Similar Characteristics of siRNAs of Plant Viruses Which Replicate in Plant and Fungal Hosts
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Fungal Strains
2.2. Plant Virus Inoculation
2.3. Total RNA Extractions
2.4. sRNA Sequencing
2.5. Bioinformatics Analysis
3. Results
3.1. Characteristics of CMV- and Mycovirus-Derived siRNAs in R. solani
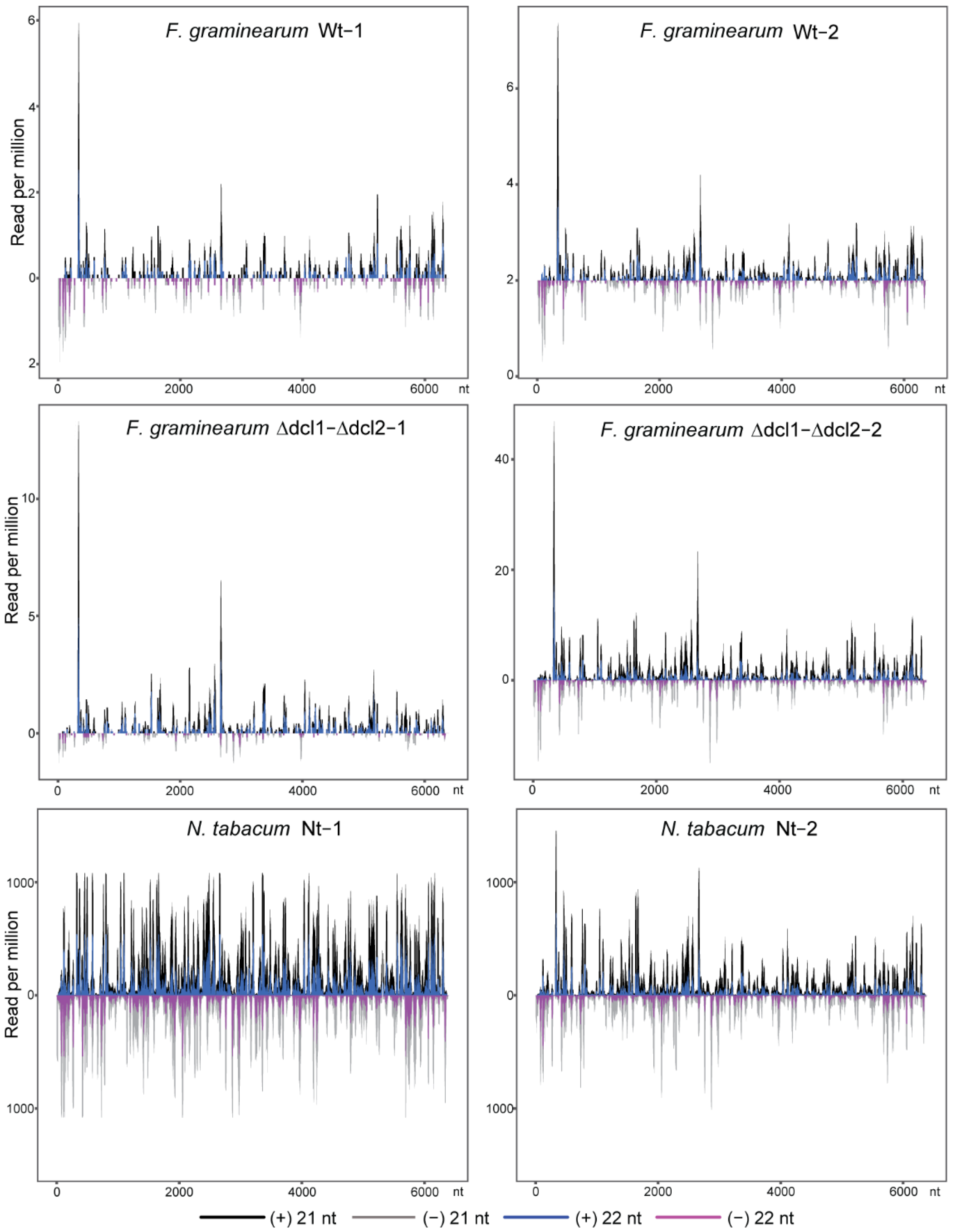
3.2. Characteristics of TMV siRNAs in Wild-Type and Dcl Mutants of F. graminearum
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Baulcombe, D. RNA silencing. Trends Biochem. Sci. 2005, 30, 290–293. [Google Scholar] [CrossRef]
- Sen, G.L.; Blau, H.M. A brief history of RNAi: The silence of the genes. FASEB J. 2006, 20, 1293–1299. [Google Scholar] [CrossRef] [Green Version]
- Lax, C.; Tahiri, G.; Patiño-Medina, J.A.; Cánovas-Márquez, J.T.; Pérez-Ruiz, J.A.; Osorio-Concepción, M.; Navarro, E.; Calo, S. The evolutionary significance of Rnai in the fungal kingdom. Int. J. Mol. Sci. 2020, 21, 9348. [Google Scholar] [CrossRef]
- Vogel, E.; Santos, D.; Mingels, L.; Verdonckt, T.-W.; Broeck, J.V. RNA interference in insects: Protecting beneficials and controlling pests. Front. Physiol. 2019, 9, 1912. [Google Scholar] [CrossRef] [Green Version]
- Bologna, N.G.; Voinnet, O. The diversity, biogenesis, and activities of endogenous silencing small RNAs in Arabidopsis. Annu. Rev. Plant Biol. 2014, 65, 473–503. [Google Scholar] [CrossRef]
- Zhuang, J.J.; Hunter, C.P. RNA interference in Caenorhabditis elegans: Uptake, mechanism, and regulation. Parasitology 2012, 139, 560–573. [Google Scholar] [CrossRef] [Green Version]
- Huntzinger, E.; Izaurralde, E. Gene silencing by microRNAs: Contributions of translational repression and mRNA decay. Nat. Rev. Genet. 2011, 12, 99–110. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Liu, Y. Diverse small non-coding RNAs in RNA interference pathways. Ther. Oligonucleotides 2011, 764, 169–182. [Google Scholar]
- Volpe, T.; Martienssen, R.A. RNA interference and heterochromatin assembly. Cold Spring Harb. Perspect. Biol. 2011, 3, a003731. [Google Scholar] [CrossRef] [Green Version]
- Ding, S.-W.; Han, Q.; Wang, J.; Li, W.-X. Antiviral RNA interference in mammals. Curr. Opin. Immunol. 2018, 54, 109–114. [Google Scholar] [CrossRef] [PubMed]
- Bonning, B.C.; Saleh, M.-C. The interplay between viruses and RNAi pathways in insects. Annu. Rev. Entomol. 2021, 66, 61–79. [Google Scholar] [CrossRef]
- Sarkies, P.; Miska, E.A. RNAi pathways in the recognition of foreign RNA: Antiviral responses and host–parasite interactions in nematodes. Biochem. Soc. Trans. 2013, 41, 876–880. [Google Scholar] [CrossRef] [PubMed]
- Llave, C. Virus-derived small interfering RNAs at the core of plant–virus interactions. Trends Plant Sci. 2010, 15, 701–707. [Google Scholar] [CrossRef] [PubMed]
- Segers, G.C.; Zhang, X.; Deng, F.; Sun, Q.; Nuss, D.L. Evidence that RNA silencing functions as an antiviral defense mechanism in fungi. Proc. Natl. Acad. Sci. USA 2007, 104, 12902–12906. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lopez-Gomollon, S.; Baulcombe, D.C. Roles of RNA silencing in viral and non-viral plant immunity and in the crosstalk between disease resistance systems. Nat. Rev. Mol. Cell Biol. 2022, 23, 645–662. [Google Scholar] [CrossRef]
- Hua, C.; Zhao, J.-H.; Guo, H.-S. Trans-kingdom RNA silencing in plant–fungal pathogen interactions. Mol. Plant 2018, 11, 235–244. [Google Scholar] [CrossRef] [Green Version]
- Nicolás, F.E.; Garre, V. RNA interference in fungi: Retention and loss. Microbiol. Spectr. 2016, 4, 657–671. [Google Scholar] [CrossRef]
- Nunes, C.C.; Sailsbery, J.K.; Dean, R.A. Characterization and application of small RNAs and RNA silencing mechanisms in fungi. Fungal Biol. Rev. 2011, 25, 172–180. [Google Scholar] [CrossRef]
- Chang, S.-S.; Zhang, Z.; Liu, Y. RNA interference pathways in fungi: Mechanisms and functions. Annu. Rev. Microbiol. 2012, 66, 305–323. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nicolás, F.E.; Ruiz-Vázquez, R.M. Functional diversity of RNAi-associated sRNAs in fungi. Int. J. Mol. Sci. 2013, 14, 15348–15360. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Feretzaki, M.; Billmyre, R.B.; Clancey, S.A.; Wang, X.; Heitman, J. Gene network polymorphism illuminates loss and retention of novel RNAi silencing components in the Cryptococcus pathogenic species complex. PLoS Genet. 2016, 12, e1005868. [Google Scholar] [CrossRef]
- Drinnenberg, I.A.; Fink, G.R.; Bartel, D.P. Compatibility with killer explains the rise of RNAi-deficient fungi. Science 2011, 333, 1592. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nicolas, F.E.; Torres-Martinez, S.; Ruiz-Vazquez, R.M. Loss and retention of RNA interference in fungi and parasites. PLoS Pathog. 2013, 9, e1003089. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, X.; Hsueh, Y.-P.; Li, W.; Floyd, A.; Skalsky, R.; Heitman, J. Sex-induced silencing defends the genome of Cryptococcus neoformans via RNAi. Genes Dev. 2010, 24, 2566–2582. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nakayashiki, H.; Kadotani, N.; Mayama, S. Evolution and diversification of RNA silencing proteins in fungi. J. Mol. Evol. 2006, 63, 127–135. [Google Scholar] [CrossRef] [Green Version]
- Cogoni, C.; Macino, G. Isolation of quelling-defective (qde) mutants impaired in posttranscriptional transgene-induced gene silencing in Neurospora crassa. Proc. Natl. Acad. Sci. USA 1997, 94, 10233–10238. [Google Scholar] [CrossRef] [Green Version]
- Cogoni, C.; Macino, G. Gene silencing in Neurospora crassa requires a protein homologous to RNA-dependent RNA polymerase. Nature 1999, 399, 166–169. [Google Scholar] [CrossRef]
- Yang, Q.; Ye, Q.A.; Liu, Y. Mechanism of siRNA production from repetitive DNA. Genes Dev. 2015, 29, 526–537. [Google Scholar] [CrossRef] [Green Version]
- Wang, X.; Wang, P.; Sun, S.; Darwiche, S.; Idnurm, A.; Heitman, J. Transgene induced co-suppression during vegetative growth in Cryptococcus neoformans. PLoS Genet. 2012, 8, e1002885. [Google Scholar] [CrossRef]
- Nicolás, F.E.; Torres-Martínez, S.; Ruiz-Vázquez, R.M. Two classes of small antisense RNAs in fungal RNA silencing triggered by non-integrative transgenes. EMBO J. 2003, 22, 3983–3991. [Google Scholar] [CrossRef] [Green Version]
- Shiu, P.K.; Metzenberg, R.L. Meiotic silencing by unpaired DNA: Properties, regulation and suppression. Genetics 2002, 161, 1483–1495. [Google Scholar] [CrossRef] [PubMed]
- Volpe, T.A.; Kidner, C.; Hall, I.M.; Teng, G.; Grewal, S.I.; Martienssen, R.A. Regulation of heterochromatic silencing and histone H3 lysine-9 methylation by RNAi. Science 2002, 297, 1833–1837. [Google Scholar] [CrossRef] [Green Version]
- Verdel, A.; Jia, S.; Gerber, S.; Sugiyama, T.; Gygi, S.; Grewal, S.I.; Moazed, D. RNAi-mediated targeting of heterochromatin by the RITS complex. Science 2004, 303, 672–676. [Google Scholar] [CrossRef] [Green Version]
- Lee, H.-C.; Li, L.; Gu, W.; Xue, Z.; Crosthwaite, S.K.; Pertsemlidis, A.; Lewis, Z.A.; Freitag, M.; Selker, E.U.; Mello, C.C. Diverse pathways generate microRNA-like RNAs and Dicer-independent small interfering RNAs in fungi. Mol. Cell 2010, 38, 803–814. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ghabrial, S.A.; Suzuki, N. Viruses of plant pathogenic fungi. Annu. Rev. Phytopathol. 2009, 47, 353–384. [Google Scholar] [CrossRef]
- Ghabrial, S.A.; Castón, J.R.; Jiang, D.; Nibert, M.L.; Suzuki, N. 50-plus years of fungal viruses. Virology 2015, 479, 356–368. [Google Scholar] [CrossRef] [Green Version]
- King, A.M.; Lefkowitz, E.J.; Mushegian, A.R.; Adams, M.J.; Dutilh, B.E.; Gorbalenya, A.E.; Harrach, B.; Harrison, R.L.; Junglen, S.; Knowles, N.J. Changes to taxonomy and the International Code of Virus Classification and Nomenclature ratified by the International Committee on Taxonomy of Viruses (2018). Arch. Virol. 2018, 163, 2601–2631. [Google Scholar] [CrossRef] [Green Version]
- Xie, J.; Jiang, D. New insights into mycoviruses and exploration for the biological control of crop fungal diseases. Annu. Rev. Phytopathol. 2014, 52, 45–68. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hillman, B.I.; Annisa, A.; Suzuki, N. Viruses of plant-interacting fungi. Adv. Virus Res. 2018, 100, 99–116. [Google Scholar]
- Campo, S.; Gilbert, K.B.; Carrington, J.C. Small RNA-based antiviral defense in the phytopathogenic fungus Colletotrichum higginsianum. PLoS Pathog. 2016, 12, e1005640. [Google Scholar] [CrossRef] [Green Version]
- Yu, J.; Lee, K.-M.; Cho, W.K.; Park, J.Y.; Kim, K.-H. Differential contribution of RNA interference components in response to distinct Fusarium graminearum virus infections. J. Virol. 2018, 92, e01756-17. [Google Scholar] [CrossRef] [Green Version]
- Honda, S.; Eusebio-Cope, A.; Miyashita, S.; Yokoyama, A.; Aulia, A.; Shahi, S.; Kondo, H.; Suzuki, N. Establishment of Neurospora crassa as a model organism for fungal virology. Nat. Commun. 2020, 11, 5627. [Google Scholar] [CrossRef]
- Mochama, P.; Jadhav, P.; Neupane, A.; Lee Marzano, S.-Y. Mycoviruses as triggers and targets of RNA silencing in white mold fungus Sclerotinia sclerotiorum. Viruses 2018, 10, 214. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Neupane, A.; Feng, C.; Mochama, P.K.; Saleem, H.; Marzano, S.-Y.L. Roles of argonautes and dicers on Sclerotinia sclerotiorum antiviral RNA silencing. Front. Plant Sci. 2019, 10, 976. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sun, Q.; Choi, G.H.; Nuss, D.L. A single Argonaute gene is required for induction of RNA silencing antiviral defense and promotes viral RNA recombination. Proc. Natl. Acad. Sci. USA 2009, 106, 17927–17932. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Muñoz-Adalia, E.J.; Diez, J.J.; Fernández, M.M.; Hantula, J.; Vainio, E.J. Characterization of small RNAs originating from mitoviruses infecting the conifer pathogen Fusarium circinatum. Arch. Virol. 2018, 163, 1009–1018. [Google Scholar] [CrossRef] [PubMed]
- Vainio, E.J.; Jurvansuu, J.; Streng, J.; Rajamäki, M.-L.; Hantula, J.; Valkonen, J.P. Diagnosis and discovery of fungal viruses using deep sequencing of small RNAs. J. Gen. Virol. 2015, 96, 714–725. [Google Scholar] [CrossRef] [Green Version]
- Hammond, T.; Andrewski, M.; Roossinck, M.; Keller, N. Aspergillus mycoviruses are targets and suppressors of RNA silencing. Eukaryot. Cell 2008, 7, 350–357. [Google Scholar] [CrossRef] [Green Version]
- Wang, Q.; Cheng, S.; Xiao, X.; Cheng, J.; Fu, Y.; Chen, T.; Jiang, D.; Xie, J. Discovery of two mycoviruses by high-throughput sequencing and assembly of mycovirus-derived small silencing RNAs from a hypovirulent strain of Sclerotinia sclerotiorum. Front. Microbiol. 2019, 10, 1415. [Google Scholar] [CrossRef]
- Himeno, M.; Maejima, K.; Komatsu, K.; Ozeki, J.; Hashimoto, M.; Kagiwada, S.; Yamaji, Y.; Namba, S. Significantly low level of small RNA accumulation derived from an encapsidated mycovirus with dsRNA genome. Virology 2010, 396, 69–75. [Google Scholar] [CrossRef] [Green Version]
- Andika, I.B.; Jamal, A.; Kondo, H.; Suzuki, N. SAGA complex mediates the transcriptional up-regulation of antiviral RNA silencing. Proc. Natl. Acad. Sci. USA 2017, 114, E3499–E3506. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yaegashi, H.; Shimizu, T.; Ito, T.; Kanematsu, S. Differential inductions of RNA silencing among encapsidated double-stranded RNA mycoviruses in the white root rot fungus Rosellinia necatrix. J. Virol. 2016, 90, 5677–5692. [Google Scholar] [CrossRef] [PubMed]
- Donaire, L.; Ayllón, M.A. Deep sequencing of mycovirus-derived small RNAs from Botrytis species. Mol. Plant Pathol. 2017, 18, 1127–1137. [Google Scholar] [CrossRef] [PubMed]
- Özkan, S.; Mohorianu, I.; Xu, P.; Dalmay, T.; Coutts, R.H. Profile and functional analysis of small RNAs derived from Aspergillus fumigatus infected with double-stranded RNA mycoviruses. BMC Genom. 2017, 18, 416. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, S.; Li, P.; Zhang, J.; Qiu, D.; Guo, L. Generation of a high resolution map of sRNAs from Fusarium graminearum and analysis of responses to viral infection. Sci. Rep. 2016, 6, 26151. [Google Scholar] [CrossRef] [Green Version]
- Fonseca, P.; Ferreira, F.; da Silva, F.; Oliveira, L.S.; Marques, J.T.; Goes-Neto, A.; Aguiar, E.; Gruber, A. Characterization of a novel mitovirus of the sand fly Lutzomyia longipalpis using genomic and virus–host interaction signatures. Viruses 2020, 13, 9. [Google Scholar] [CrossRef]
- Khalifa, M.E.; MacDiarmid, R.M. A Mechanically Transmitted DNA Mycovirus Is Targeted by the Defence Machinery of Its Host, Botrytis cinerea. Viruses 2021, 13, 1315. [Google Scholar] [CrossRef]
- Shahi, S.; Eusebio-Cope, A.; Kondo, H.; Hillman, B.I.; Suzuki, N. Investigation of host range of and host defense against a mitochondrially replicating mitovirus. J. Virol. 2019, 93, e01503-18. [Google Scholar] [CrossRef] [Green Version]
- Janda, M.; Ahlquist, P. RNA-dependent replication, transcription, and persistence of brome mosaic virus RNA replicons in S. cerevisiae. Cell 1993, 72, 961–970. [Google Scholar] [CrossRef]
- Nagy, P.D. Yeast as a model host to explore plant virus-host interactions. Annu. Rev. Phytopathol. 2008, 46, 217–242. [Google Scholar] [CrossRef] [Green Version]
- Mascia, T.; Nigro, F.; Abdallah, A.; Ferrara, M.; De Stradis, A.; Faedda, R.; Palukaitis, P.; Gallitelli, D. Gene silencing and gene expression in phytopathogenic fungi using a plant virus vector. Proc. Natl. Acad. Sci. USA 2014, 111, 4291–4296. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mascia, T.; Vučurović, A.; Minutillo, S.; Nigro, F.; Labarile, R.; Savoia, M.; Palukaitis, P.; Gallitelli, D. Infection of Colletotrichum acutatum and Phytophthora infestans by taxonomically different plant viruses. Eur. J. Plant Pathol. 2019, 153, 1001–1017. [Google Scholar] [CrossRef]
- Bian, R.; Andika, I.B.; Pang, T.; Lian, Z.; Wei, S.; Niu, E.; Wu, Y.; Kondo, H.; Liu, X.; Sun, L. Facilitative and synergistic interactions between fungal and plant viruses. Proc. Natl. Acad. Sci. USA 2020, 117, 3779–3788. [Google Scholar] [CrossRef]
- Wei, S.; Bian, R.; Andika, I.B.; Niu, E.; Liu, Q.; Kondo, H.; Yang, L.; Zhou, H.; Pang, T.; Lian, Z. Symptomatic plant viroid infections in phytopathogenic fungi. Proc. Natl. Acad. Sci. USA 2019, 116, 13042–13050. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Afanasenko, O.; Khiutti, A.; Mironenko, N.; Lashina, N. Transmission of potato spindle tuber viroid between Phytophthora infestans and host plants. Vavilov J. Genet. Breed. 2022, 26, 272. [Google Scholar] [CrossRef] [PubMed]
- Andika, I.B.; Wei, S.; Cao, C.; Salaipeth, L.; Kondo, H.; Sun, L. Phytopathogenic fungus hosts a plant virus: A naturally occurring cross-kingdom viral infection. Proc. Natl. Acad. Sci. USA 2017, 114, 12267–12272. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cao, X.; Liu, J.; Pang, J.; Kondo, H.; Chi, S.; Zhang, J.; Sun, L.; Andika, I.B. Common but Nonpersistent Acquisitions of Plant Viruses by Plant-Associated Fungi. Viruses 2022, 14, 2279. [Google Scholar] [CrossRef]
- Chen, M.-H.; Roossinck, M.; Kao, C. Efficient and specific initiation of subgenomic RNA synthesis by cucumber mosaic virus replicase in vitro requires an upstream RNA stem-loop. J. Virol. 2000, 74, 11201–11209. [Google Scholar] [CrossRef] [Green Version]
- Margis, R.; Fusaro, A.F.; Smith, N.A.; Curtin, S.J.; Watson, J.M.; Finnegan, E.J.; Waterhouse, P.M. The evolution and diversification of Dicers in plants. FEBS Lett. 2006, 580, 2442–2450. [Google Scholar] [CrossRef]
- Deleris, A.; Gallego-Bartolome, J.; Bao, J.; Kasschau, K.D.; Carrington, J.C.; Voinnet, O. Hierarchical action and inhibition of plant Dicer-like proteins in antiviral defense. Science 2006, 313, 68–71. [Google Scholar] [CrossRef]
- Wang, X.-B.; Jovel, J.; Udomporn, P.; Wang, Y.; Wu, Q.; Li, W.-X.; Gasciolli, V.; Vaucheret, H.; Ding, S.-W. The 21-nucleotide, but not 22-nucleotide, viral secondary small interfering RNAs direct potent antiviral defense by two cooperative argonautes in Arabidopsis thaliana. Plant Cell 2011, 23, 1625–1638. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bouché, N.; Lauressergues, D.; Gasciolli, V.; Vaucheret, H. An antagonistic function for Arabidopsis DCL2 in development and a new function for DCL4 in generating viral siRNAs. EMBO J. 2006, 25, 3347–3356. [Google Scholar] [CrossRef] [PubMed]
- Qin, C.; Li, B.; Fan, Y.; Zhang, X.; Yu, Z.; Ryabov, E.; Zhao, M.; Wang, H.; Shi, N.; Zhang, P. Roles of Dicer-like proteins 2 and 4 in intra-and intercellular antiviral silencing. Plant Physiol. 2017, 174, 1067–1081. [Google Scholar] [CrossRef] [PubMed]
- Andika, I.B.; Maruyama, K.; Sun, L.; Kondo, H.; Tamada, T.; Suzuki, N. Differential contributions of plant Dicer-like proteins to antiviral defences against potato virus X in leaves and roots. Plant J. 2015, 81, 781–793. [Google Scholar] [CrossRef]
- Andika, I.B.; Maruyama, K.; Sun, L.; Kondo, H.; Tamada, T.; Suzuki, N. Different Dicer-like protein components required for intracellular and systemic antiviral silencing in Arabidopsis thaliana. Plant Signal. Behav. 2015, 10, e1039214. [Google Scholar] [CrossRef] [Green Version]
- Raja, P.; Jackel, J.N.; Li, S.; Heard, I.M.; Bisaro, D.M. Arabidopsis double-stranded RNA binding protein DRB3 participates in methylation-mediated defense against geminiviruses. J. Virol. 2014, 88, 2611–2622. [Google Scholar] [CrossRef] [Green Version]
- Blevins, T.; Rajeswaran, R.; Shivaprasad, P.V.; Beknazariants, D.; Si-Ammour, A.; Park, H.-S.; Vazquez, F.; Robertson, D.; Meins Jr, F.; Hohn, T. Four plant Dicers mediate viral small RNA biogenesis and DNA virus induced silencing. Nucleic Acids Res. 2006, 34, 6233–6246. [Google Scholar] [CrossRef] [Green Version]
- de Haro, J.P.; Calo, S.; Cervantes, M.; Nicolás, F.E.; Torres-Martínez, S.; Ruiz-Vázquez, R.M. A single dicer gene is required for efficient gene silencing associated with two classes of small antisense RNAs in Mucor circinelloides. Eukaryot. Cell 2009, 8, 1486–1497. [Google Scholar] [CrossRef] [Green Version]
- Ding, S.-W. RNA-based antiviral immunity. Nat. Rev. Immunol. 2010, 10, 632. [Google Scholar] [CrossRef]
- Andika, I.B.; Kondo, H.; Suzuki, N. Dicer functions transcriptionally and posttranscriptionally in a multilayer antiviral defense. Proc. Natl. Acad. Sci. USA 2019, 116, 2274–2281. [Google Scholar] [CrossRef] [Green Version]
- Trieu, T.A.; Calo, S.; Nicolas, F.E.; Vila, A.; Moxon, S.; Dalmay, T.; Torres-Martinez, S.; Garre, V.; Ruiz-Vazquez, R.M. A non-canonical RNA silencing pathway promotes mRNA degradation in basal fungi. PLoS Genet. 2015, 11, e1005168. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dang, Y.; Li, L.; Guo, W.; Xue, Z.; Liu, Y. Convergent transcription induces dynamic DNA methylation at disiRNA loci. PLoS Genet. 2013, 9, e1003761. [Google Scholar] [CrossRef] [PubMed]
- Lo Presti, L.; Lanver, D.; Schweizer, G.; Tanaka, S.; Liang, L.; Tollot, M.; Zuccaro, A.; Reissmann, S.; Kahmann, R. Fungal effectors and plant susceptibility. Annu. Rev. Plant Biol. 2015, 66, 513–545. [Google Scholar] [CrossRef] [PubMed]
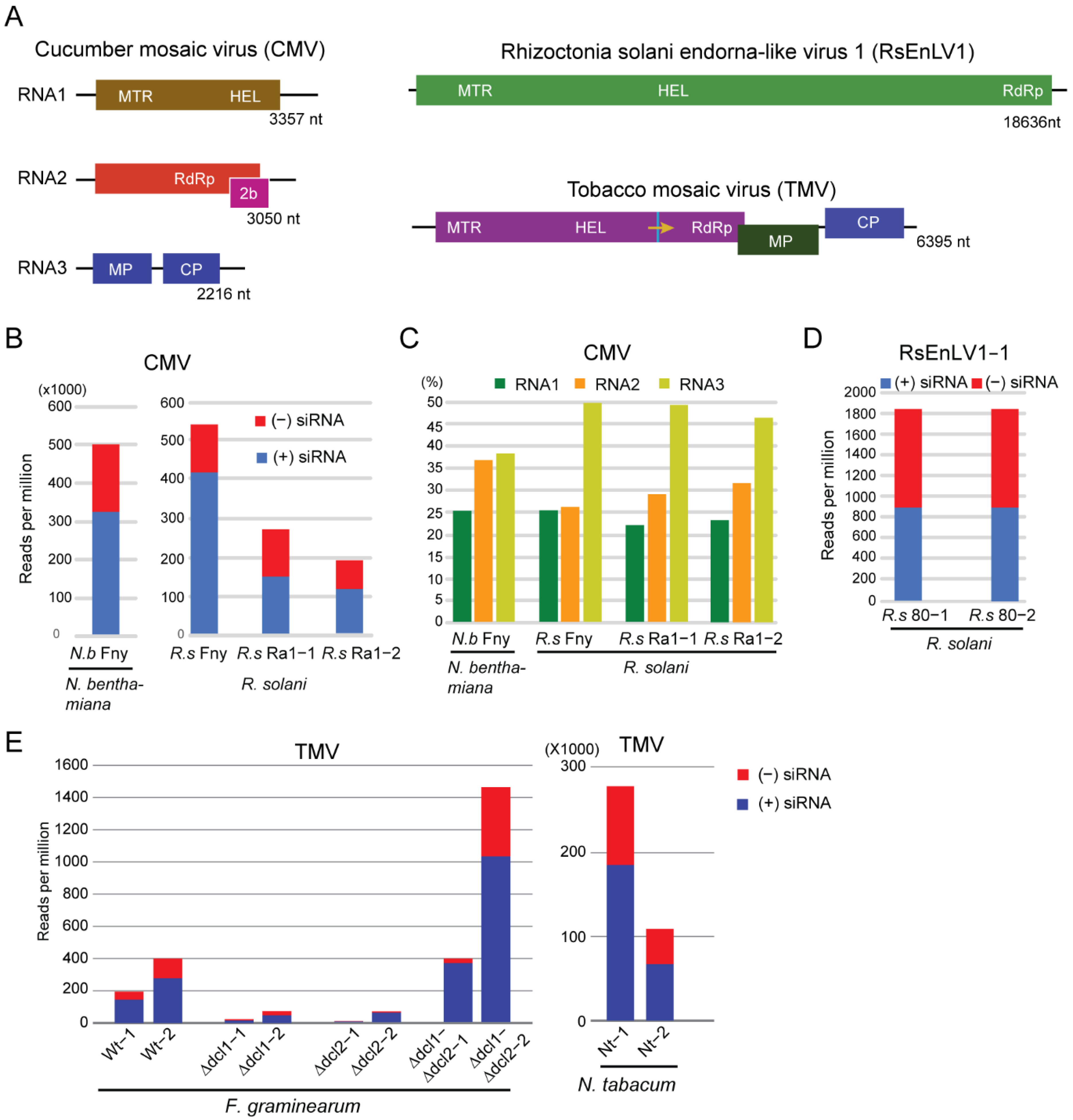
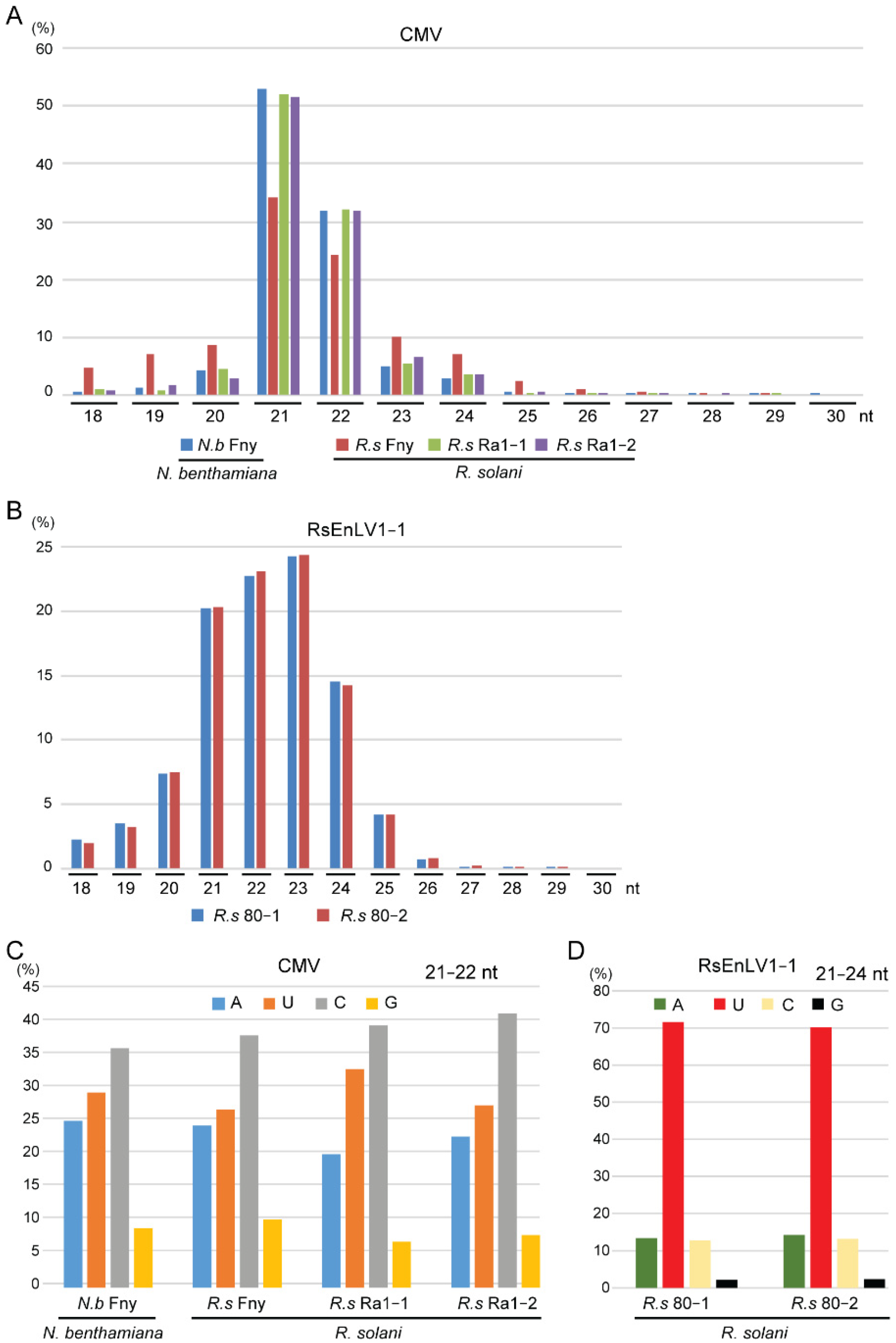
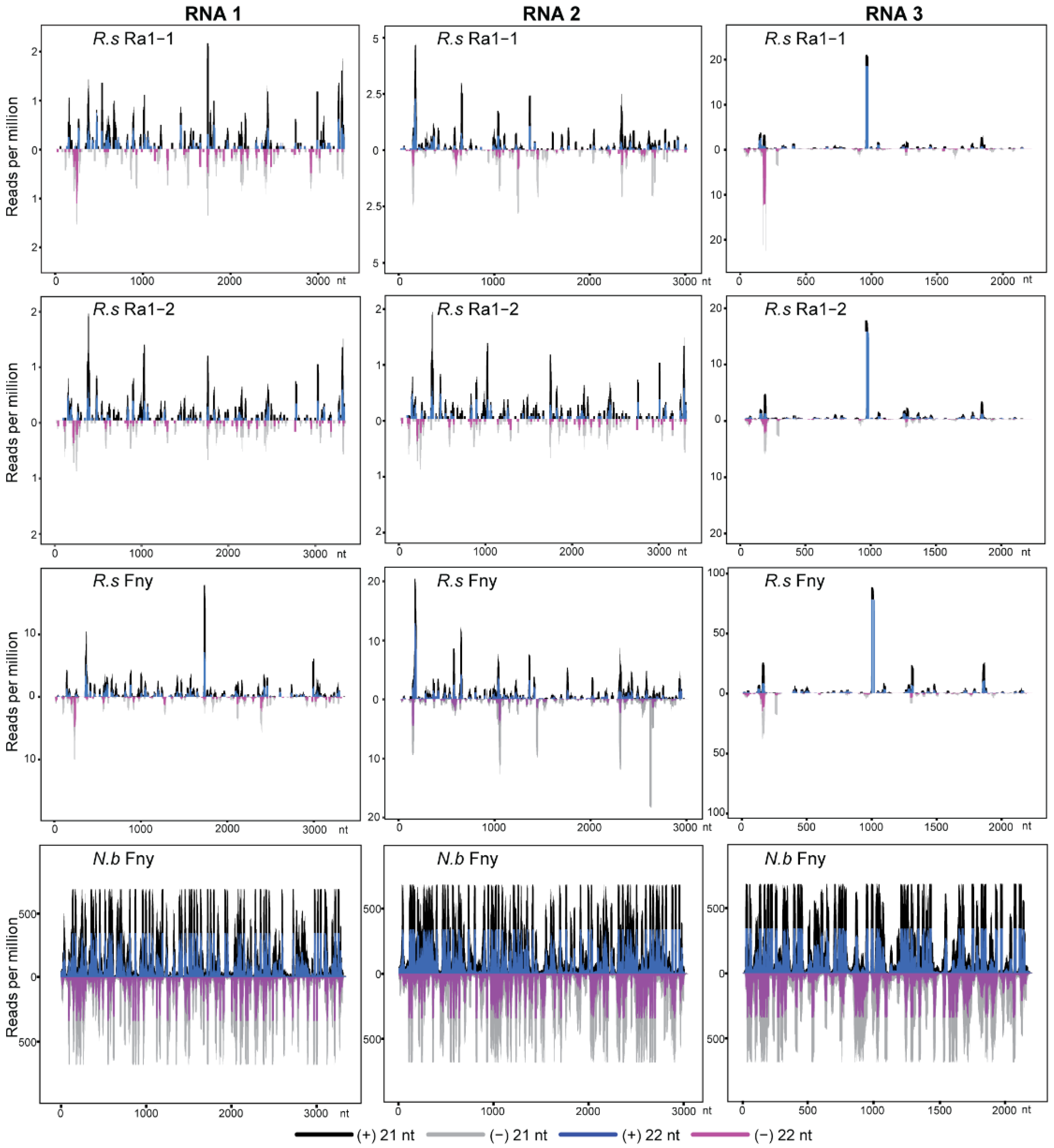
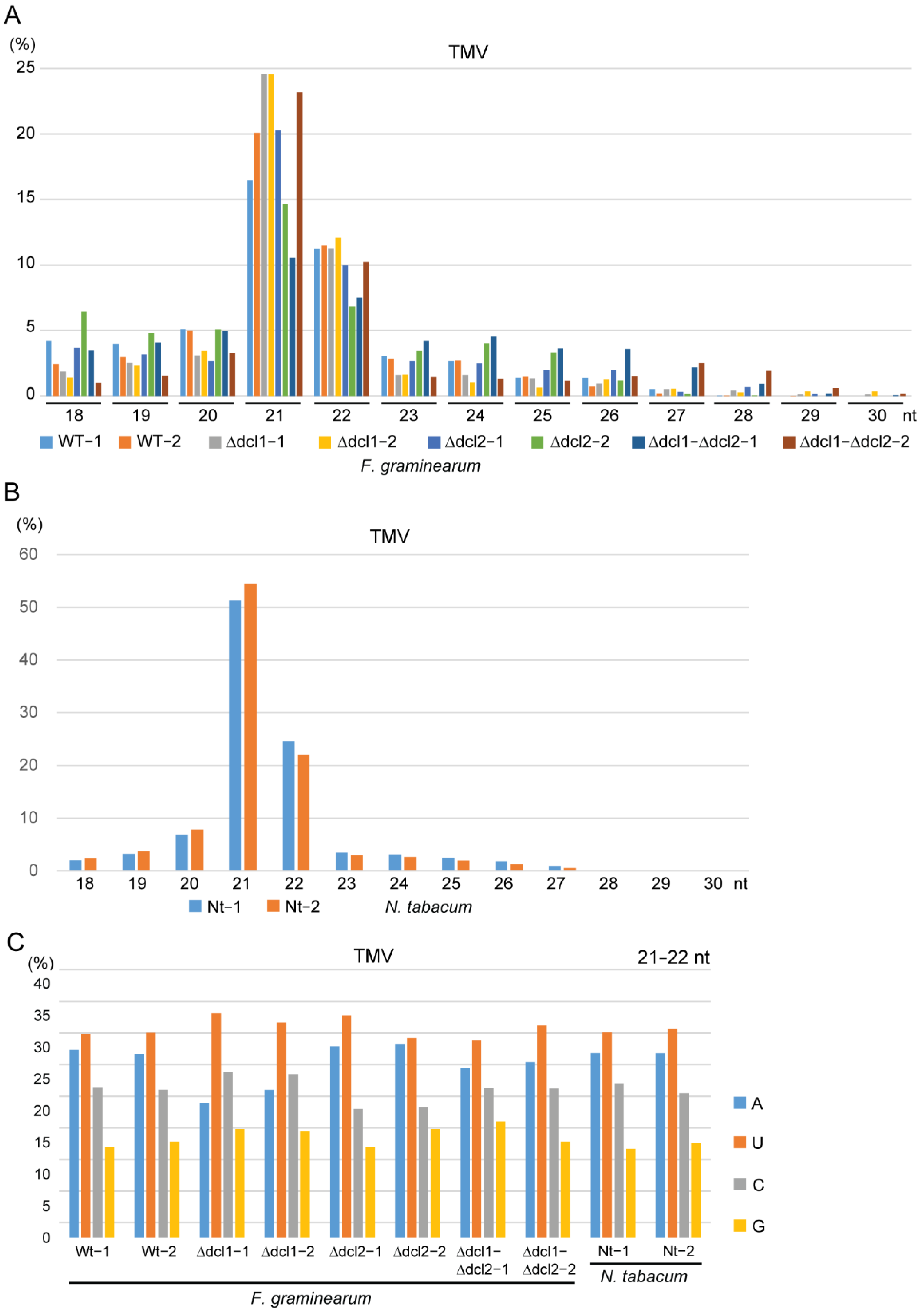
| Sample (Library) | R. solani Ra1-1 | R. solani Ra1-2 | R. solani Ra1/CMV-Cured-1 | R. solani Ra1/CMV-Cured-2 | R. solani /CMV Fny | N. benth- amiana/CMV Fny | R. Solani 80-1 | R. Solani 80-2 | R. Solani 80 /Virus-Cured-1 | R. Solani 80 /Virus-Cured-2 |
|---|---|---|---|---|---|---|---|---|---|---|
| Total sRNA | 16,482,261 | 20,302,809 | 21,988,288 | 21,827,663 | 17,824,097 | 23,621,525 | 18,828,697 | 18,258,925 | 25,667,410 | 25,075,982 |
| Virus | CMV Rs | CMV Rs | CMV Rs | CMV Rs | CMV Fny | CMV Fny | RsEnLV1 | RsEnLV1 | RsEnLV1 | RsEnLV1 |
| siRNAs | 4455 | 3885 | 0 | 2 | 11,738 | 11,817,040 | 34,391 | 33,911 | 131 | 107 |
| Percentage | 0.03% | 0.02% | 0.00% | 0.00% | 0.07% | 50.03% | 0.18% | 0.19% | 0.00% | 0.00% |
| Sample (Library) | F. graminearum | N. tabacum | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Wt/Virus Free-1 | Wt/Virus Free-2 | Wt-1 | Wt-2 | Δdcl1-1 | Δdcl1-2 | Δdcl2-1 | Δdcl2-2 | Δdcl1-Δdcl2-1 | Δdcl1-Δdcl2-2 | Nt-1 | Nt-2 | |
| Total sRNA | 7,874,233 | 9,424,347 | 12,282,580 | 11,953,464 | 11,791,300 | 9,358,452 | 16,637,401 | 12,346,680 | 11,849,385 | 12,552,172 | 14,797,683 | 11,033,408 |
| siRNAs | 0 | 0 | 2386 | 4787 | 374 | 707 | 301 | 935 | 4792 | 18,604 | 4,043,468 | 1,184,759 |
| Percentage | 0.00% | 0.00% | 0.02% | 0.04% | 0.00% | 0.01% | 0.00% | 0.01% | 0.04% | 0.15% | 27.33% | 10.74% |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pang, T.; Peng, J.; Bian, R.; Liu, Y.; Zhang, D.; Andika, I.B.; Sun, L. Similar Characteristics of siRNAs of Plant Viruses Which Replicate in Plant and Fungal Hosts. Biology 2022, 11, 1672. https://doi.org/10.3390/biology11111672
Pang T, Peng J, Bian R, Liu Y, Zhang D, Andika IB, Sun L. Similar Characteristics of siRNAs of Plant Viruses Which Replicate in Plant and Fungal Hosts. Biology. 2022; 11(11):1672. https://doi.org/10.3390/biology11111672
Chicago/Turabian StylePang, Tianxing, Jianping Peng, Ruiling Bian, Yu Liu, Dong Zhang, Ida Bagus Andika, and Liying Sun. 2022. "Similar Characteristics of siRNAs of Plant Viruses Which Replicate in Plant and Fungal Hosts" Biology 11, no. 11: 1672. https://doi.org/10.3390/biology11111672
APA StylePang, T., Peng, J., Bian, R., Liu, Y., Zhang, D., Andika, I. B., & Sun, L. (2022). Similar Characteristics of siRNAs of Plant Viruses Which Replicate in Plant and Fungal Hosts. Biology, 11(11), 1672. https://doi.org/10.3390/biology11111672






