Historical Biogeography of Earwigs
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
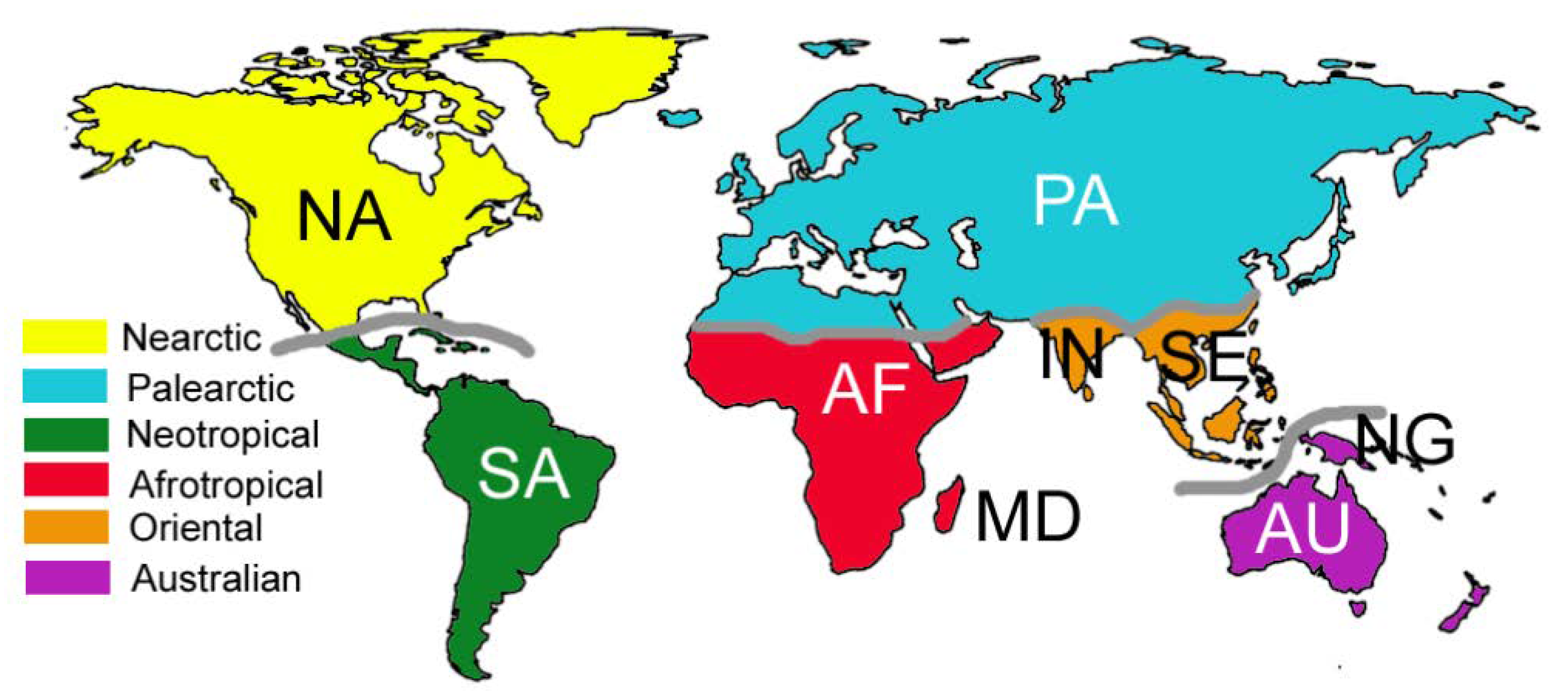
2.1. Areas of Endemism
2.2. Taxa
2.3. Data Analysis
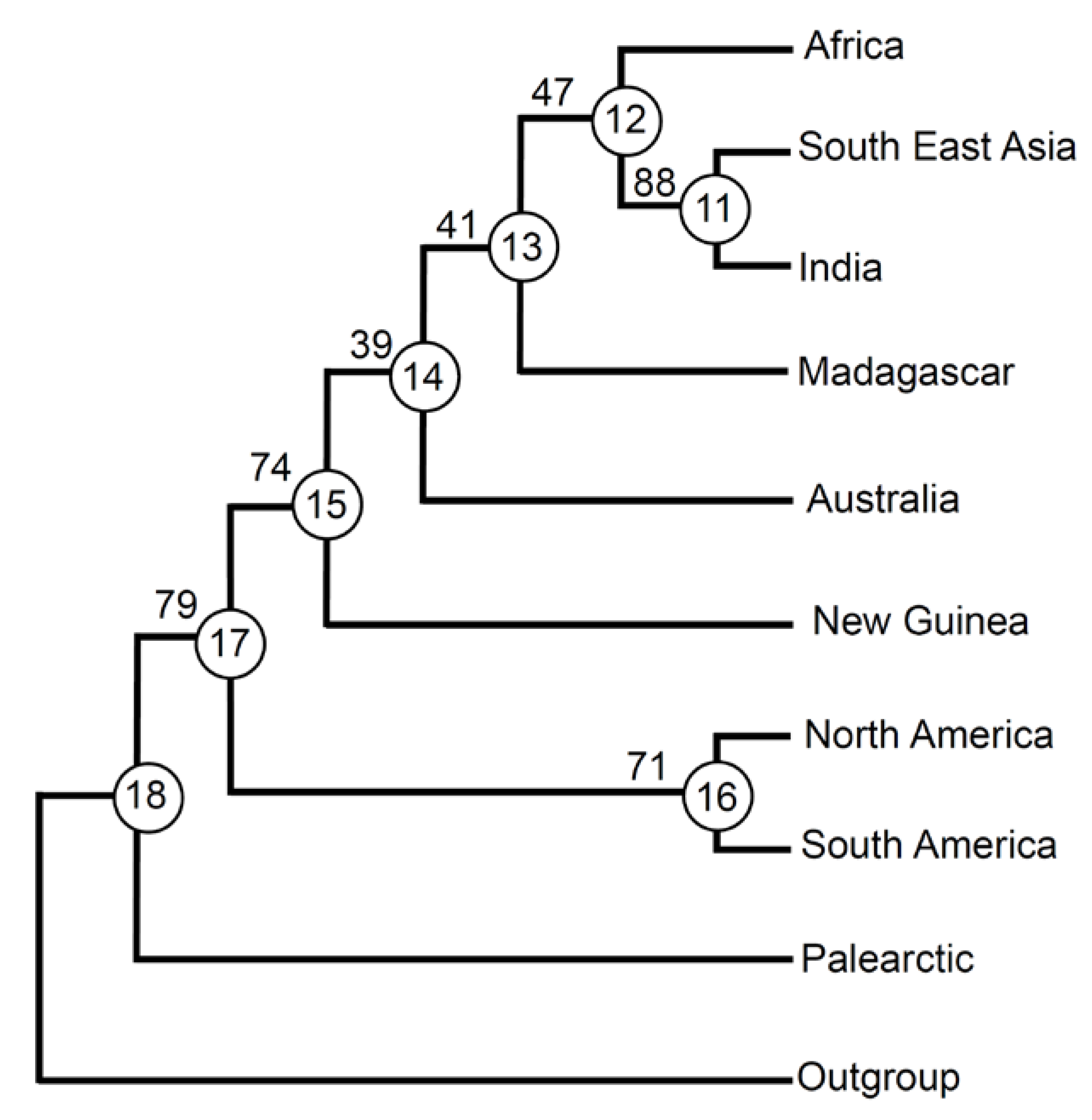
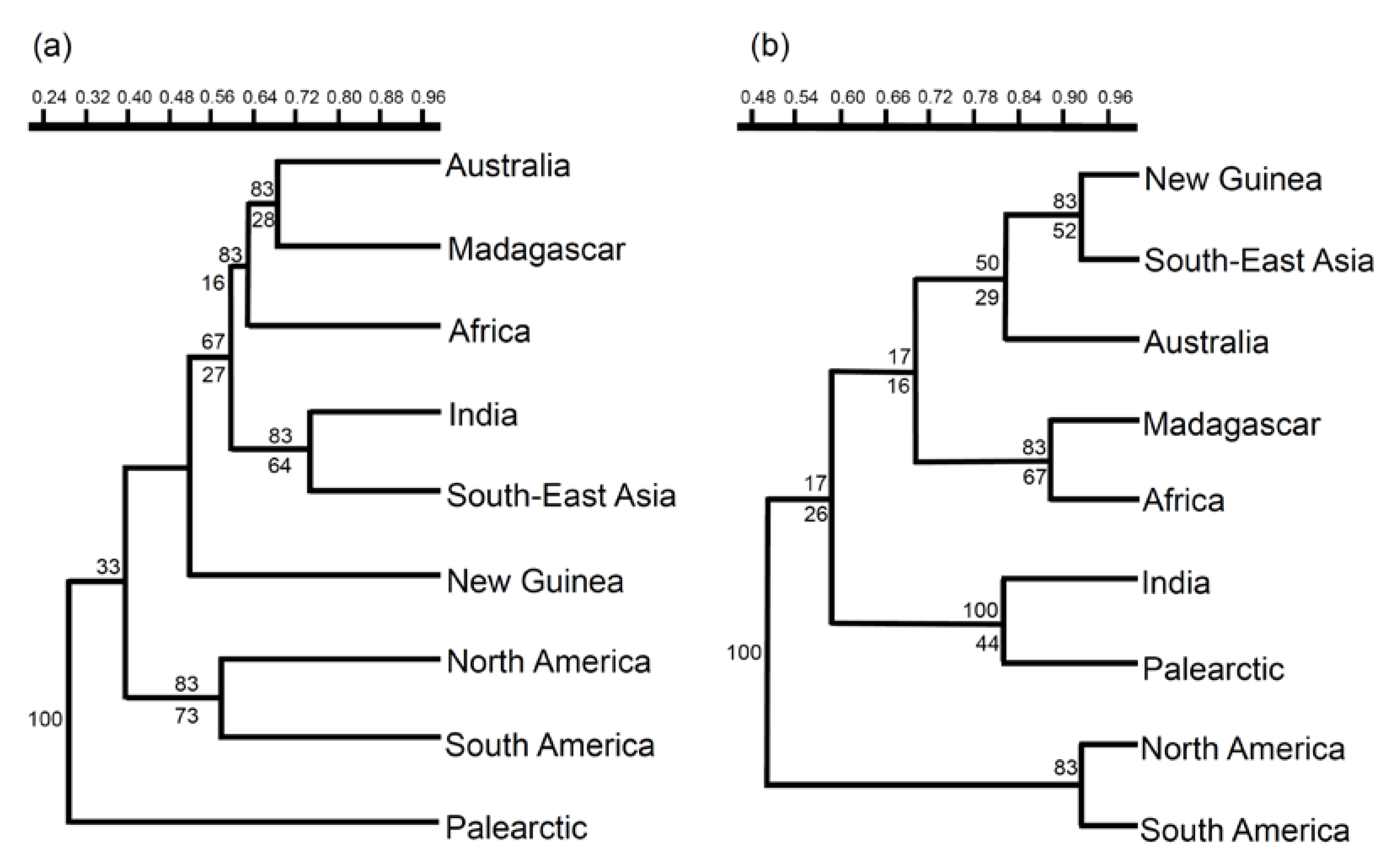
3. Results
4. Discussion
5. Conclusions
Supplementary Materials
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Haas, F. Biodiversity of Dermaptera. In Insect Biodiversity and Society; Foottit, R.G., Adler, P.H., Eds.; Wiley–Blackwell: Hoboken, NJ, USA, 2018; pp. 315–334. [Google Scholar]
- Misof, B.; Liu, S.; Meusemann, K.; Peters, R.S.; Donath, A.; Mayer, C.; Frandsen, P.B.; Ware, J.; Flouri, T.; Beutel, R.G.; et al. Phylogenomics resolves the timing and pattern of insect evolution. Science 2014, 346, 763–767. [Google Scholar] [CrossRef] [PubMed]
- Zhao, J.; Zhao, Y.; Shih, C.; Ren, D.; Wang, Y. Transitional fossil earwigs—A missing link in Dermaptera evolution. BMC Evol. Biol. 2010, 10, 344. [Google Scholar] [CrossRef] [PubMed]
- Fattorini, S. Global patterns of earwig species richness. Diversity 2022, 14, 890. [Google Scholar] [CrossRef]
- Popham, E.J. Geographical distribution of the Dermaptera—A case for continental drift? Entomologist 1969, 102, 121–124. [Google Scholar]
- Popham, E.J.; Manly, B.F.J. Geographical distribution of the Dermaptera and the continental drift hypothesis. Nature 1969, 222, 981–982. [Google Scholar] [CrossRef]
- Popham, E.J. The geographical distribution of the Dermaptera (Insecta) with reference to continental drift. J. Nat. Hist. 2000, 34, 2007–2027. [Google Scholar] [CrossRef]
- Ball, I.R. Nature and formulation of biogeographical hypotheses. Syst. Zool. 1975, 24, 407–430. [Google Scholar] [CrossRef]
- Sakai, S. A quantitative approach to the problem of distribution of Dermaptera. Contrib. Dep. Biol. Chem. Daito Bunka Univ. Spec. Bull. 1971, 4, 1–210. [Google Scholar]
- Sakai, S. Multivariate analysis of the zoogeographical distribution of the world Dermaptera with special reference to Labiidae. Bull. Daito Bunka Univ. 1978, 16, 1–53. [Google Scholar]
- Sakai, S.; Roomi, M.W. A quantitative trial of zoogeographical distribution of Dermaptera of the World with special reference to rapid and practical computergraphy. Contrib. Inst. Biol. Chem. Daito Bunka Univ. Spec. Bull. 1978, 23, 1–7. [Google Scholar]
- Steinmann, H. A zoogeographical checklist of world Dermaptera. Folia Entomol. Hung. 1972, 26, 145–154. [Google Scholar]
- Bey–Bienko, G.J. The Dermapteran Insects; The Fauna of USSR, New Series; Academy of Sciences USSR: Moscow/Leningrad, Russia, 1936; 239p. (In Russian) [Google Scholar]
- Semenov Tian-Shanskij, A. Les limites et les subdivisions zoogeographiques de la région paléarctique pour les animaux terrestres, basées sur la distribution géographique des insectes Coléoptères. (Avec une carte géographique). Travaux de l’Institut Zoologique de l’Academie des Sciences de I’URSS 1935, 2–3, 397–410, (In Russian, with summary in French). [Google Scholar]
- Crisci, J.V. The voice of historical biogeography. J. Biogeogr. 2001, 28, 157–168. [Google Scholar] [CrossRef]
- Fattorini, S. Endemism in historical biogeography and conservation biology: Concepts and implications. Biogeographia 2017, 32, 47–75. [Google Scholar] [CrossRef]
- Fattorini, S. Hovenkamp’s ostracized vicariance analysis: Testing new methods of historical biogeography. Cladistics 2008, 24, 611–622. [Google Scholar] [CrossRef]
- Porzecanski, A.L.; Cracraft, J. Cladistic analysis of distributions and endemism (CADE): Using raw distributions of birds to unravel the biogeography of the South American aridlands. J. Biogeogr. 2005, 32, 261–275. [Google Scholar] [CrossRef]
- Melo Santos, A.M.; Cavalcanti, D.R.; da Silva, J.M.C.; Tabarelli, M. Biogeographical relationships among tropical forests in north-eastern Brazil. J. Biogeogr. 2006, 34, 437–446. [Google Scholar] [CrossRef]
- Fattorini, S. Historical relationships of African mountains based on cladistic analysis of distributions and endemism of flightless insects. Afr. Entomol. 2007, 15, 340–355. [Google Scholar] [CrossRef]
- Vázquez-Miranda, H.; Navarro-Sigüenza, A.G.; Morrone, J.J. Biogeographical patterns of the avifaunas of the Caribbean Basin Islands: A parsimony perspective. Cladistics 2007, 23, 180–200. [Google Scholar] [CrossRef]
- Sánchez-González, L.A.; Morrone, J.J.; Navarro-Sigüenza, A.G. Distributional patterns of the Neotropical humid montane forest avifaunas. Biol. J. Linn. Soc. 2008, 94, 175–194. [Google Scholar] [CrossRef]
- Gelfo, J.N.; Goin, F.J.; Woodburne, M.O.; De Muizon, C. Biochronological relationships of the earliest South American Paleogene mammalian faunas. Palaeontology 2008, 52, 251–269. [Google Scholar] [CrossRef]
- Juen, L.; De Marco, P. Dragonfly endemism in the Brazilian Amazon: Competing hypotheses for biogeographical patterns. Biodivers. Conserv. 2012, 21, 3507–3521. [Google Scholar] [CrossRef]
- Martínez-Aquino, A.; Vigliano-Relva, J.; Brusa, F.; Damborenea, C. Historical biogeography of Temnocephalida (Platyhelminthes, Rhabdocoela): Testing the Gondwanan hypothesis. Syst. Biodivers. 2016, 15, 327–345. [Google Scholar] [CrossRef]
- Dagosta, F.C.P.; de Pinna, M. Biogeography of Amazonian fishes: Deconstructing river basins as biogeographic units. Neotrop. Ichthyol. 2017, 15, e170034. [Google Scholar] [CrossRef]
- Prieto-Torres, D.A.; Rojas-Soto, O.R.; Bonaccorso, E.; Santiago-Alarcon, D.; Navarro-Sigüenza, A.G. Distributional patterns of Neotropical seasonally dry forest birds: A biogeographical regionalization. Cladistics 2018, 35, 446–460. [Google Scholar] [CrossRef]
- Castro, D.P.; Rodrigues, J.F.M.; Borges-Leite, M.J.; Lima, D.C.; Borges-Nojosa, D.M. Anuran diversity indicates that Caatinga relictual Neotropical forests are more related to the Atlantic Forest than to the Amazon. PeerJ 2019, 6, e6208. [Google Scholar] [CrossRef]
- Paz-Ríos, C.E.; Pech, D.; Carrera-Parra, L.F.; Simões, N. Biodiversity and biogeographic affinity of benthic amphipods from the Yucatan Shelf: An analysis across the warm Northwest Atlantic ecoregions. Syst. Biodivers. 2021, 19, 928–939. [Google Scholar] [CrossRef]
- Lomolino, M.V.; Riddle, B.R.; Whittaker, R.J.; Brown, J.H. Biogeography, 4th ed.; Sinauer Associates Inc.: Sunderland, MA, USA, 2010. [Google Scholar]
- Brindle, A. The Dermaptera of Madagascar. Trans. R. Entomol. Soc. Lond. 1966, 118, 221–259. [Google Scholar] [CrossRef]
- Haas, F. Earwigs of Australia. Available online: http://www.earwigs-online.de/AU/au.html (accessed on 1 March 2021).
- Stuart, O.P.; Binns, M.; Umina, P.A.; Holloway, J.; Severtson, D.; Nash, M.; Heddle, T.; van Helden, M.; Hoffmann, A.A. Morphological and molecular analysis of Australian earwigs (Dermaptera) points to unique species and regional endemism in the Anisolabididae Family. Insects 2019, 10, 72. [Google Scholar] [CrossRef]
- Srivastava, G.K. Fauna of India and Adjacent Countries. Dermaptera. Part-I. Superfamily: Pygidicranoidea; Zoological Survey of India: Calcutta, India, 1988; 268p. [Google Scholar]
- Srivastava, G.K. Fauna of India and adjacent Countries. Dermaptera. Part-II. Superfamily: Anisolaboidea; Zoological Survey of India: Kolkata, India, 2003; 235p. [Google Scholar]
- Srivastava, G.K. Fauna of India and the Adjacent Countries. Dermaptera. Part-III. Superfamilies Apachyoidea and Forficuloidea; Zoological Survey of India: Kolkata, India, 2013; 469p. [Google Scholar]
- Wipfler, B.; Koehler, W.; Frandsen, P.B.; Donath, A.; Liu, S.; Machida, R.; Misof, B.; Peters, R.S.; Shimizu, S.; Zhou, X.; et al. Phylogenomics changes our understanding about earwig evolution. Syst. Entomol. 2020, 45, 516–526. [Google Scholar] [CrossRef]
- Engel, M.S.; Haas, F. Family-Group Names for Earwigs (Dermaptera). Am. Mus. Novit. 2007, 3567, 1–20. [Google Scholar] [CrossRef]
- Engel, M.S.; Huang, D.; Thomas, J.C.; Cai, C. A new genus and species of pygidicranid earwigs from the Upper Cretaceous of southern Asia (Dermaptera: Pygidicranidae). Cretac. Res. 2017, 69, 178–183. [Google Scholar] [CrossRef]
- Liu, H.-L.; Chen, S.; Chen, Q.-D.; Pu, D.-Q.; Chen, Z.-T.; Liu, Y.-Y.; Liu, X. The first mitochondrial genomes of the family Haplodiplatyidae (Insecta: Dermaptera) reveal intraspecific variation and extensive gene rearrangement. Biology 2022, 11, 807. [Google Scholar] [CrossRef]
- Rosen, B.R. From fossils to Earth history: Applied historical biogeography. In Analytical Biogeography; Myers, A.A., Giller, P.S., Eds.; Chapman & Hall: London, UK, 1988; pp. 437–481. [Google Scholar]
- Cracraft, J. Patterns of diversification within continental biotas: Hierarchical congruence among the areas of endemism of Australian vertebrates. Aust. Syst. Bot. 1991, 4, 211–227. [Google Scholar] [CrossRef]
- Swofford, D.L. PAUP*. Phylogenetic Analysis Using Parsimony (*and Other Methods). Version 4; Sainauer Associates: Sunderland, MA, USA. Available online: http://phylosolutions.com/paup-test/ (accessed on 15 January 2022).
- Stanely, L.; Murray, C.M.; Murray, J.J.; Crother, B.I. Areas of endemism of Jamaica: Inferences from Parsimony Analysis of Endemism based on amphibian and reptile distributions. Biogeogr. J. Integr. Biogeogr. 2021, 36, a006. [Google Scholar] [CrossRef]
- Carpenter, J.M. Uninformative bootstrapping. Cladistics 1996, 12, 177–181. [Google Scholar] [CrossRef] [PubMed]
- Smith, V.S. Avian louse phylogeny (Phthiraptera: Ischnocera): A cladistic study based on morphology. Zool. J. Linn. Soc. 2001, 132, 81–144. [Google Scholar] [CrossRef]
- Di Giulio, A.; Fattorini, S.; Kaupp, A.; Vigna Taglianti, A.; Nagel, P. Review of competing hypotheses of phylogenetic relationships of Paussinae (Coleoptera: Carabidae) based on larval characters. Syst. Entomol. 2003, 28, 509–537. [Google Scholar] [CrossRef]
- Graham, C.H.; Smith, T.B.; Languy, M. Current and historical factors influencing patterns of species richness and turnover of birds in the Gulf of Guinea highlands. J. Biogeogr. 2005, 32, 1371–1384. [Google Scholar] [CrossRef]
- Guerrero, J.C.; Vargas, J.; Real, R. A hypothetico-deductive analysis of the environmental factors involved in the current reptile distribution pattern in the Canary Islands. J. Biogeogr. 2005, 32, 1343–1351. [Google Scholar] [CrossRef]
- Dapporto, L.; Wolf, H.; Strumia, F. Recent geography determines the distribution of some flying Hymenoptera in the Tuscan Archipelago. J. Zool. 2007, 272, 37–44. [Google Scholar] [CrossRef]
- López, H.L.; Menni, R.C.; Donato, M.; Miquelarena, A.M. Biogeographical revision of Argentina (Andean and Neotropical regions): An analysis using freshwater fishes. J. Biogeogr. 2008, 35, 1564–1579. [Google Scholar] [CrossRef]
- Fattorini, S. Biogeography of the tenebrionid beetles (Coleoptera, Tenebrionidae) on the Aegean Islands (Greece). J. Biogeogr. 2002, 29, 49–67. [Google Scholar] [CrossRef]
- Fattorini, S. Faunal patterns in tenebrionids (Coleoptera: Tenebrionidae) on the Tuscan Islands: The dominance of paleogeography over recent geography. Eur. J. Entomol. 2009, 277, 291–301. [Google Scholar] [CrossRef]
- Fattorini, S. Both Recent and Pleistocene geography determine animal distributional patterns in the Tuscan Archipelago. J. Zool. 2009, 277, 291–301. [Google Scholar] [CrossRef]
- Fattorini, S. Influence of recent geography and paleogeography on the structure of reptile communities in a landbridge archipelago. J. Herpetol. 2010, 44, 242–252. [Google Scholar] [CrossRef]
- Fattorini, S. Biogeography of tenebrionid beetles (Coleoptera: Tenebrionidae) on the circum-Sicilian islands (Italy, Sicily): Multiple biogeographical patterns require multiple explanations. Eur. J. Entomol. 2011, 108, 659–672. [Google Scholar] [CrossRef]
- Fattorini, S. Tenebrionid beetle distributional patterns in Italy: Multiple colonization trajectories in a biogeographical crossroad. Insect Conserv. Divers. 2014, 7, 144–160. [Google Scholar] [CrossRef]
- Heiser, M.; Dapporto, L.; Schmitt, T. Coupling impoverishment analysis and partitioning of beta diversity allows a comprehensive description of Odonata biogeography in the Western Mediterranean. Org. Divers. Evol. 2014, 14, 203–214. [Google Scholar] [CrossRef]
- Koleff, P.; Gaston, K.J.; Lennon, J.J. Measuring beta diversity for presence–absence data. J. Anim. Ecol. 2003, 72, 367–382. [Google Scholar] [CrossRef]
- Baselga, A. Partitioning the turnover and nestedness components of beta diversity. Glob. Ecol. Biogeogr. 2010, 19, 134–143. [Google Scholar] [CrossRef]
- Kreft, H.; Jetz, W. A framework for delineating biogeographical regions based on species distributions. J. Biogeogr. 2010, 37, 2029–2053. [Google Scholar] [CrossRef]
- Baselga, A. The relationship between species replacement, dissimilarity derived from nestedness, and nestedness. Glob. Ecol. Biogeogr. 2012, 21, 1223–1232. [Google Scholar] [CrossRef]
- Baselga, A.; Leprieur, F. Comparing methods to separate components of beta diversity. Methods Ecol. Evol. 2015, 6, 1069–1079. [Google Scholar] [CrossRef]
- Castro-Insua, A.; Gómez-Rodríguez, C.; Baselga, A. Dissimilarity measures affected by richness differences yield biased delimitations of biogeographic realms. Nat. Commun. 2018, 9, 5084. [Google Scholar] [CrossRef]
- Smith, S.A.; Bermingham, E. The biogeography of lower Mesoamerican freshwater fishes. J. Biogeogr. 2005, 32, 1835–1854. [Google Scholar] [CrossRef]
- Holt, B.; Lessard, J.P.; Borregaard, M.K.; Fritz, S.A.; Araujo, M.B.; Dimitrov, D.; Fabre, P.H.; Graham, C.H.; Graves, G.R.; Jonsson, K.A.; et al. An update of Wallace’s zoogeographic regions of the world. Science 2013, 339, 74–78. [Google Scholar] [CrossRef] [PubMed]
- Costello, M.J.; Tsai, P.; Wong, P.S.; Cheung, A.K.L.; Basher, Z.; Chaudhary, C. Marine biogeographic realms and species endemicity. Nat. Commun. 2017, 8, 1057. [Google Scholar] [CrossRef]
- Podani, J. On the sensitivity of ordination and classification methods to variation in the input order of data. J. Veg. Sci. 1997, 8, 153–156. [Google Scholar] [CrossRef]
- Dapporto, L.; Ramazzotti, M.; Fattorini, S.; Talavera, G.; Vila, R.; Dennis, R.L.H. Recluster: An unbiased clustering procedure for beta-diversity turnover. Ecography 2013, 36, 1070–1075. [Google Scholar] [CrossRef]
- Dapporto, L.; Ramazzotti, M.; Fattorini, S.; Vila, R.; Talavera, G.; Dennis, R.L.H. Recluster: Ordination Methods for the Analysis of Beta-Diversity Indices. Available online: https://CRAN.R-project.org/package=recluster (accessed on 15 March 2022).
- R Core Team. R: A Language and Environment for Statistical Computing; R Core Team: Vienna, Austria, 2020; Available online: https://www.R-project.org (accessed on 31 August 2021).
- Dapporto, L.; Ciolli, G.; Dennis, R.L.H.; Fox, R.; Shreeve, T.G. A new procedure for extrapolating turnover regionalization at mid-small spatial scales, tested on British butterflies. Methods Ecol. Evol. 2015, 6, 1287–1297. [Google Scholar] [CrossRef]
- Colbert, E.H. Wandering Lands and Animals; Hutchinson: London, UK, 1974; 326p. [Google Scholar]
- Van Andel, T.H. New Views on an Old Planet: Continental Drift and the History of the Earth; Cambridge University Press: Cambridge, UK, 1985; 439p. [Google Scholar]
- Jeannel, R. Le Genèse des faunes terrestres. Éléments de biogéographie; Presses Universitaires de France: Paris, France, 1942; p. viii + 513. [Google Scholar]
- Darlington, P.J. Beetles and continents. A review of “La genese des faunas terrestres” by R. Jeannel. Q. Rev. Biol. 1949, 24, 342–345. [Google Scholar] [CrossRef]
- Rehn, J.A.G. Review and critique of recent work on biogeography by Jeannel. Ecology 1950, 31, 307–311. [Google Scholar] [CrossRef]
- Darlington, P.J., Jr. Zoogeography: The Geographical Distribution of Animals; John Wiley & Sons, Inc.: New York, NY, USA, 1957; p. xiii + 675. [Google Scholar]
- George, W. Animal Geography; Heinemann: London, UK, 1962; 142p. [Google Scholar]
- Sereno, P.C. The Evolution of Dinosaurs. Science 1999, 284, 2137–2147. [Google Scholar] [CrossRef] [PubMed]
- Cox, C.B.; Moore, P.D. Biogeography: An Ecological and Evolutionary Approach, 8th ed.; Wiley: Hoboken, NJ, USA, 2010; 498p. [Google Scholar]
- Krause, D.W.; Sertich, J.J.W.; O’Connor, P.M.; Curry Rogers, K.; Rogers, R.R. The Mesozoic biogeographic history of Gondwanan terrestrial vertebrates: Insights from Madagascar’s fossil record. Annu. Rev. Earth Planet. Sci. 2019, 47, 519–553. [Google Scholar] [CrossRef]
- Davis, A.L.V.; Scholtz, C.H.; Philips, T.K. Historical biogeography of scarabaeine dung beetles. J. Biogeogr. 2002, 29, 1217–1256. [Google Scholar] [CrossRef]
- Royden, L.; Burchfiel, B.; Hilst, R.D. The geological evolution of the Tibetan Plateau. Science 2008, 321, 1054–1058. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fattorini, S. Historical Biogeography of Earwigs. Biology 2022, 11, 1794. https://doi.org/10.3390/biology11121794
Fattorini S. Historical Biogeography of Earwigs. Biology. 2022; 11(12):1794. https://doi.org/10.3390/biology11121794
Chicago/Turabian StyleFattorini, Simone. 2022. "Historical Biogeography of Earwigs" Biology 11, no. 12: 1794. https://doi.org/10.3390/biology11121794
APA StyleFattorini, S. (2022). Historical Biogeography of Earwigs. Biology, 11(12), 1794. https://doi.org/10.3390/biology11121794






