1. Introduction
The enhancement of sheep growth traits has become a major social demand in China. Muscle growth and development are closely related to sheep growth traits. Some studies have found that the growth and development of skeletal muscle is a complex biological process, in which the varying signaling pathways and genes that regulate muscle fiber proliferation and differentiation are related to the oxygen content and energy metabolism [
1], and the
HIF-1 gene has the potential function of regulating muscle cell growth and development via the Wnt signaling pathway [
2,
3,
4].
HIF-1 consists of two subunits, including HIF-1α and HIF-1β (
ARNT), which is not only an important transcription factor but also a transcription regulation factor of hypoxia stress in mammals, and HIF-1 can regulate the activity of multiple types of protein, such as cell surface receptors, transcription factors, cytoskeletal proteins, angiogenic growth factors, cytokines, survival factors, glucose transporters, and glycolytic enzymes [
5,
6,
7]. HIF-1α and ARNT belong to the bHLH-PER-ARNT-SIM(PAS) protein family. Among them, the HIF-α exists in the cytoplasm, and the HIF-1α, HIF-2α, and HIF-3α are composed of it. Additionally, the expression of HIF-1α, HIF-2α, and HIF-3α is regulated by the oxygen content in the body. Under a normal oxygen content, these subunits are degraded, while under low oxygen content conditions, the transcription activity and stability of the three subunits are increased, and they combine with
ARNT to form a dimer so as to regulate the transcription and expression of pre-emptive target genes. The three subunits have different functions under different oxygen conditions [
8,
9,
10,
11].
In contrast to HIF-α,
ARNT is a binding partner of the aryl hydrocarbon receptor (Ahr) and HIF-α subunit. It is stable in the cytoplasm and nucleus under normal oxygen, hypoxic conditions have a nuclear localization signal that can enter the nucleus through an import/dependent pathway [
12]. Meanwhile, some researchers reported that
ARNT is only expressed in certain types of cells, and it has the same regulation function as the HIF-1α [
13,
14,
15], but the function of
ARNT in skeletal muscle cell proliferation and differentiation is not clear. Currently, it is believed that
ARNT regulates glucose metabolism and myofibroblast proliferation and differentiation [
16].
At the cellular level,
ARNT can interact with the transcription factor Miz-1 of Myc, which can directly regulate several biological processes, such as the metabolism, cell cycle, and proliferation [
17]. Deng et al. established a hypoxic rat model at the model biology level, showing that Microrna-103/107 participates in hypoxia-induced pulmonary artery smooth muscle cell proliferation by targeting
ARNT [
18]. The results of Deng et al. are similar to the results of Maktepe et al., and they all indicated that in
ARNT knockout mice, the number of smooth muscle fibers was reduced [
19]. Moreover,
ARNT depletion inhibits muscle growth and function, and recent studies have shown that the knockdown of
ARNT in primary muscle cell lines impinges on differentiation in vitro, whereas skeletal-muscle-specific
ARNT depletion in young mice results in decreased whole-muscle
N1ICD expression levels and limits muscle growth and regeneration, and the knockdown of HIF-1 in embryonic mice does not affect muscle growth and development but has a regulatory effect on postnatal skeletal muscle satellite cells [
20]. Furthermore,
ARNT may be involved in the regulation of muscle growth and development. The protein interacts with some pathways that are involved in muscle growth and development. It also forms a myofiber regulatory network that regulates sheep growth. In addition, our previous TMT proteomic analysis found that a key protein,
ARNT, may influence growth traits in sheep. Thus, whatever the molecular function or regulatory mechanism of
ARNT is, it is worth studying further [
21,
22].
In this study, the single-nucleotide polymorphisms (SNPs) of the sheep ARNT gene were discovered and genotyped, and the relationship between each SNP and the growth characteristics was examined. Using PCR techniques, the CDS cDNA sequence of the ARNT gene was cloned from the skeletal muscle of Hu sheep. The gene and potential protein products of the gene were studied using bioinformatics. qRT-PCR was used to assess the expression of the ARNT gene in two sheep muscle tissues and myoblasts at various developmental stages. This study lays the groundwork for future research investigating how sheep ARNT gene function and expression. Additionally, it serves as a foundation for additional research on the molecular processes controlling sheep meat output.
4. Discussion
Research on the
ARNT gene has mainly focused on its roles in mouse and human tumors and its molecular mechanisms [
33,
34,
35,
36]. Meanwhile, some research on the
ARNT gene has focused on myofiber type determination and muscle regeneration [
20,
21], whereas research on its roles in sheep growth traits is relatively rare. As a result of our previous proteomic studies, we found that the ARNT protein and its potential functions might affect the growth and development of sheep embryonic muscle. However, the function and regulation mechanism of this gene have not been reported in regard to sheep growth traits. To reveal the association between the
ARNT gene and sheep growth and development, we performed a quantitative analysis of the muscle tissues of different breeds of sheep. These Chinese Merino sheep were meat and wool breeds from Xinjiang, China, and the
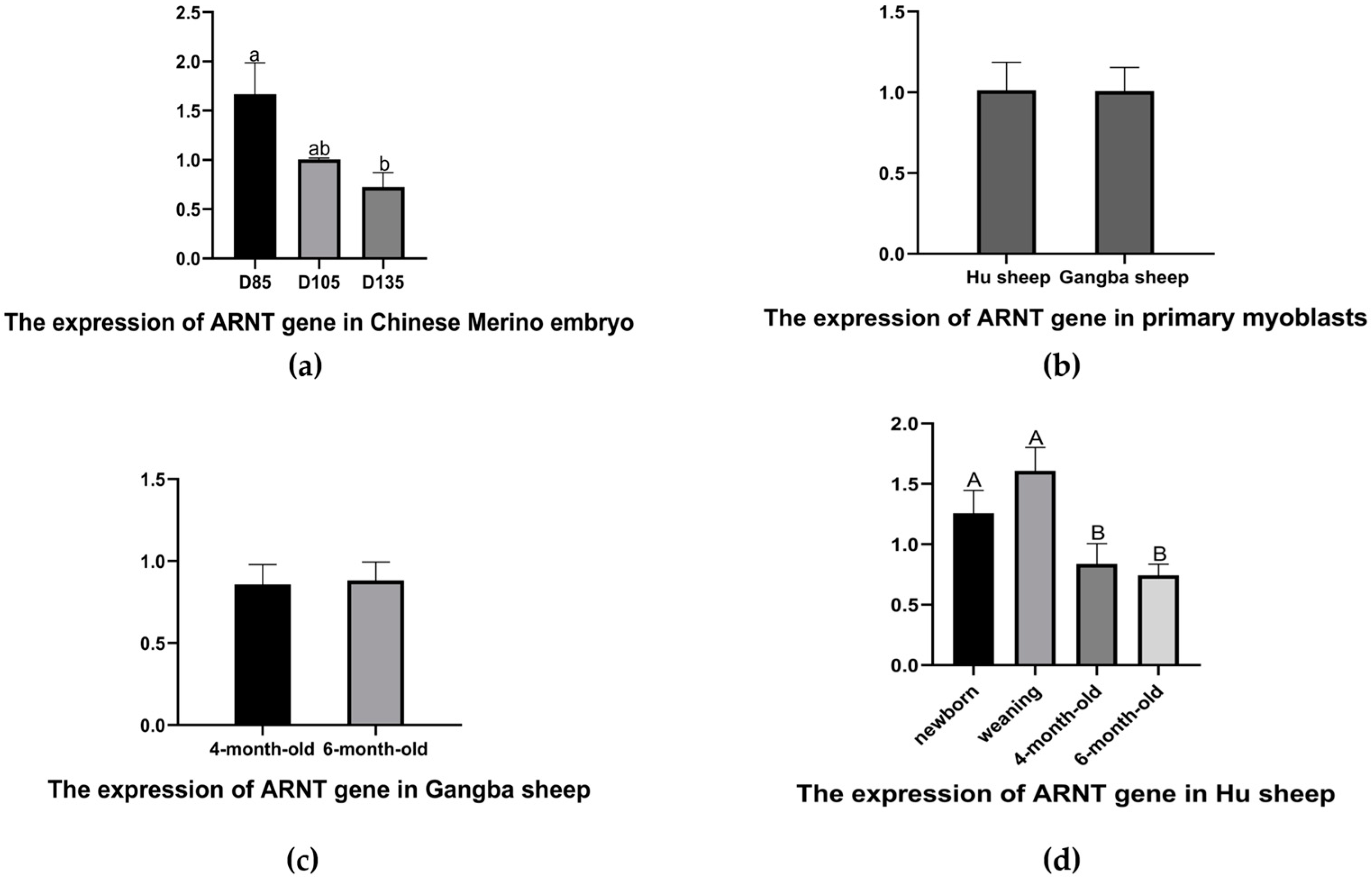
ARNT expression at D85 was significantly higher than at D135 (
Figure 1a), and the degree of gene expression decreased gradually with muscle maturation, suggesting that a negative association may exist between the
ARNT expression and muscle growth and development in Chinese Merino embryos. Furthermore, the
ARNT gene is an essential component of HIF, and the different oxygen levels in sheep can affect the ability of the
ARNT gene to regulate muscle growth and development. To explore whether there are differences in the
ARNT gene function among sheep at different altitudes, the Hu sheep, a meat sheep breed that is famous in China, and the Gangba sheep, a special meat sheep breed from Tibet, were used to reveal the potential functions. At the level of the myoblasts, there was no significant difference between the Hu and Gangba sheep myoblasts (
Figure 1b). Although the expression of the
ARNT gene showed no significant difference in the afterborn Gangba sheep (
Figure 1c), this expression of the gene showed a significant difference between the pre- and post-weaning afterborn Hu sheep (
Figure 1d). These results confirm that the
ARNT gene functions differently in sheep at different altitudes. However, this gene may be a key factor in the growth and development of sheep.
To date, there have been few studies analyzing the SNPs in the ARNT gene. To better understand the polymorphism and expression of the ARNT gene, our study analyzed the polymorphism of the ARNT gene in the Hu and Ujimqin sheep populations. The polymorphism analysis showed that the ARNT gene SNP loci in the Hu sheep were less numerous than in the Ujimqin sheep. In addition, there was only one SNP locus (rs413597480 A > G) in the Hu sheep. The genetic variation at this locus was small in the Hu sheep population. However, this locus had a favorable relationship with the Hu sheep growth traits. The BW at weaning was strongly associated with rs413597480 A > G. The rs413597480 A > G in the Hu sheep also had a strong association with the BFT at 5-month-old, as was the case for the CC and CW in the developmental stages of 5- and 6-month-old in the Hu sheep (p < 0.05). It is worth noting that the GG genotype was the dominant genotype at the rs413597480 A > G mutation locus in the Hu sheep, with substantial values for all the statistically linked growth characteristics, while the expression of the ARNT gene was significantly different at the weaning stage in the Hu sheep compared to other developmental stages (p < 0.01). These results demonstrated that the rs413597480 A > G of the ARNT gene has the potential to regulate growth and development during the weaning period and to affect the BFT of Hu sheep during the 5-month-old stage.
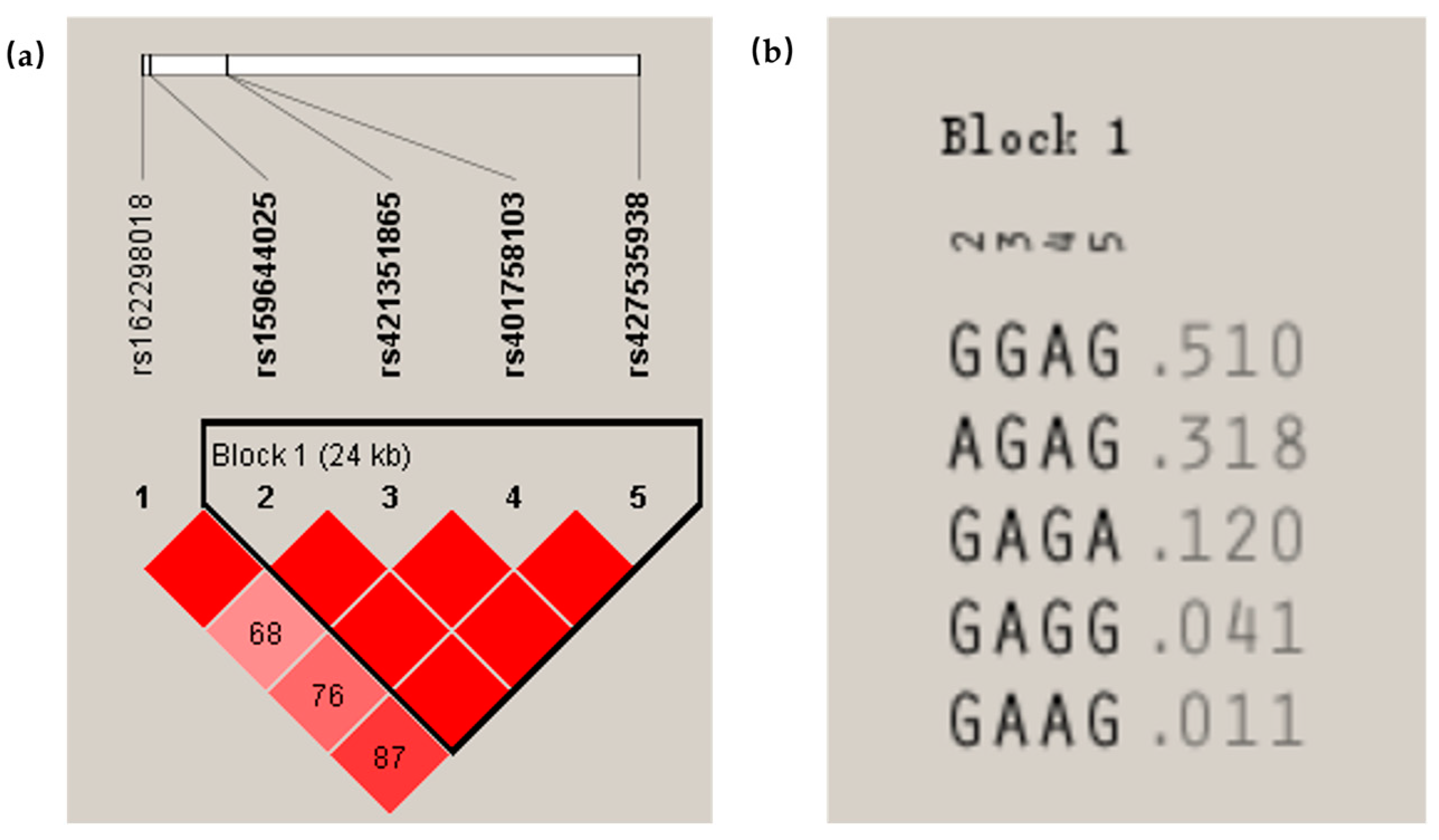
As for the
ARNT gene in the Ujimqin sheep, there were five SNPs, of which the rs162298018 G > C SNP was a G/C transversion, and this SNP had a good statistical association with the BW and CC of 4-month-old sheep and CC and the TC of 6-month-old Ujimqin sheep, respectively. Furthermore, this SNP had high heterozygosity and a large amount of polymorphic information. In addition, there was strong linkage disequilibrium among rs162298018 G > C and rs159644025 G > A. According to this result, the rs162298018 G > C mutation was ruled out by chance. The remaining three SNP loci of the
ARNT gene in the Ujimqin sheep were also associated with the growth traits, except for the rs427535938 G > A, while a haplotype domain was detected between rs159644025 G > A, rs421351865 G > A, rs401758103 A > G, and rs427535938 G > A, and the GGAG was the dominant haplotype. Based on the above results, both the analysis of the
ARNT gene expression and the analysis of the genetic stability and the association of the mutant loci with the growth traits suggest that this gene may have some relevance to sheep growth and development, and this conclusion is similar to that of the studies of the influence of
ARNT on the muscle in human and animal models [
18,
19,
20].
Then, we further investigated the ARNT CDS sequence and protein. At present, there is no complete report of the ARNT gene CDS sequence in sheep. The Hu sheep is bred for meat in China, and there is no CDS cDNA sequence of the ARNT gene in Hu sheep. In this study, the complete CDS sequences of the Hu sheep ARNT gene were successfully obtained. The phylogenetic tree showed that the Hu sheep ARNT protein had the closest relationship with goat and sheep. The similarity was over 99%, indicating that the ARNT protein has been highly conserved throughout evolution. Additionally, the analysis of the CDS sequence showed that a G > C of the ARNT CDS sequence resulted in a nonconservative missense point mutation, leading to a change from proline to alanine at position 1948 bp. Notably, the non-conserved missense point mutation in the ARNT CDS sequence that we cloned from the Hu sheep was the same single-base mutation as the rs162298018 G > C in the Ujimqin sheep, and the GG genotype of this mutation site was the dominant genotype that was significantly associated with some growth traits in the Hu and Ujimqin sheep. The Hu sheep and Ujimqin sheep with this genotype had greater body weight and body size traits. These findings not only confirm the ability of the ARNT gene to control growth and development in sheep, but they also raise the possibility that the G > C mutation at 1948 bp in the CDS region may be a significant candidate SNP influencing the growth and development of Hu and Ujimqin sheep.
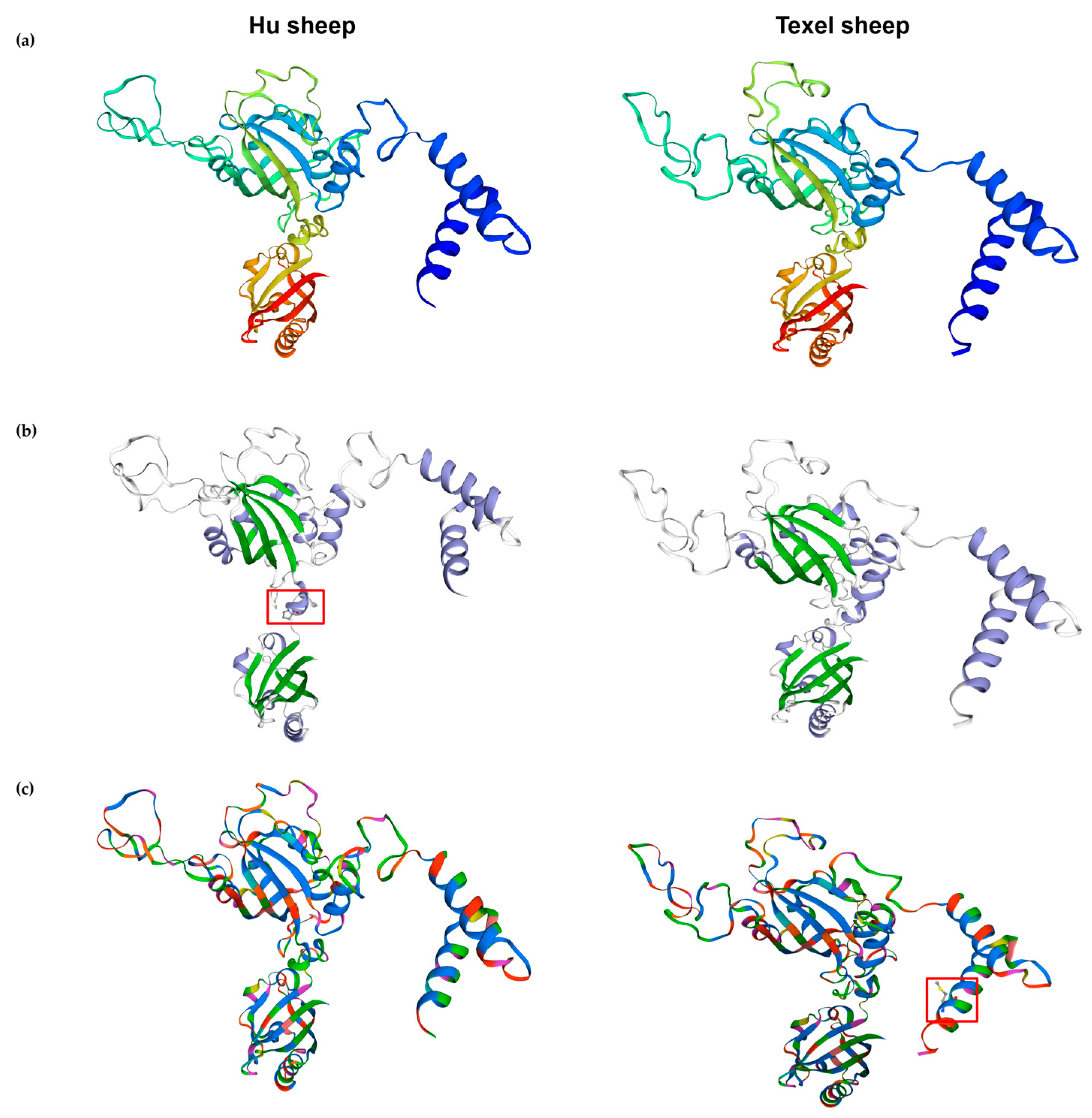
Meanwhile, the secondary and tertiary structure analysis of the ARNT protein of the Hu sheep revealed that this nonconservative missense has significant influences on the secondary and tertiary structures of ARNT. Compared to the Texel sheep, there are more DNA- and protein-binding sites in the ARNT protein in the Hu sheep. Some researchers reported that specific SNPs can affect the secondary structures of mRNAs and proteins [
37,
38], indicating that the DNA- or protein-binding sites of the Hu sheep ARNT protein may play a crucial role in gene function and that this can change the gene function in regard to sheep growth and development [
39,
40,
41,
42]. While the changes in the tertiary conformation of the ARNT protein and the increase in the α-helix after the G > C mutation suggest that the G > C mutation at 1948 bp in the CDS region of the
ARNT gene may affect the function of the ARNT protein and, thus, the regulation of growth traits in Hu and Ujimqin sheep, they also suggest that the G > C mutation at 1948 bp in the CDS region of the
ARNT gene may be a potential molecular regulatory locus for the growth traits of sheep.
Based on the results of our analysis, we suggest that the ARNT gene, with its key potential function, regulates the growth traits of sheep. The G > C mutation at 1948 bp in the cloned ARNT CDS sequence of the Hu sheep was the same locus mutation as rs162298018 G > C in the Ujimqin sheep, which resulted in a nonconservative missense point mutation, leading to a change from proline to alanine, and this SNP locus was significantly associated with the growth traits of the Ujimqin sheep. Meanwhile, we also found that the results of the ARNT protein function analysis are consistent with the quantitative results for the ARNT gene in the Hu sheep muscle tissues, indicating that the ARNT gene may play an important functional role in affecting the muscle growth and development of Hu and Ujimqin sheep. At present, we do not know the specific function of the DNA- or protein-binding regions in the ARNT protein. However, it can be seen that the growth traits are associated with the functionality of ARNT in sheep. In the future, we will perform more functional analyses to clarify our doubts. For example, we will create an overexpression vector containing the rs162298018 G > C mutation site of ARNT. We will also investigate the changes in the ARNT gene function before and after the mutation, as well as their effects on myoblast growth and development. Additionally, the key factors that bind to DNA- and protein-binding sites will also be predicted.