data, as follows: (1) asymptomatic, pre-symptomatic infection—positive test and no symptoms; (2) mild illness with symptoms—fever, cough, sore throat, malaise, headache, muscle pain, nausea, vomiting, diarrhoea, and loss of taste and smell—but without shortness of breath, dyspnoea, or abnormal chest imaging; (3) moderate illness with lower respiratory disease and oxygen saturation ≥ 94%; (4) severe illness with oxygen saturation under 94%, the ratio of arterial partial pressure of oxygen to fraction of inspired oxygen < 300 mm, a respiratory rate > 30 breaths/min, or lung infiltrates > 50%; (5) critical illness with respiratory failure, septic shock, and/or multiple organ dysfunction [11].
COVID-19 symptoms are fever, a dry cough, dyspnoea, myalgia, and pneumonia. Some patients also reported sore throat, rhinorrhoea, headache, and hyposmia [12]. COVID-19 patients that needed hospitalization or intensive care unit (ICU) presented with pneumonia with fever, lymphopenia, highly elevated pro-inflammatory cytokines, C-reactive protein (CRP), serum ferritin, and D-dimers [13]. Plasma levels of IL2, IL7, IL10, GSCF, IP10, MCP1, MIP1A, and TNFα were higher in ICU patients compared with non-ICU patients [14].
Genetic predisposition for viral infection or disease progression has been proposed/suggested. Biomarkers can alarm the medical doctors to susceptibility or resistance of the patient towards SARS-CoV-2. Human biomarkers can be used to detect and predict the severity or life-threatening condition of COVID-19 disease. Disease severity can be foreseen prior to infection with COVID-19.
In this review, we have performed a literature and database search to summarize potential viral and host-related genomic and epigenomic biomarkers, such as genetic variability, miRNA, and DNA methylation in the molecular pathway of SARS-CoV-2 entry into the host cell, that may be related to COVID-19 susceptibility and severity.
2. Virus Mutations
2.1. Virus Strains
Viral mutations and recombination gave birth to new strains that may have different reproduction rates or infectivity and may impact the course and severity of the disease.
The COVID-19 virus strains were named after Greek alphabetical letters, and the designation is based on the positions and number of mutations. There are some disagreements regarding mutations belonging to specific strain groups, probably because different mutations evolved and spread further on different continents and states. Mutations labeled with * are present in some strains [15]. The Alpha variant has mutations of sites E484K*, D614G, delH69V70, and N501Y. Next, the Beta strain, characterized by E484K, D614G, A701V, N501Y, L242_244L, and K417N mutations, and the Gamma strain, with mutations of sites E484K, D614G, K417T, N501Y, and T20, evolved, and both of them outcompeted the wild-type strain [16]. The Delta variant, with mutations of sites D614G, L452R, P681R, and T478K emerged in April 2021. In July 2021, the Delta variant outcompeted all the other strains [16]. Iota has mutations of sites A701V*, E484K*, L452R*, and D614G; Epsilon has mutations of sites L452R and D614G; Eta has mutations of sites E484K, D614G, and delH69V70; Kappa has mutations of sites E484Q, D614G, L452R, and P681R.
Another classification was made with slightly different mutations: Beta (B.1.351) has mutations of sites N501Y, E484K, K417N; Alpha (B.1.1.7) has mutations of sites N501Y, E484K, and HV69/70del; Gamma (P.1) has mutations of sites N501Y, E484K, K417T, and V1176F; Delta (B.1.617.2) has mutations of sites N501, E484, and (L452R, P681R, T478K) [17]. The third classification defines that the Delta strain holds L452R and T478K mutations, the Epsilon (B.1.427 and B.1.429) strain holds L452R mutations, and the Kappa (B.1.617.1) strain holds L452R and E484Q mutations [18].
A new Omicron variant (B.1.1.529) emerged at the end of November 2021 [19]. It is classified as a variant of concern (next to Beta, Gamma, and Delta) and holds 33 spike protein mutations, many of which were found in the Alpha and Delta strains [19].
2.2. Virus Mutations Position and Their Influence on SARS 2 Disease Development
SARS-CoV, which has a similar structure and RNA sequence to SARS-CoV2, had an estimated mutation rate of approximately 0.80–2.38 × 10–3 nucleotide substitutions per site per year, and the non-synonymous and synonymous substitution of approximately 1.16–3.30 × 10–3 and 1.67–4.67 × 10–3 per site per year, respectively, which is similar to other RNA viruses [20]. The large CoV RNA genome allows modification by introducing ‘’non-lethal’’ mutations and recombination, leading to increased probability for intraspecies variability, interspecies “host jump”, and novel CoVs to emerge [20]. SARS-CoV-2 has a higher fidelity in its transcription and replication process than other single-stranded RNA viruses because it has a proofreading mechanism, regulated by NSP14. However, despite this mechanism, the mutation rate is very high [21].
The mutation rate of SARS-CoV-2 is so high that it may impact diagnostic test accuracy [21]. In summary, the target spike and other SARS-CoV-2 proteins have numerous mutations. In total, 13,402 single mutations were found among 31,421 virus isolates, many of them located in coding regions currently used for COVID-19 diagnostic tests [21].
Out of 400 distinct mutation sites of spike protein, 10 mutation sites are most commonly mutated: D614(7859), L5(109), L54(105), P1263(61), P681(51), S477(57), T859(30), S221(28), V483(28), and A845(24) [22]. In Figure 2, mutations positions are presented on the 3D structure of activated mono-trimer spike protein (Figure 2A). Spike protein extracellular domain is divided into the S1 and S2 domains, on which RBD resides, and HR is divided into two domains, which connect S domains with the transmembrane region [23]. Mutations 483 and 477 are located on the loop close to the ACE2 binding site (Figure 2B). The rest are spread throughout the 3D structure of the spike protein, as shown in Figure 2C. Figures were prepared as described in the Supplementary Materials (Supplementary File S1).
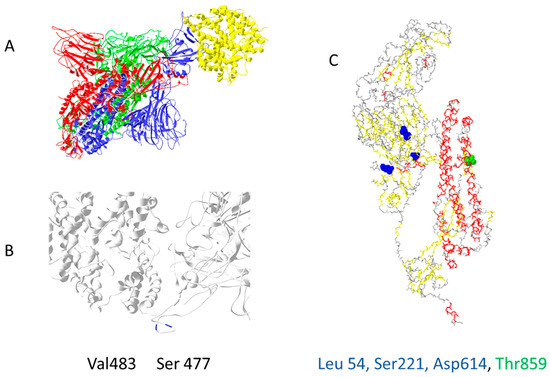
Figure 2.
The most common mutation sites on activated spike protein: (A) trimer of activated spike protein (green, blue, red) interacts with ACE2 receptor (yellow); (B) two common mutation sites, V483 and S477 (dark blue), are close to the ACE2 interaction site; (C) three mutations should impact the 3D structure of spike proteins L54, S221, D614; T859 is located near spike trimer interaction site, PDB file—7DF4 [24].
Spike protein D614G mutation increases virus entry (Figure 3A) by enhancing the binding properties to the ACE2 protein [25]. Increased transmission is predicted for N501Y mutation [26] (Figure 3B). Residues 452, 489, 500, 501, and 505 on the RBM of spike protein (Figure 3C) have high chances of mutating into more infective strains [27]. Suleman et al. computed three mutations, N439K, S477 N, and T478K, shown in Figure 3D, to increase binding with ACE2 [28]. These mutations are localized on, or close to, the ACE2 binding site, except for D614G, which indirectly enhances ACE2 interaction through spike trimer binding enhancement (Figure 3). Bioinformatic predictions data match strain infectivity data. Mutations in strains overlap with the mutations predicted by bioinformatics tools [26,27,28].
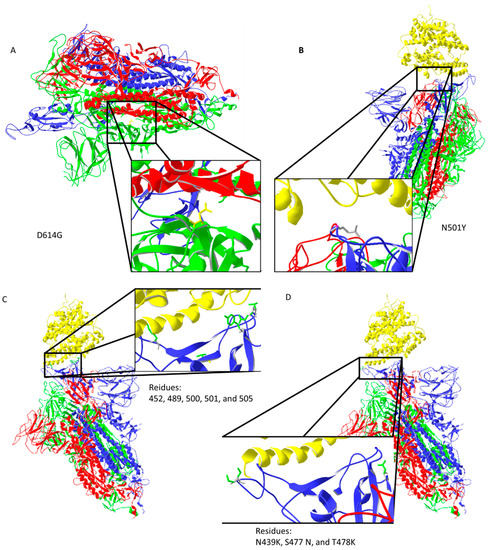
Figure 3.
Important mutation sites of activated spike protein that increase or could increase the infectivity of the virus: (A) D614G mutation may increase interaction between trimer of spike protein (the negatively charged amino acid is replaced by neutral glycine that interacts with the opposite chain, where aliphatic amino acids are present); (B) N501Y mutation should reduce repulsive forces with ACE2; (C) mutations in the ACE2 interaction region could potentially increase the infectiveness of the virus; (D) predicted amino acid replacements that should increase binding with ACE2 are localized on the ACE2 binding site. Protein data bank (PDB) file: 7DF4 [24].
Information regarding 3D structure and mutagenesis is getting more accurate, which helps in drug development. Fast mutagenesis helps the virus evolve much faster than the human defense system can adapt to.
3. Virus–Host Interactions Affecting Viral Replication and Transcription
Viral infection triggers several mechanisms that are both virus- and host-dependent. On the one hand, viral replication affects transcription factors that promote viral replication; on the other hand, the host defense mechanism tries to activate factors to stop the virus from replicating itself.
SARS-CoV-2 targets transcription factors E2F1, SP1, EIF4A1, and TBP, and tumour suppressor genes, including PTEN, AKT1, and RB1, to influence the expression of different host genes (MAPK1, MAPK3, MAPK4, MAPK6, MAPK7, PIK3CA, and CAMK). For regulation, SARS-CoV-2 uses miRNAs [29]. Virus influence on RELA (NF-κB activation), E2F1, STAT3, TP53, NFKB1, GATA3, and CREB1 may impact the regulation of disease progression [30].
One CpG island with 18 CpG sites was detected at the start of viral RNA (UCSC Genome Browser). This site could impact the replication process and translation of proteins coded at the start of the RNA. If COVID-19 integrates into human DNA, this CpG island could impact when and how virus reactivation would start.
4. Human Polymorphism May Influence SARS-CoV-2 Viral Entry and Replication
Natural genome diversity contributes to the survival of the species. Polymorphisms in human genes responsible for viral entry and later replication may contribute to disease prognosis and outcome (Figure 1). These polymorphic genes, which may influence the course of COVID-19 disease, have different roles in viral entry and replication (Table 1).

Table 1.
Updated list of polymorphisms that influence on SARS2 susceptibility, severity, and mortality: information regarding gene name, variation, effect, reference, and function (NCBI subsection gene) are stated in the table. Stated genes: OAS [31], ACE2 [32,33,34,35], TMPRSS2 [36,37,38], TNFRSF1B (ClinVar database [39]), TNF [39], TNFRSF1A (ClinVar database [39]), TBK1 (ClinVar database [39]), TNFRSF13B (ClinVar database [39,40]), TLR7 [41], IFITM3 [42,43], ACE [44,45,46], TMEM189–UBE2V1 [47], HLA [48,49,50,51], TLR3 [48,52,53], IRF7 [48,52], STXBP5/STXBP5-AS1 [48,54], CPQ [48,54], CLUAP1 [48,54], WSB1 [48,54], DNAH7/SLC39A10 [48,54], DES/SPEG [48,54], TOMM7 [48,54], PCDH15 [48,54], TLR4 [55], ABO [48,56,57,58], APOE [59], RIMBP3 [47,48], GOLGA8B [47,48], C3 [60,61], CCR5 [62], IL37 [63], IFNAR2 [31], DPP9 [31], IFNL4 [64] GE [65], NADSYN1 [65], VDR [65] AGT [46].
The most investigated genes are the ones that directly interact with spike protein. Single nucleotide polymorphisms (SNPs) in ACE2 [30,31,32,33,34,35] and TMPRSS2 may contribute to selective binding of SARS-CoV-2 [36,37,38]. Interestingly, only the intron variant of ACE2 and two same sense variants of TMPRSS2 should influence SARS-CoV-2 entry (Table 1). Polymorphisms in the ADAM17 gene were suggested to have a role in the outcome of the disease [37]. It was proposed that the higher frequency of ACE D allele contributed to higher numbers of infected patients/million and mortality rate in the Asian population [66].
Vitamin D deficiency was also associated with a more severe course of COVID-19. Polymorphisms in genes that may lead to this condition, such as vitamin D transporter (GE), receptor (VDR), and NAD synthase gene (NADSYN1), were associated with the critical condition [65].
It was discovered that blood type might also contribute to susceptibility to the COVID-19 disease. HLA types also contribute to susceptibility and severity. Increased susceptibility to SARS-CoV-2 was discovered in ABO A- type patients (ABO (A, B, and O)), e4e4 genotype (APOE (e3 and e4)), HLA B, DRB1, DQB1, and DRB1 alleles, 3p21.31 region minor allele and novel missense variant in GOLGA8B rs200975425 and RIMBP3 rs200584390 [48].
Polymorphisms in Toll-like receptors TLR3 [48,52], TLR4 [55], and TLR7 [41] influence COVID-19 susceptibility and severity as these receptors play a fundamental role in innate immunity through recognition of different pathogen molecules. TLR3 recognizes double-stranded RNA in endosomes [53], TLR4 is activated by lipopolysaccharide and some viral proteins [67], while TLR7 binds to single-stranded RNA [53]. Specific polymorphisms in these receptors attenuate innate immunity response.
Polymorphisms in genes that are directly or indirectly involved in the immune defense system contribute in different ways to infectivity severity and mortality of the SARS-CoV-2 disease (Table 1).
Increased susceptibility to SARS-CoV-2 was associated with carriers of several alleles in HLA-A, HLA-DRB1, minor allele carriers ACE2 rs61735794, and rs61735792 [48].
Several variations were associated with increased severity of SARS-CoV-2 disease—IFITM3 (rs12252), ACE1 (rs4646994), and TMEM189–UBE2V1 (rs6020298)—while multiple alleles of HLA-A, HLA-B, HLA-C, and HLA-DRB1 were associated with decreased severity [48].
Increased mortality was reported in carriers of variants in some loci—STXBP5/STXBP5-AS1, CPQ, CLUAP1, WSB1, DNAH7/SLC39A10, DES/SPEG, TOMM7, PCDH15—and in 10 variants in TLR3 and IRF7 [48].
Several additional polymorphisms were associated with COVID-19: PCDH15 [48,53], ABO [48,56,57,58], APOE [59], RIMBP3 [47,48], GOLGA8B [47,48], C3 [60,61], CCR5 [62], IL37 [63], IFNAR2 [31], DPP9 [31], IFNL4 [64] TNFRSF13B [39,40], TNF [39,68], TNFRSF1A [39], TBK1 [39], IFITM3 [42,43], ACE [44,45,46], IRF7 [48,52], TMPRSS2 [36,37,38], HLA [48,49,50,51], and AGT [46] (Table 1).
Several polymorphic genes were investigated for differences in susceptibility and protection against SARS-CoV-1, which virus has similar structure, RNA sequence, and receptor binding properties to SARS-CoV-2 [69]. Important human polymorphisms were also reported in HLA, ACE1, OAS-1, MxA, PKR, MBL, E-CR1, FcγRIIA, MBL2, L-SIGN (CLEC4M), IFNG, CD14, ICAM3, RANTES, IL-12 RB1, TNFA, CXCL10/IP-10, CD209 (DC-SIGN), AHSG, CYP4F3, and CCL2 [69].
Gene ontology analysis (WEBGESTALT) (Supplementary Materials (Supplementary File S1)) shows that proteins encoded by genes stated in Table 1 are highly interconnected in regulation of defense response (p = 4.0406e-10, enrichment ratio = 9.1490) (mostly through regulation of interferon alpha and beta (p = 5.1619 × 10−10, enrichment ratio = 277.89)). Mostly, they are localized on the cellular membrane (p = 0.0000054530, enrichment ration = 4.3801) or endosomal membranes (p = 0.00010358, enrichment ratio = 7.6820), where they combat against virus entry with the help of signal receptor activity (p = 0.00068370, enrichment analysis = 3.4852) and exopeptidase activity (p = 0.0010791, enrichment ratio = 14.740). Data were accessed as described in Supplementary Materials (Supplementary File S1). Genetic polymorphisms may change cell defense parameters and contribute to different susceptibility and later severity of disease.
Engineered ACE2 Mutations May Predict Virus–Host Interactions
Due to the main interaction with spike protein, ACE2 protein was thoroughly investigated for increased/decreased interaction with SARS-CoV-2 spike protein. They predicted an important role of mutations in regions: 19–42; 69–92; 324–330 (Table 2). Human ACE2 receptor with Y27, L330, and L386 triple mutation showed the highest increase in interaction with RBD of SARS-CoV-2 spike protein (Uniprot database section: “Mutagenesis”). The opposite effect was observed for ACE2 D355N mutation both in vitro and in vivo [70]. ACE2 D355A mutation has a similar effect when interacting with the spike protein of SARS-CoV-1 (Uniprot database section: “Mutagenesis”). These mutations are set in or close to the spike binding site (Figure 4).

Table 2.
List of mutations engineered into ACE2 protein. Mutations importantly impact interaction with spike protein, as stated in the table. Three mutations on the same molecule—Y27, L330, and L386—increase interaction with the RBD domain of the SARS-CoV-2 spike protein (Uniprot database section: “Mutagenesis”).
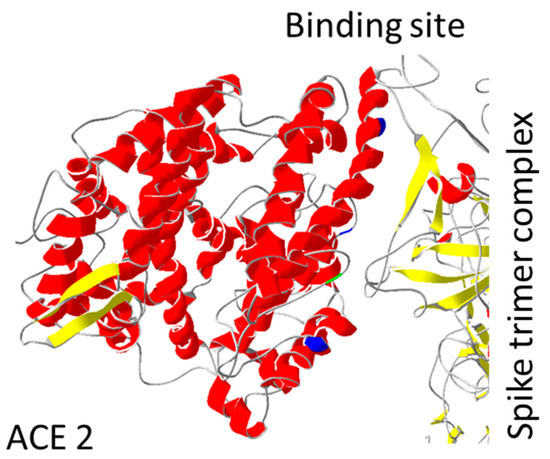
Figure 4.
ACE2 mutations position: Mutations important for interaction with spike protein are shown. Mutation D355A is coloured green, while the trio of Y27, L330, and L386 are coloured dark blue. All mutations are localized close to spike binding site. PDB file: 7DF4 [24].
The diversity of polymorphisms in genes with different functions indicates that the virus replication is affected in all stages, from entry, to transcription, to exocytosis (Figure 1). Knowledge of a patient’s genetic background may support informed choice of treatment.
5. Changes in mRNA Expression—miRNA ‘’Silencing’’ Interference
siRNA are small RNAs that interfere with RNA translation as they bind to mRNA. miRNA have a similar role and regulate gene expression in the cytoplasm. They are involved in transcriptional gene regulation and alternative splicing [71]. miRNA binding slows down replication of viral RNA and slows translation of viral proteins. Genetic variations may change either miRNA or target site sequence and thus change the expression pattern of several genes.
The virus may interfere with RNA expression in host cells by exploiting endogenous miRNA and its viral RNA.
The SARS-CoV-2 genome targets a large number of different miRNA, as follows: transcripts of interleukins IL2, IL5, IL7, IL8, IL10, IL13, IL15, IL16, IL17, IL21, IL22, IL24, IL25, and IL33, histone demethylase genes JARID1A, JARID1C, and JARID2, and histone deacetylase genes, including HDAC1, HDAC2, and HDAC3 [29].
Four key miRNAs (hsa-miR-342-5p, hsa-miR-432-5p, hsa-miR-98-5p, and hsa-miR-17-5p) are believed to be involved in antiviral SARS-CoV-2 gene silencing (ORF1ab) [72]. For three miRNA (hsa-miR-17-5p, hsa-miR-20b-5p, and hsa-miR-323a-5p), there is experimental evidence of having antiviral roles during infections against SARS-CoV-1 and SARS-CoV-2 (Table 3) [73].

Table 3.
List of miRNA found in the literature and its function in SARS-CoV-2 disease.
7c-5p, miR-27b-3p, miR-98-5p and miR-125a-5p target COVID-19 genome; let-7b-5p, miR-155-5p, miR-186-5p, miR-16-5p, miR-27b-3p, miR-29a-3p, and miR-30a-5p are associated with development of COVID-19 symptoms (Table 3) [30].
Importantly, seven key microRNAs were identified (miRNAs 8066, 5197, 3611, 3934-3p, 1307-3p, 3691-3p, and 1468-5p) that may be linked to human response and virus pathogenicity (Table 3) [40].
A total of 22 potential SARS-CoV-2 miRNAs, found in 5 different genomes, targeted 12 human miRNAs. Binding sites for human miRNAs hsa-mir-1267, hsa-mir-1-3p, and hsa-mir-5683 were predicted in all 5 viral SARS-CoV-2 strain genomes (Table 3) [74].
Many studies have shown that miRNAs were significantly dysregulated in COVID-19 [75]. The most important miRNAs that can be used as COVID-19 biomarkers are miR-21-5p, miR-144, and miR-155 [75]. miRNA-146a can be used as a biomarker for the severity of COVID-19 [75].
Our search of miRBASE (mirbase.org) identified a new set of 24 miRNAs that may bind to viral RNA: hsa-miR-1468-5p, hsa-miR-378c, hsa-miR-3611, hsa-miR-3914, hsa-miR-3120-5p, hsa-miR-10397-5p, hsa-miR-515-5p, hsa-miR-584-3p, hsa-miR-4502, hsa-miR-190b-5p, hsa-miR-3691-3p, hsa-miR-597-3p, hsa-miR-1287-5p, hsa-miR-148b-3p, hsa-miR-3672, hsa-miR-5197-3p, hsa-miR-3934-3p, hsa-miR-129-2-3p, hsa-miR-3085-3p, hsa-miR-8076, hsa-miR-8066, hsa-miR-1307-3p, hsa-miR-3613-5p, and hsa-miR-1270 (Table 4). miR-3914, 515-5p, 5197-3p, 4502, 584-3p, 3120-5p, 8066, 5197-3p, and 1287-5p have more than one binding site. miR-3914, 515-5p, 3934-3p, 8076, 4502, and 584-3p have a higher chance of duplexing (https://sfold.wadsworth.org/cgi-bin/index.pl (accessed 15 December 2021)). miRNA regulate numerous human genes (www.TargetScan.org (accessed 15 December 2021)), only a selected few are stated in Table 4. Data were collected as described in Supplementary Materials (Supplementary File S1). During infection, host miRNA may be silenced by binding to viral RNA, disrupting normal cell function through up-regulation of gene expression.

Table 4.
miRNAs binding to SARS-CoV-2 RNA (miRBASE): probability, duplex position, dG, and miRNA-influenced genes are stated.
Analysis of miRNA data with sFOLD shows that several RNAs may have more than one binding site on viral RNA (Table 4). The duplex formation was evaluated in probability and bond energy enthalpy (Figure 5).
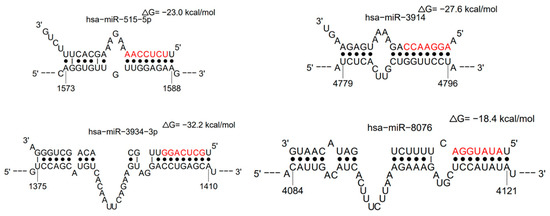
Figure 5.
Examples of alignments of miRNAs duplexes with viral RNA—hsa-miR-515-5p, hsa-miR-3914, hsa-miR-3934-3p, and hsa-miR-8076—have the highest probability of duplex formation and high bond enthalpy (sFOLD- presentation of duplexes). High bond energy needs to be overcome to allow viral RNA duplication and translation of proteins.
The virus attacks the cell through miRNA and subdues it for faster viral replication.
Published data show that virus RNA highly interacts with human miRNAs to change the expression of important defense molecules [29,72,73]. Our analysis showed that numerous human miRNA may form duplexes with different coding and non-coding regions of viral RNA. These interactions could disturb important virus proteins’ replication and/or translation. RNA polymerase and ribosomes require additional energy to remove duplexes during RNA duplication or protein synthesis.
Changes in DNA Methylation Profile
Coronaviruses can delay pathogen recognition and block interferon-stimulated genes [76]. Several known viral proteins associated with viral pathogenesis are controlled epigenetically [76]. Viruses such as Epstein–Barr virus and SARS-CoV-2 can demethylate the syncytin 1 and 2 genes, resulting in the augmentation of gene transcription [76]. Hypo-methylation of ACE2 coupled with demethylation of interferon- and cytokine-regulated genes and enhanced NF-κB axis have been shown to contribute to SARS-CoV-2 disease severity [76]. Critically ill COVID-19 patients had hypermethylation of IFN related genes and hypomethylation of inflammatory genes [76].
An epigenome-wide COVID-19 study reported 51 CpG sites with different methylation profiles between moderate and severe cases [77]. After thorough analysis, 44 CpGs were marked as important, as follows: 15 CpG sites were located in human genomic regions with no currently described gene sequence; 6 CpG sites were associated with non-coding RNA; 23 CpG sites were located within 20 known coding genes. In 17 out of 20 coding genes (85%), the presence of hyper-methylation was significantly associated with transcript down-regulation—7 out of 20 were effectors of interferon signalling, as follows: AIM2, HLA-C, IFI44L, CXCR2, KIFAP3, SGMS1, and VIM [77]. Two CpG methylation sites were found in PM20D1, AIM2, and HLA-C [77].
Methylation of DNA may influence virus replication, and vice versa; the infection may start to change the methylation profile of patients. Better knowledge of epigenetic markers could perhaps help predict the course of the disease.
6. Extracellular Vesicles as Biomarkers
Extracellular vesicles (EVs), such as exosomes and microvesicles, could be used as biomarkers if sufficiently up- or down-regulated in COVID-19 patients. Several lipid molecules, GM3, and sphingomyelins were enriched in exosomes, while diacylglycerol levels were decreased [78].
Several RNA species were detected in large quantity in exosomes (exRNAs), such as snRNAs SNORD33, RNU2-29P; transcripts AL732437.2 and AL365184.1; non-coding RNAs CDKN2B-AS1, and miRNAs miR-122-5p [79], hsa-miR-146a and hsa-miR126-3p [80].
Proteins such as fibrinogen, fibronectin, complement C1r subcomponent, and serum amyloid P-component were downregulated in EVs [81].
EVs have numerous receptors on the surface: CD9, CD63, CD81, ESCRT, TSG101, Alix, Hsp60, Hsp70, Hsp90, and RAB27a/b—among them, ACE2. ACE2 on EVs may divert the virus from entering the cell, modulating or mediating SARS-CoV-2 infection [79].
EVs research is new, and it is expected that several new diagnostic methods will be developed, especially in virology and immunology.
7. Conclusions
This review described COVID-19 cell viral and host parameters that may influence SARS-CoV-2 entry and infectivity, along with factors that influence the susceptibility and severity of the COVID-19 disease. Virus mutations, strains, changes in transcriptome, miRNA ‘’silencing’’ interference, methylation profiles (epigenetics), and individual polymorphisms were reviewed.
COVID-19 appeared at the end of 2019, and several new strains have already been discovered. The new strain can overcome other strains in a few months. New mutations in the RBD domain can develop the virus into more infectious strains that cause diseases with more severe symptoms during infection. Sequencing analysis shows that beneficial mutations remain in the sequence of virus via outcompeting the wild-type
Virus replication may also be influenced by host genetic variability during all phases from the entry and transcription to the final stage. Several human polymorphisms, miRNAs, and methylation profiles led to different susceptibility, severity, and even mortality of the disease. In the future, complete genome sequence information may support a professional, personalized medicine approach to treat and diagnose SARS-CoV-2.
Knowledge of genetic and epigenetic biomarkers may help predict the course of the disease and the response to treatment. Research of extracellular vesicles is also on the rise, so new COVID-19 biomarkers are anticipated due to differences in vesicle composition.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/biology11020178/s1, Supplementary File S1: Methodology.
Author Contributions
Conceptualization, V.D. and J.G.; methodology, J.G.; software, J.G.; writing—original draft preparation, J.G.; writing—review and editing, V.D.; visualization, J.G.; supervision, V.D.; project administration, V.D.; funding acquisition, V.D. All authors have read and agreed to the published version of the manuscript.
Funding
This study was supported by the Slovenian Research Agency, grant number P1-0170.
Data Availability Statement
All the data are included in the paper; any additional data is available from the authors upon request.
Acknowledgments
We are grateful to David Vogrinc for helpful comments on the article.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Silhol, F.; Marlinge, M.; Guiol, C.; Chefrour, M.; Mace, P.; Criado, C.; Kipson, N.; Vaisse, B.; Vairo, D.; Sarlon, G.; et al. Characterization of adenosine A2 receptors in peripheral blood mononuclear cells of patients with fibromuscular dysplasia. Hypertens Res. 2020, 43, 466–469. [Google Scholar] [CrossRef] [PubMed]
- Shang, J.; Wan, Y.; Luo, C.; Ye, G.; Geng, Q.; Auerbach, A.; Li, F. Cell entry mechanisms of SARS-CoV-2. Proc. Natl. Acad. Sci. USA 2020, 117, 11727–11734. [Google Scholar] [CrossRef] [PubMed]
- Bestle, D.; Heindl, M.R.; Limburg, H.; Van Lam van, T.; Pilgram, O.; Moulton, H.; Stein, D.A.; Hardes, K.; Eickmann, M.; Dolnik, O.; et al. TMPRSS2, and furin are both essential for proteolytic activation of SARS-CoV-2 in human airway cells. Life Sci. Alliance 2020, 3, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Hoffmann, M.; Kleine-Weber, H.; Pohlmann, S. A Multibasic Cleavage Site in the Spike Protein of SARS-CoV-2 Is Essential for Infection of Human Lung Cells. Mol. Cell 2020, 78, 779–784.e5. [Google Scholar] [CrossRef]
- Sarker, J.; Das, P.; Sarker, S.; Roy, A.K.; Momen, A. A Review on Expression, Pathological Roles, and Inhibition of TMPRSS2, the Serine Protease Responsible for SARS-CoV-2 Spike Protein Activation. Scientifica 2021, 2021, 2706789. [Google Scholar] [CrossRef]
- Zhang, L.; Richards, A.; Barrasa, M.I.; Hughes, S.H.; Young, R.A.; Jaenisch, R. Reverse-transcribed SARS-CoV-2 RNA can integrate into the genome of cultured human cells and can be expressed in patient-derived tissues. Proc. Natl. Acad. Sci. USA 2021, 118, e2105968118. [Google Scholar] [CrossRef] [PubMed]
- Saraste, J.; Prydz, K. Assembly and Cellular Exit of Coronaviruses: Hijacking an Unconventional Secretory Pathway from the Pre-Golgi Intermediate Compartment via the Golgi Ribbon to the Extracellular Space. Cells 2021, 10, 503. [Google Scholar] [CrossRef]
- Kadam, S.B.; Sukhramani, G.S.; Bishnoi, P.; Pable, A.A.; Barvkar, V.T. SARS-CoV-2, the pandemic coronavirus: Molecular and structural insights. J. Basic. Microbiol. 2021, 61, 180–202. [Google Scholar] [CrossRef]
- Liu, J.; Li, Y.; Liu, Q.; Yao, Q.; Wang, X.; Zhang, H.; Chen, R.; Ren, L.; Min, J.; Deng, F.; et al. SARS-CoV-2 cell tropism and multiorgan infection. Cell Discov. 2021, 7, 17. [Google Scholar] [CrossRef]
- Ricci, D.; Etna, M.P.; Rizzo, F.; Sandini, S.; Severa, M.; Coccia, E.M. Innate Immune Response to SARS-CoV-2 Infection: From Cells to Soluble Mediators. Int. J. Mol. Sci. 2021, 22, 7017. [Google Scholar] [CrossRef]
- NIH. Clinical Spectrum of SARS-CoV-2 Infection. 2021. Available online: https://www.covid19treatmentguidelines.nih.gov/overview/clinical-spectrum/ (accessed on 15 December 2021).
- Tammam, S.N.; El Safy, S.; Ramadan, S.; Arjune, S.; Krakor, E.; Mathur, S. Repurpose but also (nano)-reformulate! The potential role of nanomedicine in the battle against SARS-CoV2. J. Control. Release 2021, 337, 258–284. [Google Scholar] [CrossRef] [PubMed]
- Shakaib, B.; Zohra, T.; Ikram, A.; Shakaib, M.B.; Ali, A.; Bashir, A.; Salman, M.; Khan, M.A.; Ansari, J. A comprehensive review on clinical and mechanistic pathophysiological aspects of COVID-19 Malady: How far have we come? Virol. J. 2021, 18, 120. [Google Scholar] [CrossRef] [PubMed]
- Huang, C.; Wang, Y.; Li, X.; Ren, L.; Zhao, J.; Hu, Y.; Zhang, L.; Fan, G.; Xu, L.; Gu, X.; et al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet 2020, 395, 497–506. [Google Scholar] [CrossRef] [Green Version]
- Neopane, P.; Nypaver, J.; Shrestha, R.; Beqaj, S.S. SARS-CoV-2 Variants Detection Using TaqMan SARS-CoV-2 Mutation Panel Molecular Genotyping Assays. Infect. Drug Resist. 2021, 14, 4471–4479. [Google Scholar] [CrossRef]
- Fisman, D.N.; Tuite, A.R. Evaluation of the relative virulence of novel SARS-CoV-2 variants: A retrospective cohort study in Ontario, Canada. CMAJ 2021, 193, E1619–E1625. [Google Scholar] [CrossRef]
- Buchta, C.; Camp, J.V.; Jovanovic, J.; Radler, U.; Benka, B.; Puchhammer-Stockl, E.; Müller, M.M.; Griesmacher, A.; Aberle, S.W.; Görzer, I. Inadequate design of mutation detection panels prevents interpretation of variants of concern: Results of an external quality assessment for SARS-CoV-2 variant detection. Clin. Chem. Lab. Med. 2021, 60, 291–298. [Google Scholar] [CrossRef]
- Aoki, A.; Adachi, H.; Mori, Y.; Ito, M.; Sato, K.; Okuda, K.; Sakakibara, T.; Okamoto, Y.; Jinno, H. A rapid screening assay for L452R and T478K spike mutations in SARS-CoV-2 Delta variant using high-resolution melting analysis. J. Toxicol. Sci. 2021, 46, 471–476. [Google Scholar] [CrossRef]
- Callaway, E. Heavily mutated Omicron variant puts scientists on alert. Nature 2021, 600, 21. [Google Scholar] [CrossRef]
- Su, S.; Wong, G.; Shi, W.; Liu, J.; Lai, A.C.K.; Zhou, J.; Liu, W.; Bi, Y.; Gao, G.F. Epidemiology, Genetic Recombination, and Pathogenesis of Coronaviruses. Trends Microbiol. 2016, 24, 490–502. [Google Scholar] [CrossRef] [Green Version]
- Wang, R.; Hozumi, Y.; Yin, C.; Wei, G.W. Mutations on COVID-19 diagnostic targets. Genomics. 2020, 112, 5204–5213. [Google Scholar] [CrossRef]
- Guruprasad, L. Human SARS CoV-2 spike protein mutations. Proteins 2021, 89, 569–576. [Google Scholar] [CrossRef] [PubMed]
- Ruiz-Rodriguez, P.; Frances-Gomez, C.; Chiner-Oms, A.; Lopez, M.G.; Jimenez-Serrano, S.; Cancino-Munoz, I.; Ruiz-Hueso, P.; Torres-Puente, M.; Bracho, M.A.; D’Auria, G.; et al. Evolutionary and Phenotypic Characterization of Two Spike Mutations in European Lineage 20E of SARS-CoV-2. Mbio 2021, 12, e0231521. [Google Scholar] [CrossRef] [PubMed]
- Xu, C.; Wang, Y.; Liu, C.; Zhang, C.; Han, W.; Hong, X.; Wang, Y.; Hong, Q.; Wang, S.; Zhao, Q.; et al. Conformational dynamics of SARS-CoV-2 trimeric spike glycoprotein in complex with receptor ACE2 revealed by cryo-EM. Sci. Adv. 2021, 7, eabe5575. [Google Scholar] [CrossRef] [PubMed]
- Ozono, S.; Zhang, Y.; Ode, H.; Sano, K.; Tan, T.S.; Imai, K.; Miyoshi, K.; Kishigami, S.; Ueno, T.; Iwatani, Y.; et al. SARS-CoV-2 D614G spike mutation increases entry efficiency with enhanced ACE2-binding affinity. Nat. Commun. 2021, 12, 848. [Google Scholar] [CrossRef] [PubMed]
- Erol, A. Are the emerging SARS-COV-2 mutations friend or foe? Immunol. Lett. 2021, 230, 63–64. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Wang, R.; Wang, M.; Wei, G.W. Mutations Strengthened SARS-CoV-2 Infectivity. J. Mol. Biol. 2020, 432, 5212–5226. [Google Scholar] [CrossRef]
- Suleman, M.; Yousafi, Q.; Ali, J.; Ali, S.S.; Hussain, Z.; Ali, S.; Waseem, M.; Iqbal, A.; Ahmad, S.; Khan, A.; et al. Bioinformatics analysis of the differences in the binding profile of the wild-type and mutants of the SARS-CoV-2 spike protein variants with the ACE2 receptor. Comput. Biol. Med. 2021, 138, 104936. [Google Scholar] [CrossRef]
- Aydemir, M.N.; Aydemir, H.B.; Korkmaz, E.M.; Budak, M.; Cekin, N.; Pinarbasi, E. Computationally predicted SARS-COV-2 encoded microRNAs target NFKB, JAK/STAT and TGFB signaling pathways. Gene Rep. 2021, 22, 101012. [Google Scholar] [CrossRef]
- Nain, Z.; Rana, H.K.; Lio, P.; Islam, S.M.S.; Summers, M.A.; Moni, M.A. Pathogenetic profiling of COVID-19 and SARS-like viruses. Brief. Bioinform. 2021, 22, 1175–1196. [Google Scholar] [CrossRef]
- Pairo-Castineira, E.; Clohisey, S.; Klaric, L.; Bretherick, A.D.; Rawlik, K.; Pasko, D.; Walker, S.; Parkinson, N.; Fourman, M.H.; Russell, C.D.; et al. Genetic mechanisms of critical illness in COVID-19. Nature 2021, 591, 92–98. [Google Scholar] [CrossRef]
- Mohlendick, B.; Schonfelder, K.; Breuckmann, K.; Elsner, C.; Babel, N.; Balfanz, P.; Dahl, E.; Dreher, M.; Fistera, D.; Herbstreit, F.; et al. ACE2 polymorphism and susceptibility for SARS-CoV-2 infection and severity of COVID-19. Pharm. Genom. 2021, 31, 165–171. [Google Scholar] [CrossRef] [PubMed]
- Hamet, P.; Pausova, Z.; Attaoua, R.; Hishmih, C.; Haloui, M.; Shin, J.; Paus, T.; Abrahamowicz, M.; Gaudet, D.; Santucci, L.; et al. SARS-CoV-2 Receptor ACE2 Gene Is Associated with Hypertension and Severity of COVID 19: Interaction with Sex, Obesity, and Smoking. Am. J. Hypertens 2021, 34, 367–376. [Google Scholar] [CrossRef]
- Benetti, E.; Tita, R.; Spiga, O.; Ciolfi, A.; Birolo, G.; Bruselles, A.; Doddato, G.; Giliberti, A.; Marconi, C.; Musacchia, F.; et al. ACE2 gene variants may underlie interindividual variability and susceptibility to COVID-19 in the Italian population. Eur. J. Hum. Genet. 2020, 28, 1602–1614. [Google Scholar] [CrossRef]
- Hou, Y.; Zhao, J.; Martin, W.; Kallianpur, A.; Chung, M.K.; Jehi, L.; Sharifi, N.; Erzurum, S.; Eng, C.; Cheng, F.; et al. New insights into genetic susceptibility of COVID-19: An ACE2 and TMPRSS2 polymorphism analysis. BMC Med. 2020, 18, 216. [Google Scholar] [CrossRef] [PubMed]
- Torre-Fuentes, L.; Matias-Guiu, J.; Hernandez-Lorenzo, L.; Montero-Escribano, P.; Pytel, V.; Porta-Etessam, J.; Gómez-Pinedo, U.; Matías-Guiu, J.A. ACE2, TMPRSS2, and Furin variants and SARS-CoV-2 infection in Madrid, Spain. J. Med. Virol. 2021, 93, 863–869. [Google Scholar] [CrossRef]
- Brest, P.; Refae, S.; Mograbi, B.; Hofman, P.; Milano, G. Host Polymorphisms May Impact SARS-CoV-2 Infectivity. Trends Genet. 2020, 36, 813–815. [Google Scholar] [CrossRef] [PubMed]
- Jeon, S.; Blazyte, A.; Yoon, C.; Ryu, H.; Jeon, Y.; Bhak, Y.; Bolser, D.; Manica, A.; Shin, E.S.; Cho, Y.S.; et al. Regional TMPRSS2 V197M Allele Frequencies Are Correlated with COVID-19 Case Fatality Rates. Mol. Cells 2021, 44, 680–687. [Google Scholar] [CrossRef]
- Landrum, M.J.; Lee, J.M.; Benson, M.; Brown, G.; Chao, C.; Chitipiralla, S.; Gu, B.; Hart, J.; Hoffman, D.; Hoover, J.; et al. ClinVar: Public archive of interpretations of clinically relevant variants. Nucleic Acids Res. 2016, 44, D862–D868. [Google Scholar] [CrossRef] [Green Version]
- Arisan, E.D.; Dart, A.; Grant, G.H.; Arisan, S.; Cuhadaroglu, S.; Lange, S.; Uysal-Onganer, P. The Prediction of miRNAs in SARS-CoV-2 Genomes: Hsa-miR Databases Identify 7 Key miRs Linked to Host Responses and Virus Pathogenicity-Related KEGG Pathways Significant for Comorbidities. Viruses 2020, 12, 614. [Google Scholar] [CrossRef]
- van der Made, C.I.; Simons, A.; Schuurs-Hoeijmakers, J.; van den Heuvel, G.; Mantere, T.; Kersten, S.; van Deuren, R.C.; Steehouwer, M.; van Reijmersdal, S.V.; Jaeger, M.; et al. Presence of Genetic Variants Among Young Men With Severe COVID-19. JAMA 2020, 324, 663–673. [Google Scholar] [CrossRef]
- Pati, A.; Padhi, S.; Suvankar, S.; Panda, A.K. Minor Allele of Interferon-Induced Transmembrane Protein 3 Polymorphism (rs12252) Is Covered Against Severe Acute Respiratory Syndrome Coronavirus 2 Infection and Mortality: A Worldwide Epidemiological Investigation. J. Infect. Dis. 2021, 223, 175–178. [Google Scholar] [CrossRef] [PubMed]
- Cuesta-Llavona, E.; Albaiceta, G.M.; Garcia-Clemente, M.; Duarte-Herrera, I.D.; Amado-Rodriguez, L.; Hermida-Valverde, T.; Enríquez-Rodriguez, A.I.; Hernández-González, C.; Melón, S.; Alvarez-Argüelles, M.E.; et al. Association between the interferon-induced transmembrane protein 3 gene (IFITM3) rs34481144 / rs12252 haplotypes and COVID-19. Curr. Res. Virol. Sci. 2021, 2, 100016. [Google Scholar] [CrossRef] [PubMed]
- Hubacek, J.A.; Dusek, L.; Majek, O.; Adamek, V.; Cervinkova, T.; Dlouha, D.; Adamkova, V. ACE I/D polymorphism in Czech first-wave SARS-CoV-2-positive survivors. Clin. Chim. Acta 2021, 519, 206–209. [Google Scholar] [CrossRef] [PubMed]
- Jacobs, M.; Lahousse, L.; Van Eeckhoutte, H.P.; Wijnant, S.R.A.; Delanghe, J.R.; Brusselle, G.G.; Bracke, K.R. Effect of ACE1 polymorphism rs1799752 on protein levels of ACE2, the SARS-CoV-2 entry receptor, in alveolar lung epithelium. ERJ Open Res. 2021, 7, 206–209. [Google Scholar] [CrossRef]
- Kouhpayeh, H.R.; Tabasi, F.; Dehvari, M.; Naderi, M.; Bahari, G.; Khalili, T.; Clark, C.; Ghavami, S.; Taheri, M. Association between angiotensinogen (AGT), angiotensin-converting enzyme (ACE) and angiotensin-II receptor 1 (AGTR1) polymorphisms and COVID-19 infection in the southeast of Iran: A preliminary case-control study. Transl. Med. Commun. 2021, 6, 26. [Google Scholar] [CrossRef]
- Wang, F.; Huang, S.; Gao, R.; Zhou, Y.; Lai, C.; Li, Z.; Xian, W.; Qian, X.; Zhiyu Li, Z.; Huang, Y.; et al. Initial whole-genome sequencing and analysis of the host genetic contribution to COVID-19 severity and susceptibility. Cell Discov. 2020, 6, 83. [Google Scholar] [CrossRef]
- Ferreira de Araujo, J.L.; Menezes, D.; Saraiva-Duarte, J.M.; de Lima Ferreira, L.; Santana de Aguiar, R.; Pedra de Souza, R. Systematic review of host genetic association with Covid-19 prognosis and susceptibility: What have we learned in 2020? Rev. Med Virol. 2021, 2021, e2283. [Google Scholar]
- Tomita, Y.; Ikeda, T.; Sato, R.; Sakagami, T. Association between HLA gene polymorphisms and mortality of COVID-19: An in silico analysis. Immun. Inflamm. Dis. 2020, 8, 684–694. [Google Scholar] [CrossRef]
- Littera, R.; Campagna, M.; Deidda, S.; Angioni, G.; Cipri, S.; Melis, M.; Firinu, D.; Santus, S.; Lai, A.; Porcella, R.; et al. Human Leukocyte Antigen Complex and Other Immunogenetic and Clinical Factors Influence Susceptibility or Protection to SARS-CoV-2 Infection and Severity of the Disease Course. The Sardinian Experience. Front. Immunol. 2020, 11, 605688. [Google Scholar] [CrossRef]
- Novelli, A.; Andreani, M.; Biancolella, M.; Liberatoscioli, L.; Passarelli, C.; Colona, V.L.; Rogliani, P.; Leonardis, F.; Campana, A.; Carsetti, R.; et al. HLA allele frequencies and susceptibility to COVID-19 in a group of 99 Italian patients. HLA 2020, 96, 610–614. [Google Scholar] [CrossRef]
- Zhang, Q.; Bastard, P.; Liu, Z.; Le Pen, J.; Moncada-Velez, M.; Chen, J.; Ogishi, M.; Sabli, I.K.D.; Hodeib, S.; Korol, C.; et al. Inborn errors of type I IFN immunity in patients with life-threatening COVID-19. Science 2020, 370, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Kuznik, A.; Bencina, M.; Svajger, U.; Jeras, M.; Rozman, B.; Jerala, R. Mechanism of endosomal TLR inhibition by antimalarial drugs and imidazoquinolines. J. Immunol. 2011, 186, 4794–4804. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hu, J.; Li, C.; Wang, S.; Li, T.; Zhang, H. Genetic variants are identified to increase risk of COVID-19 related mortality from UK Biobank data. Hum. Genom. 2021, 15, 10. [Google Scholar] [CrossRef] [PubMed]
- Taha, S.I.; Shata, A.K.; Baioumy, S.A.; Fouad, S.H.; Anis, S.G.; Mossad, I.M.; Moustafa, N.M.; Abdou, D.M.; Youssef, M.K. Toll-Like Receptor 4 Polymorphisms (896A/G and 1196C/T) as an Indicator of COVID-19 Severity in a Convenience Sample of Egyptian Patients. J. Inflamm. Res. 2021, 14, 6293–6303. [Google Scholar] [CrossRef]
- Zhao, J.; Yang, Y.; Huang, H.; Li, D.; Gu, D.; Lu, X.; Zhang, Z.; Liu, L.; Liu, T.; Liu, Y.K.; et al. Relationship Between the ABO Blood Group and the Coronavirus Disease 2019 (COVID-19) Susceptibility. Clin. Infect. Dis. 2021, 73, 328–331. [Google Scholar] [CrossRef]
- Fan, Q.; Zhang, W.; Li, B.; Li, D.J.; Zhang, J.; Zhao, F. Association Between ABO Blood Group System and COVID-19 Susceptibility in Wuhan. Front. Cell Infect. Microbiol. 2020, 10, 404. [Google Scholar] [CrossRef]
- Zhang, Y.; Garner, R.; Salehi, S.; La Rocca, M.; Duncan, D. Association between ABO blood types and coronavirus disease 2019 (COVID-19), genetic associations, and underlying molecular mechanisms: A literature review of 23 studies. Ann. Hematol. 2021, 100, 1123–1132. [Google Scholar] [CrossRef] [PubMed]
- Kuo, C.L.; Pilling, L.C.; Atkins, J.L.; Masoli, J.A.H.; Delgado, J.; Kuchel, G.A.; Melzer, D. ApoE e4e4 Genotype and Mortality With COVID-19 in UK Biobank. J. Gerontol. A Biol. Sci. Med. Sci. 2020, 75, 1801–1803. [Google Scholar] [CrossRef]
- Delanghe, J.R.; De Buyzere, M.L.; Speeckaert, M.M. C3 and ACE1 polymorphisms are more important confounders in the spread and outcome of COVID-19 in comparison with ABO polymorphism. Eur. J. Prev. Cardiol. 2020, 27, 1331–1332. [Google Scholar] [CrossRef] [PubMed]
- Delanghe, J.R.; De Buyzere, M.L.; Speeckaert, M.M. Genetic Polymorphisms in the Host and COVID-19 Infection. Adv. Exp. Med. Biol. 2021, 1318, 109–118. [Google Scholar]
- Mehlotra, R.K. Chemokine receptor gene polymorphisms and COVID-19: Could knowledge gained from HIV/AIDS be important? Infect. Genet. Evol. 2020, 85, 104512. [Google Scholar] [CrossRef]
- Ahmed, A.A.; Ad’hiah, A.H. Interleukin-37 gene polymorphism and susceptibility to coronavirus disease 19 among Iraqi patients. Meta. Gene. 2022, 31, 100989. [Google Scholar] [CrossRef] [PubMed]
- Saponi-Cortes, J.M.R.; Rivas, M.D.; Calle-Alonso, F.; Sanchez, J.F.; Costo, A.; Martin, C.; Zamorano, J. IFNL4 genetic variant can predispose to COVID-19. Sci. Rep. 2021, 11, 21185. [Google Scholar] [CrossRef]
- Al-Anouti, F.; Mousa, M.; Karras, S.N.; Grant, W.B.; Alhalwachi, Z.; Abdel-Wareth, L.; Uddin, M.; Alkaabi, N.; Tay, G.K.; Mahboub, B.; et al. Associations between Genetic Variants in the Vitamin D Metabolism Pathway and Severity of COVID-19 among UAE Residents. Nutrients 2021, 13, 3680. [Google Scholar] [CrossRef] [PubMed]
- Pati, A.; Mahto, H.; Padhi, S.; Panda, A.K. ACE deletion allele is associated with susceptibility to SARS-CoV-2 infection and mortality rate: An epidemiological study in the Asian population. Clin. Chim. Acta. 2020, 510, 455–458. [Google Scholar] [CrossRef] [PubMed]
- Avbelj, M.; Horvat, S.; Jerala, R. The role of intermediary domain of MyD88 in cell activation and therapeutic inhibition of TLRs. J. Immunol. 2011, 187, 2394–2404. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Heidari Nia, M.; Rokni, M.; Mirinejad, S.; Kargar, M.; Rahdar, S.; Sargazi, S.; Sarhadi, M.; Saravani, R. Association of polymorphisms in tumor necrosis factors with SARS-CoV-2 infection and mortality rate: A case-control study and in silico analyses. J. Med. Virol. 2021, 1–11. [Google Scholar] [CrossRef] [PubMed]
- dos Santos, A.C.M.; dos Santos, B.R.C.; Dos Santos, B.B.; de Moura, E.L.; Ferreira, J.M.; Dos Santos, L.K.C.; Oliveira, S.P.; Dias, R.B.F.; Pereira e Silva, A.C.; de Farias, K.F.; et al. Genetic polymorphisms as multi-biomarkers in severe acute respiratory syndrome (SARS) by coronavirus infection: A systematic review of candidate gene association studies. Infect Genet Evol. 2021, 93, 104846. [Google Scholar] [CrossRef] [PubMed]
- Ren, W.; Zhu, Y.; Lan, J.; Chen, H.; Wang, Y.; Shi, H.; Feng, F.; Chen, D.Y.; Close, B.; Zhao, X.; et al. Susceptibilities of human ACE2 genetic variants in coronavirus infection. J. Virol. 2022, 96, e01492-21. [Google Scholar] [CrossRef]
- Turunen, T.A.; Roberts, T.C.; Laitinen, P.; Vaananen, M.A.; Korhonen, P.; Malm, T.; Ylä-Herttuala, S.; Turunen, M.P. Changes in nuclear and cytoplasmic microRNA distribution in response to hypoxic stress. Sci. Rep. 2019, 9, 10332. [Google Scholar] [CrossRef] [Green Version]
- Banaganapalli, B.; Al-Rayes, N.; Awan, Z.A.; Alsulaimany, F.A.; Alamri, A.S.; Elango, R.; Malik, Z.; Shaik, N.A. Multilevel systems biology analysis of lung transcriptomics data identifies key miRNAs and potential miRNA target genes for SARS-CoV-2 infection. Comput. Biol. Med. 2021, 135, 104570. [Google Scholar] [CrossRef] [PubMed]
- Khan, M.A.; Sany, M.R.U.; Islam, M.S.; Islam, A. Epigenetic Regulator miRNA Pattern Differences Among SARS-CoV, SARS-CoV-2, and SARS-CoV-2 World-Wide Isolates Delineated the Mystery Behind the Epic Pathogenicity and Distinct Clinical Characteristics of Pandemic COVID-19. Front. Genet. 2020, 11, 765. [Google Scholar] [CrossRef] [PubMed]
- Sarma, A.; Phukan, H.; Halder, N.; Madanan, M.G. An in-silico approach to study the possible interactions of miRNA between human and SARS-CoV2. Comput. Biol. Chem. 2020, 88, 107352. [Google Scholar] [CrossRef] [PubMed]
- Visacri, M.B.; Nicoletti, A.S.; Pincinato, E.C.; Loren, P.; Saavedra, N.; Saavedra, K.; Salazar, L.A.; Moriel, P. Role of miRNAs as biomarkers of COVID-19: A scoping review of the status and future directions for research in this field. Biomark. Med. 2021, 15, 1785–1795. [Google Scholar] [CrossRef] [PubMed]
- Saksena, N.; Bonam, S.R.; Miranda-Saksena, M. Epigenetic Lens to Visualize the Severe Acute Respiratory Syndrome Coronavirus-2 (SARS-CoV-2) Infection in COVID-19 Pandemic. Front. Genet. 2021, 12, 581726. [Google Scholar] [CrossRef]
- Castro de Moura, M.; Davalos, V.; Planas-Serra, L.; Alvarez-Errico, D.; Arribas, C.; Ruiz, M.; Aguilera-Albesa, S.; Troya, J.; Valencia-Ramos, J.; Vélez-Santamaria, V.; et al. Epigenome-wide association study of COVID-19 severity with respiratory failure. EBioMedicine 2021, 66, 103339. [Google Scholar] [CrossRef]
- Song, J.W.; Lam, S.M.; Fan, X.; Cao, W.J.; Wang, S.Y.; Tian, H.; Chua, G.H.; Zhang, C.; Meng, F.P.; Xu, Z.; et al. Omics-Driven Systems Interrogation of Metabolic Dysregulation in COVID-19 Pathogenesis. Cell Metab. 2020, 32, 188–202.e5. [Google Scholar] [CrossRef]
- Fujita, Y.; Hoshina, T.; Matsuzaki, J.; Yoshioka, Y.; Kadota, T.; Hosaka, Y.; Fujimoto, S.; Kawamoto, H.; Watanabe, N.; Sawaki, K.; et al. Early prediction of COVID-19 severity using extracellular vesicle COPB2. J. Extracell Vesicles 2021, 10, e12092. [Google Scholar] [CrossRef]
- Mitchell, M.I.; Ben-Dov, I.Z.; Liu, C.; Ye, K.; Chow, K.; Kramer, Y.; Gangadharan, A.; Park, S.; Fitzgerald, S.; Ramnauth, A.; et al. Extracellular Vesicle Capture by AnTibody of CHoice and Enzymatic Release (EV-CATCHER): A customizable purification assay designed for small-RNA biomarker identification and evaluation of circulating small-EVs. J. Extracell Vesicles 2021, 10, e12110. [Google Scholar] [CrossRef]
- Barberis, E.; Vanella, V.V.; Falasca, M.; Caneapero, V.; Cappellano, G.; Raineri, D.; Ghirimoldi, M.; De Giorgis, V.; Puricelli, C.; Vaschetto, R.; et al. Circulating Exosomes Are Strongly Involved in SARS-CoV-2 Infection. Front. Mol. Biosci. 2021, 8, 632290. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).