Genetic Structure of an East Asian Minnow (Toxabramis houdemeri) in Southern China, with Implications for Conservation
Abstract
Simple Summary
Abstract
1. Introduction
2. Methods
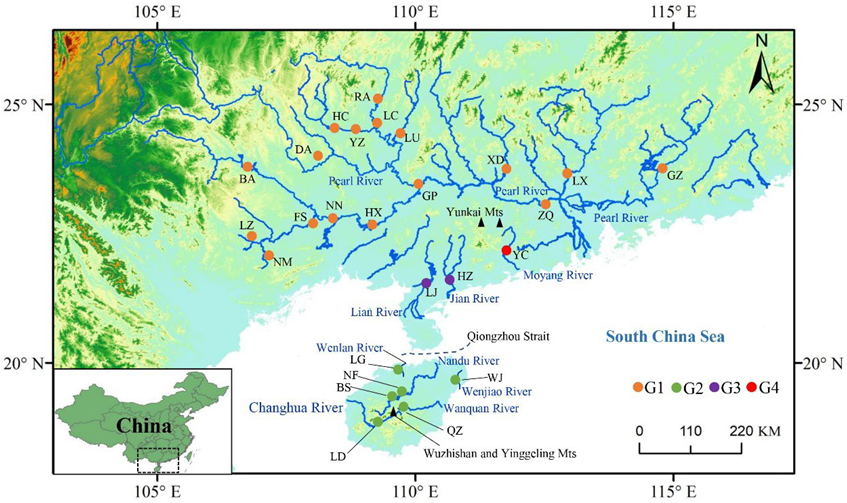
2.1. Sampling
2.2. Sequence Analyses and Haplotype Network
2.3. Molecular Diversity and Genetic Structure
2.4. Divergence Time Estimate
2.5. Demographic History and Historical Gene Flow
3. Results
3.1. Sequence Statistics
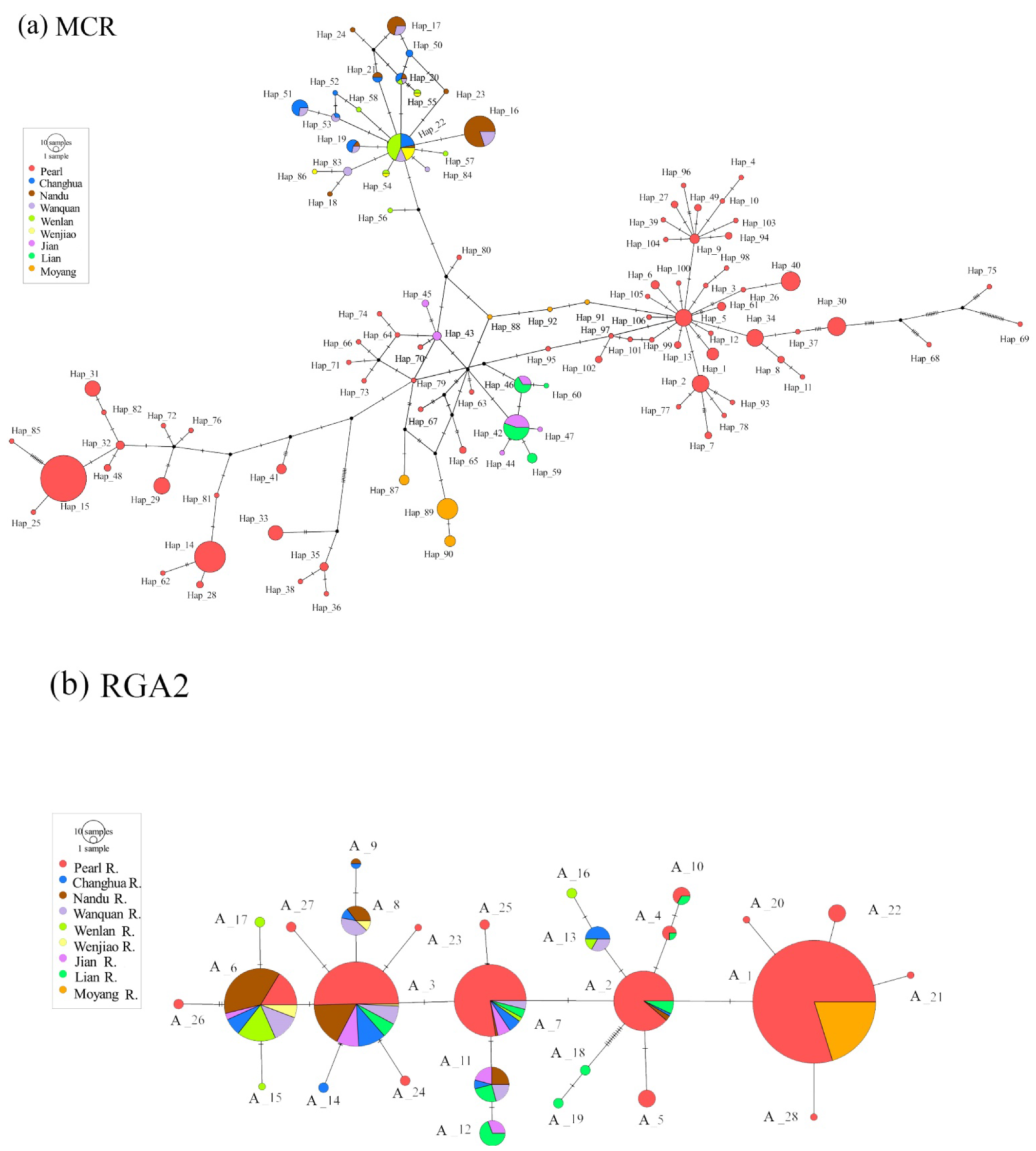
3.2. Haplotype and Allele Networks
3.3. Genetic Structure
3.4. Molecular Diversity Indexes
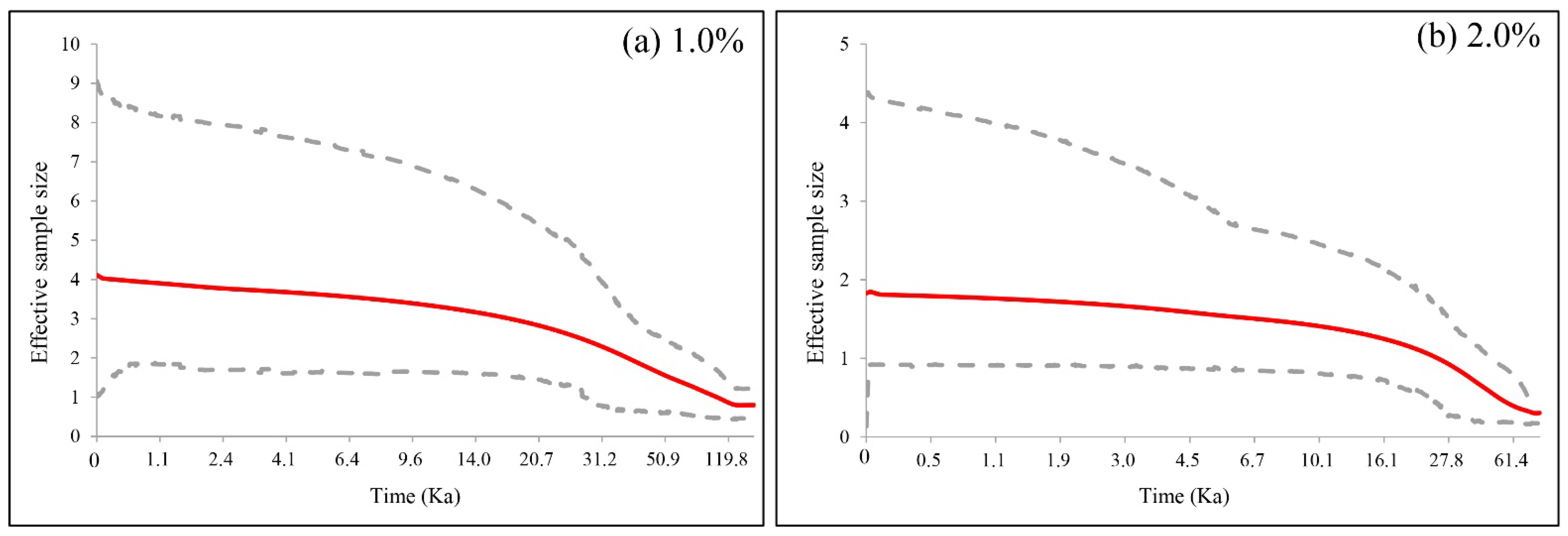
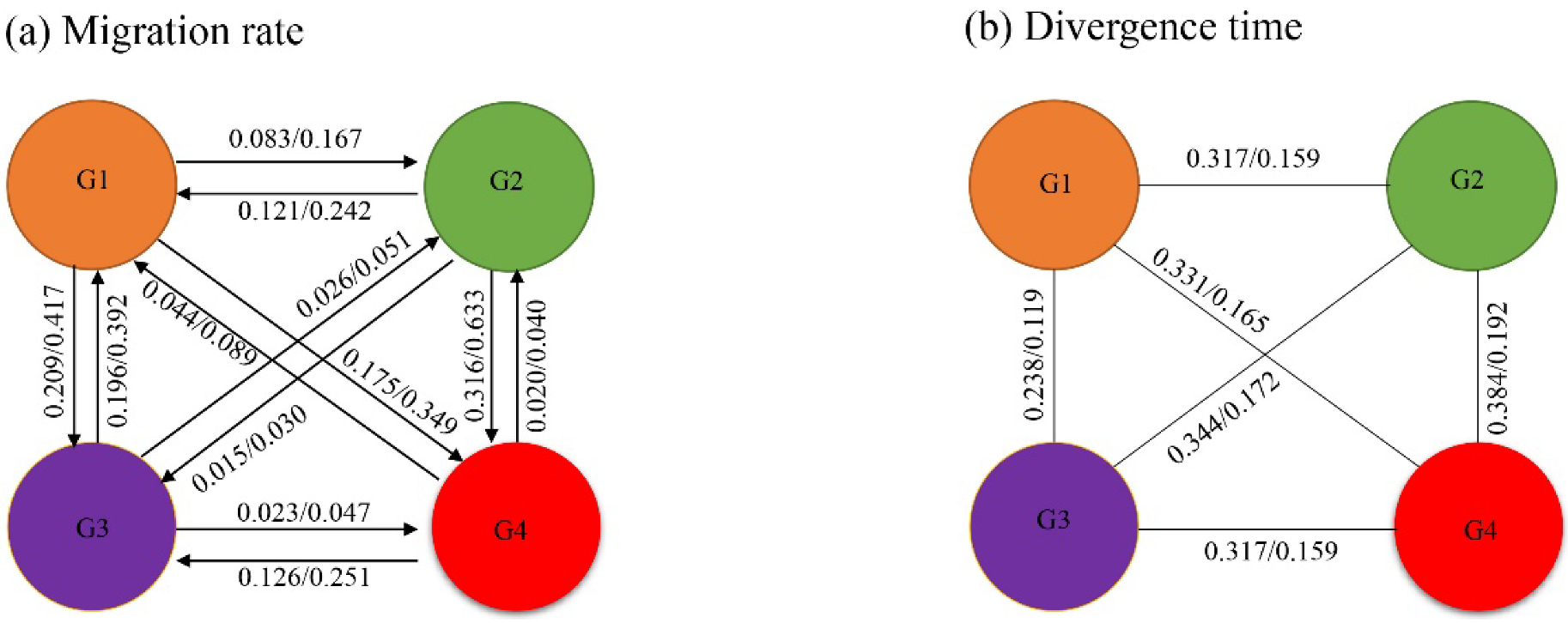
3.5. Demographic Analyses and Gene Flow
3.6. Divergence Time Estimates
4. Discussion
4.1. Genetic Structure of T. houdemeri
4.2. Demographic History
4.3. Genetic Diversity
4.4. Conservation Implications
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Burridge, C.P.; Craw, D.; Fletcher, D.; Waters, J.M. Geological dates and molecular rates: Fish DNA sheds light on time dependency. Mol. Biol. Evol. 2008, 25, 624–633. [Google Scholar] [CrossRef] [PubMed]
- Yang, L.; He, S. Phylogeography of the freshwater catfish Hemibagrus guttatus (Siluriformes, Bagridae): Implications for South China biogeography and influence of sea-level changes. Mol. Phylogenet. Evol. 2008, 49, 393–398. [Google Scholar] [CrossRef] [PubMed]
- Wong, W.; Ma, K.; Tsang, L.; Chu, K. Genetic legacy of tertiary climatic change: A case study of two freshwater loaches, Schistura fasciolata and Pseudogastromyzon myersi, in Hong Kong. Heredity 2017, 119, 360–370. [Google Scholar] [CrossRef] [PubMed]
- Perea, S.; Doadrio, I. Phylogeography, historical demography and habitat suitability modelling of freshwater fishes inhabiting seasonally fluctuating Mediterranean river systems: A case study using the Iberian cyprinid Squalius valentinus. Mol. Ecol. 2015, 24, 3706–3722. [Google Scholar] [CrossRef]
- Swartz, E.R.; Skelton, P.H.; Bloomer, P. Sea-level changes, river capture and the evolution of populations of the Eastern Cape and fiery redfins (Pseudobarbus afer and Pseudobarbus phlegethon, Cyprinidae) across multiple river systems in South Africa. J Biogeogr. 2007, 34, 2086–2099. [Google Scholar] [CrossRef]
- Dolby, G.A.; Ellingson, R.A.; Findley, L.T.; Jacobs, D.K. How sea level change mediates genetic divergence in coastal species across regions with varying tectonic and sediment processes. Mol. Ecol. 2018, 27, 994–1011. [Google Scholar] [CrossRef]
- Chen, W.; Li, C.; Chen, F.; Li, Y.; Yang, J.; Li, J.; Li, X. Phylogeographic analyses of a migratory freshwater fish (Megalobrama terminalis) reveal a shallow genetic structure and pronounced effects of sea-level changes. Gene 2020, 737, 144478. [Google Scholar] [CrossRef]
- Xiang, D.; Li, Y.; Li, X.; Chen, W.; Ma, X. Population structure and genetic diversity of Culter recurviceps revealed by multi-loci. Biodivers. Sci. 2021, 29, 1505–1512. [Google Scholar] [CrossRef]
- Yang, J.Q.; Hsu, K.C.; Liu, Z.Z.; Su, L.W.; Kuo, P.H.; Tang, W.Q.; Zhou, Z.C.; Liu, D.; Bao, B.L.; Lin, H.D. The population history of Garra orientalis (Teleostei: Cyprinidae) using mitochondrial DNA and microsatellite data with approximate Bayesian computation. BMC Evol. Biol. 2016, 16, 1–15. [Google Scholar] [CrossRef]
- Wu, T.H.; Tsang, L.M.; Chen, I.-S.; Chu, K.H. Multilocus approach reveals cryptic lineages in the goby Rhinogobius duospilus in Hong Kong streams: Role of paleodrainage systems in shaping marked population differentiation in a city. Mol. Phylogenet. Evol. 2016, 104, 112–122. [Google Scholar] [CrossRef]
- Wang, P.; Li, Q. The South China Sea: Paleoceanography and Sedimentology; Springer: Dordrecht, The Netherlands, 2009. [Google Scholar]
- Zong, Y.; Yim, W.S.; Yu, F.; Huang, G. Late Quaternary environmental changes in the Pearl River mouth region, China. Quatern. Int. 2009, 206, 35–45. [Google Scholar] [CrossRef]
- Qiu, C.; Lin, Y.; Qing, N.; Zhao, J.; Chen, X. Genetic variation and phylogeography of Micronoemacheilus pulcher populations among drainage systems between western South China and Hainan Island. Acta Entomol. Sin. 2008, 51, 1099–1128. [Google Scholar]
- Luo, Y.; Chen, Y. Culterinae. Fauna Sinica, Osteichthyes, Cypriniformes II; Science Press: Beijing, China, 1998. [Google Scholar]
- Zheng, C. Ichthyography of the Pearl River; Science Press: Beijing, China, 1989. [Google Scholar]
- Reid, A.J.; Carlson, A.K.; Creed, I.F.; Eliason, E.J.; Gell, P.A.; Johnson, P.T.; Kidd, K.A.; MacCormack, T.J.; Olden, J.D.; Ormerod, S.J. Emerging threats and persistent conservation challenges for freshwater biodiversity. Biol. Rev. 2019, 94, 849–873. [Google Scholar] [CrossRef]
- Castello, L.; Macedo, M.N. Large-scale degradation of Amazonian freshwater ecosystems. Glob. Change Biol. 2016, 22, 990–1007. [Google Scholar] [CrossRef]
- Radinger, J.; Britton, J.R.; Carlson, S.M.; Magurran, A.E.; Alcaraz-Hernández, J.D.; Almodóvar, A.; Benejam, L.; Fernández-Delgado, C.; Nicola, G.G.; Oliva-Paterna, F.E. Effective monitoring of freshwater fish. Fish Fish. 2019, 20, 729–747. [Google Scholar] [CrossRef]
- Xiao, W.; Zhang, Y.; Liu, H. Molecular systematics of Xenocyprinae (Teleostei: Cyprinidae): Taxonomy, biogeography, and coevolution of a special group restricted in East Asia. Mol. Phylogenet. Evol. 2001, 18, 163–173. [Google Scholar] [CrossRef]
- Liu, H.; Chen, Y. Phylogeny of the East Asian cyprinids inferred from sequences of the mitochondrial DNA control region. Can. J. Zool. 2003, 81, 1938–1946. [Google Scholar] [CrossRef][Green Version]
- Lovejoy, N.R.; Collette, B.B. Phylogenetic relationships of new world needlefishes (Teleostei: Belonidae) and the biogeography of transitions between marine and freshwater habitats. Copeia 2001, 2001, 324–338. [Google Scholar] [CrossRef]
- Edgar, R.C. MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004, 32, 1792–1797. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef]
- Librado, P.; Rozas, J. DnaSP v5: A software for comprehensive analysis of DNA polymorphism data. Bioinformatics 2009, 25, 1451–1452. [Google Scholar] [CrossRef] [PubMed]
- Leigh, J.W.; Bryant, D. popart: Full-feature software for haplotype network construction. Methods Ecol. Evol. 2015, 6, 1110–1116. [Google Scholar] [CrossRef]
- Woerner, A.E.; Cox, M.P.; Hammer, M.F. Recombination-filtered genomic datasets by information maximization. Bioinformatics 2007, 23, 1851–1853. [Google Scholar] [CrossRef] [PubMed]
- Weir BS, C.C. Estimating F-statistics for the analysis of population structure. Evolution 1984, 38, 1358–1370. [Google Scholar] [PubMed]
- Excoffier, L.; Lischer, H.E.L. Arlequin suite ver 3.5: A new series of programs to perform population genetics analyses under Linux and Windows. Mol. Ecol. Resour. 2010, 10, 564–567. [Google Scholar] [CrossRef] [PubMed]
- Kimura, M. A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J. Mol. Evol. 1980, 16, 111–120. [Google Scholar] [CrossRef]
- Tajima, F. Statistical method for testing the neutral mutation hypothesis by DNA polymorphism. Genetics 1989, 123, 585–595. [Google Scholar] [CrossRef]
- Fu, Y.X. Statistical tests of neutrality of mutations against population growth, hitchhiking and background selection. Genetics 1997, 147, 915–925. [Google Scholar] [CrossRef]
- Schneider, S.; Excoffier, L. Estimation of past demographic parameters from the distribution of pairwise differences when the mutation rates vary among sites: Application to human mitochondrial DNA. Genetics 1999, 152, 1079–1089. [Google Scholar] [CrossRef]
- Drummond, A.J.; Rambaut, A.; Shapiro, B.; Pybus, O.G. Bayesian coalescent inference of past population dynamics from molecular sequences. Mol. Biol. Evol. 2005, 22, 1185–1192. [Google Scholar] [CrossRef]
- Drummond, A.J.; Rambaut, A. BEAST: Bayesian evolutionary analysis by sampling trees. BMC Evol. Biol. 2007, 7, 1–8. [Google Scholar] [CrossRef]
- Nylander, J. MrModeltest v2. Program Distributed by the Author; Evolutionary Biology Centre, Uppsala University: Uppsala, Sweden, 2004. [Google Scholar]
- Meyer, A. Evolution of Mitochondrial DNA in Fishes: Biochemistry and Molecular Biology of Fishes; Hochachka, P.W., Mommsen, T.P., Eds.; Elsevier Press: Hague, Netherlands, 1993; Volume 2. [Google Scholar]
- Durand, J.D.; Tsigenopoulos, C.S.; Unlu, E.; Berrebi, P. Phylogeny and biogeography of the family Cyprinidae in the Middle East inferred from cytochrome b DNA-Evolutionary significance of this region. Mol. Phylogenet. Evol. 2002, 25, 218. [Google Scholar] [CrossRef]
- Ketmaier, V.; Bianco, P.G.; Cobollia, M.; Krivokapic, M.; Caniglia, R.; de Matthaeis, E. Molecular phylogeny of two lineages of Leuciscinae cyprinids (Telestes and Scardinius) from the peri-Mediterranean area based on cytochrome b data. Mol. Phylogenet. Evol. 2004, 32, 1061–1071. [Google Scholar] [CrossRef]
- Yu, D.; Chen, M.; Tang, Q.; Li, X.; Liu, H. Geological events and Pliocene climate fluctuations explain the phylogeographical pattern of the cold water fish Rhynchocypris oxycephalus (Cypriniformes: Cyprinidae) in China. BMC Evol. Biol. 2014, 14, 1–12. [Google Scholar] [CrossRef]
- Tracer v1.4. Available online: http://beast.bio.ed.ac.uk/Tracer (accessed on 8 August 2022).
- Beerli, P. How to Use MIGRATE or Why Are Markov Chain Montecarlo Programs Difficult to Use? Cambridge University Press: Cambridge, UK, 2009. [Google Scholar]
- Gascoyne, M.; Benjamin, G.J.; Schwarcz, H.P.; Ford, D.C. Sea-level lowering during the illinoian glaciation: Evidence from a bahama “blue hole”. Science 1979, 205, 806–808. [Google Scholar] [CrossRef]
- Liu, Z.; Trentesaux, A.; Clemens, S.C.; Colin, C.; Wang, P.; Huang, B.; Boulay, S. Clay mineral assemblages in the northern South China Sea: Implications for East Asian monsoon evolution over the past 2 million years. Mar. Geol. 2003, 201, 133–146. [Google Scholar] [CrossRef]
- Chen, X.L.; Chiang, T.Y.; Lin, H.D.; Zheng, H.S.; Shao, K.T.; Zhang, Q.; Hsu, K.C. Mitochondrial DNA phylogeography of Glyptothorax fokiensis and Glyptothorax hainanensis in Asia. J. Fish Biol. 2007, 70, 75–93. [Google Scholar] [CrossRef]
- Lin, H.D.; Kuo, P.H.; Wang, W.K.; Chiu, Y.W.; Ju, Y.M.; Lin, F.J.; Hsu, K.C. Speciation and differentiation of the genus Opsariichthys (Teleostei: Cyprinidae) in East Asia. Biochem. Syst. Ecol. 2016, 68, 92–100. [Google Scholar] [CrossRef]
- Chen, W.; Hubert, N.; Li, Y.; Xiang, D.; Cai, X.; Zhu, S.; Yang, J.; Zhou, C.; Li, X.; Li, J. Large-scale DNA barcoding of the subfamily Culterinae (Cypriniformes: Xenocyprididae) in East Asia unveils a geographical scale effect, taxonomic warnings and cryptic diversity. Mol. Ecol. 2022, 31, 3871–3887. [Google Scholar] [CrossRef]
- Wang, J.; Li, C.; Chen, J.; Wang, J.; Jin, J.; Jiang, S.; Yan, L.; Lin, H.D.; Zhao, J. Phylogeographic structure of the dwarf snakehead (Channa gachua) around Gulf of Tonkin: Historical biogeography and pronounced effects of sea-level changes. Ecol. Evol. 2021, 11, 12583–12595. [Google Scholar] [CrossRef]
- Zhou, J.; Zhang, C. Freshwater Fishes of Guangxi, China; Guangxi People’s Publishing House: Nanning, China, 2005. [Google Scholar]
- Araripe, J.; do Rêgo, P.S.; Queiroz, H.; Sampaio, I.; Schneider, H. Dispersal capacity and genetic structure of Arapaima gigas on different geographic scales using microsatellite markers. PLoS ONE 2013, 8, e54470. [Google Scholar] [CrossRef] [PubMed]
- Tzeng, C.S.; Lin, Y.S.; Lin, S.M.; Wang, T.Y.; Wang, F.Y. The phylogeography and population demographics of selected freshwater fishes in Taiwan. Zool. Stud. 2006, 45, 285–297. [Google Scholar]
- Chen, W.; Zhong, Z.; Dai, W.; Fan, Q.; He, S. Phylogeographic structure, cryptic speciation and demographic history of the sharpbelly (Hemiculter leucisculus), a freshwater habitat generalist from southern China. BMC Evol. Biol. 2017, 17, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.; Li, C.; Chen, W.; Li, Y.; Li, X. Genetic diversity and population demographic history of Ochetobius elongatus in the middle and lower reaches of the Xijiang River. Biodivers. Sci. 2018, 26, 1289–1295. [Google Scholar] [CrossRef]
- Grant, W.S.; Bowen, B.W. Shallow population histories in deep evolutionary lineages of marine fishes: Insights from sardines and anchovies and lessons for conservation. J. Hered. 1998, 89, 415–426. [Google Scholar] [CrossRef]
- Bernard, A.M.; Feldheim, K.A.; Heithaus, M.R.; Wintner, S.P.; Wetherbee, B.M.; Shivji, M.S. Global population genetic dynamics of a highly migratory, apex predator shark. Mol. Ecol. 2016, 25, 5312–5329. [Google Scholar] [CrossRef]
- Huey, J.A.; Cook, B.D.; Unmack, P.J.; Hughes, J.M. Broadscale phylogeographic structure of five freshwater fishes across the Australian Monsoonal Tropics. Freshw. Sci. 2013, 33, 273–287. [Google Scholar] [CrossRef]
- Frankham, R. Effective population size/adult population size ratios in wildlife: A review. Genet. Res. 1995, 66, 95–107. [Google Scholar] [CrossRef]
- Moritz, C. Uses of molecular phylogenies for conservation. Phil. Trans. R. Soc. B 1995, 349, 113–118. [Google Scholar]
| Locations | Codes | River | MCR | RAG2 | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| n | H | Hd | π | n | H | Hd | π | |||
| Baise | BA | Pearl R. | 28 | 3 | 0.611 ± 0.060 | 0.0029 ± 0.0003 | 56 | 20 | 0.933 ± 0.015 | 0.0027 ± 0.0002 |
| Duan | DA | Pearl R. | 2 | 1 | — | — | 2 | 1 | — | — |
| Fushui | FS | Pearl R. | 22 | 2 | 0.091 ± 0.081 | 0.0001 ± 0.0001 | 44 | 9 | 0.796 ± 0.049 | 0.0023 ± 0.0001 |
| Guiping | GP | Pearl R. | 26 | 9 | 0.868 ± 0.040 | 0.0035 ± 0.0005 | 52 | 15 | 0.880 ± 0.033 | 0.0027 ± 0.0002 |
| Guzhu | GZ | Pearl R. | 33 | 8 | 0.782 ± 0.045 | 0.0056 ± 0.0003 | 66 | 7 | 0.304 ± 0.073 | 0.0005 ± 0.0001 |
| Hechi | HC | Pearl R. | 11 | 1 | 0 | 0 | 22 | 9 | 0.896 ± 0.034 | 0.0018 ± 0.0001 |
| Hengxian | HX | Pearl R. | 6 | 4 | 0.867 ± 0.129 | 0.0041 ± 0.0008 | 12 | 5 | 0.848 ± 0.067 | 0.0032 ± 0.0003 |
| Liucheng | LC | Pearl R. | 19 | 8 | 0.830 ± 0.066 | 0.0040 ± 0.0004 | 38 | 13 | 0.902 ± 0.022 | 0.0024 ± 0.0001 |
| Lixi | LX | Pearl R. | 28 | 13 | 0.905 ± 0.034 | 0.0013 ± 0.0002 | 48 | 20 | 0.855 ± 0.043 | 0.0023 ± 0.0003 |
| Longzhou | LZ | Pearl R. | 26 | 18 | 0.954 ± 0.027 | 0.0059 ± 0.0011 | 52 | 21 | 0.956 ± 0.013 | 0.0029 ± 0.0002 |
| Luzhai | LU | Pearl R. | 11 | 8 | 0.927 ± 0.066 | 0.0047 ± 0.0004 | 22 | 13 | 0.952 ± 0.024 | 0.0026 ± 0.0002 |
| Nanning | NN | Pearl R. | 7 | 7 | 1.0 ± 0.076 | 0.0028 ± 0.0005 | 14 | 6 | 0.879 ± 0.052 | 0.0034 ± 0.0004 |
| Ningming | NM | Pearl R. | 17 | 4 | 0.331 ± 0.143 | 0.0017 ± 0.0007 | 30 | 9 | 0.703 ± 0.085 | 0.0015 ± 0.0002 |
| Rongan | RA | Pearl R. | 28 | 2 | 0.071 ± 0.065 | 0.0003 ± 0.0003 | 56 | 14 | 0.647 ± 0.073 | 0.0018 ± 0.0003 |
| Xindu | XD | Pearl R. | 1 | 1 | — | — | 2 | 2 | — | — |
| Yizhou | YZ | Pearl R. | 17 | 2 | 0.309 ± 0.122 | 0.0021 ± 0.0009 | 34 | 7 | 0.793 ± 0.040 | 0.0018 ± 0.0002 |
| Zhaoqing | ZQ | Pearl R. | 30 | 22 | 0.972 ± 0.017 | 0.0033 ± 0.0004 | 50 | 15 | 0.788 ± 0.054 | 0.0020 ± 0.0003 |
| Baisha | BS | Nandu R. | 30 | 9 | 0.632 ± 0.096 | 0.0010 ± 0.0002 | 60 | 15 | 0.811 ± 0.035 | 0.0014 ± 0.0001 |
| Nanfeng | NF | Nandu R. | 20 | 2 | 0.442 ± 0.087 | 0.0009 ± 0.0002 | 40 | 12 | 0.897 ± 0.025 | 0.0023 ± 0.0001 |
| Ledong | LD | Changhua R. | 27 | 8 | 0.835 ± 0.043 | 0.0010 ± 0.0010 | 54 | 24 | 0.942 ± 0.016 | 0.0024 ± 0.0002 |
| Lingao | LG | Wenlan R. | 20 | 7 | 0.521 ± 0.135 | 0.0005 ± 0.0002 | 30 | 9 | 0.821 ± 0.048 | 0.0023 ± 0.0004 |
| Qiongzhong | QZ | Wanquan R. | 28 | 9 | 0.873 ± 0.037 | 0.0011 ± 0.0001 | 56 | 23 | 0.944 ± 0.015 | 0.0025 ± 0.0002 |
| Wenjiao | WJ | Wenjiao R. | 9 | 4 | 0.583 ± 0.183 | 0.0007 ± 0.0003 | 14 | 7 | 0.890 ± 0.055 | 0.0021 ± 0.0002 |
| Huazhou | HZ | Jian R. | 23 | 6 | 0.700 ± 0.088 | 0.0008 ± 0.0002 | 44 | 14 | 0.895 ± 0.03- | 0.0018 ± 0.0001 |
| Lianjiang | LJ | Lian R. | 28 | 4 | 0.632 ± 0.067 | 0.0004 ± 0.0001 | 60 | 30 | 0.953 ± 0.014 | 0.0047 ± 0.0008 |
| Yangchun | YC | Moyang R. | 30 | 6 | 0.611 ± 0.088 | 0.0013 ± 0.0003 | 60 | 1 | 0 | 0 |
| G1 | — | 312 | 71 | 0.895 ± 0.013 | 0.0044 ± 0.0002 | 600 | 91 | 0.873 ± 0.012 | 0.0027 ± 0.0001 | |
| G2 | — | 134 | 21 | 0.835 ± 0.020 | 0.0010 ± 0.0001 | 254 | 56 | 0.912 ± 0.011 | 0.0023 ± 0.0001 | |
| G3 | — | 51 | 8 | 0.665 ± 0.058 | 0.0006 ± 0.0001 | 104 | 38 | 0.935 ± 0.014 | 0.0036 ± 0.0005 | |
| G4 | — | 30 | 6 | 0.611 ± 0.088 | 0.0013 ± 0.0003 | 60 | 1 | 0 | 0 | |
| Global | — | 527 | 106 | 0.948 ± 0.0049 | 0.0044 ± 0.0001 | 1018 | 168 | 0.915 ± 0.0064 | 0.0036 ± 0.0001 | |
| Source of Variation | MCR | RAG2 | ||||
|---|---|---|---|---|---|---|
| Percentage of Variation | Φ-Statistic | p-Value | Percentage of Variation | Φ-Statistic | p-Value | |
| Grouped by locations | ||||||
| Between populations | 53.94 | 0.539 | <0.001 | 41.44 | 0.414 | <0.001 |
| Within populations | 46.06 | 0.461 | 58.56 | 0.586 | ||
| Grouped by nine rivers | ||||||
| Between rivers | 25.26 | 0.253 | <0.001 | 32.15 | 0.322 | <0.001 |
| Between populations within rivers | 32.65 | 0.327 | <0.001 | 15.69 | 0.157 | <0.001 |
| Within populations | 42.09 | 0.421 | <0.001 | 52.16 | 0.522 | <0.001 |
| Grouped by genetic groups | ||||||
| Between genetic groups | 37.98 | 0.379 | <0.001 | 37.74 | 0.377 | <0.001 |
| Between populations within genetic groups | 22.91 | 0.229 | <0.001 | 12.40 | 0.124 | <0.001 |
| Within populations | 39.11 | 0.391 | <0.001 | 49.86 | 0.499 | <0.001 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chen, W.; Li, Y.; Cai, X.; Xiang, D.; Gao, S.; Li, C.; Lan, C.; Zhu, S.; Yang, J.; Li, X.; et al. Genetic Structure of an East Asian Minnow (Toxabramis houdemeri) in Southern China, with Implications for Conservation. Biology 2022, 11, 1641. https://doi.org/10.3390/biology11111641
Chen W, Li Y, Cai X, Xiang D, Gao S, Li C, Lan C, Zhu S, Yang J, Li X, et al. Genetic Structure of an East Asian Minnow (Toxabramis houdemeri) in Southern China, with Implications for Conservation. Biology. 2022; 11(11):1641. https://doi.org/10.3390/biology11111641
Chicago/Turabian StyleChen, Weitao, Yuefei Li, Xingwei Cai, Denggao Xiang, Shang Gao, Ce Li, Chun Lan, Shuli Zhu, Jiping Yang, Xinhui Li, and et al. 2022. "Genetic Structure of an East Asian Minnow (Toxabramis houdemeri) in Southern China, with Implications for Conservation" Biology 11, no. 11: 1641. https://doi.org/10.3390/biology11111641
APA StyleChen, W., Li, Y., Cai, X., Xiang, D., Gao, S., Li, C., Lan, C., Zhu, S., Yang, J., Li, X., & Li, J. (2022). Genetic Structure of an East Asian Minnow (Toxabramis houdemeri) in Southern China, with Implications for Conservation. Biology, 11(11), 1641. https://doi.org/10.3390/biology11111641





