Isotopic Niche Analysis of Long-Finned Pilot Whales (Globicephala melas edwardii) in Aotearoa New Zealand Waters
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
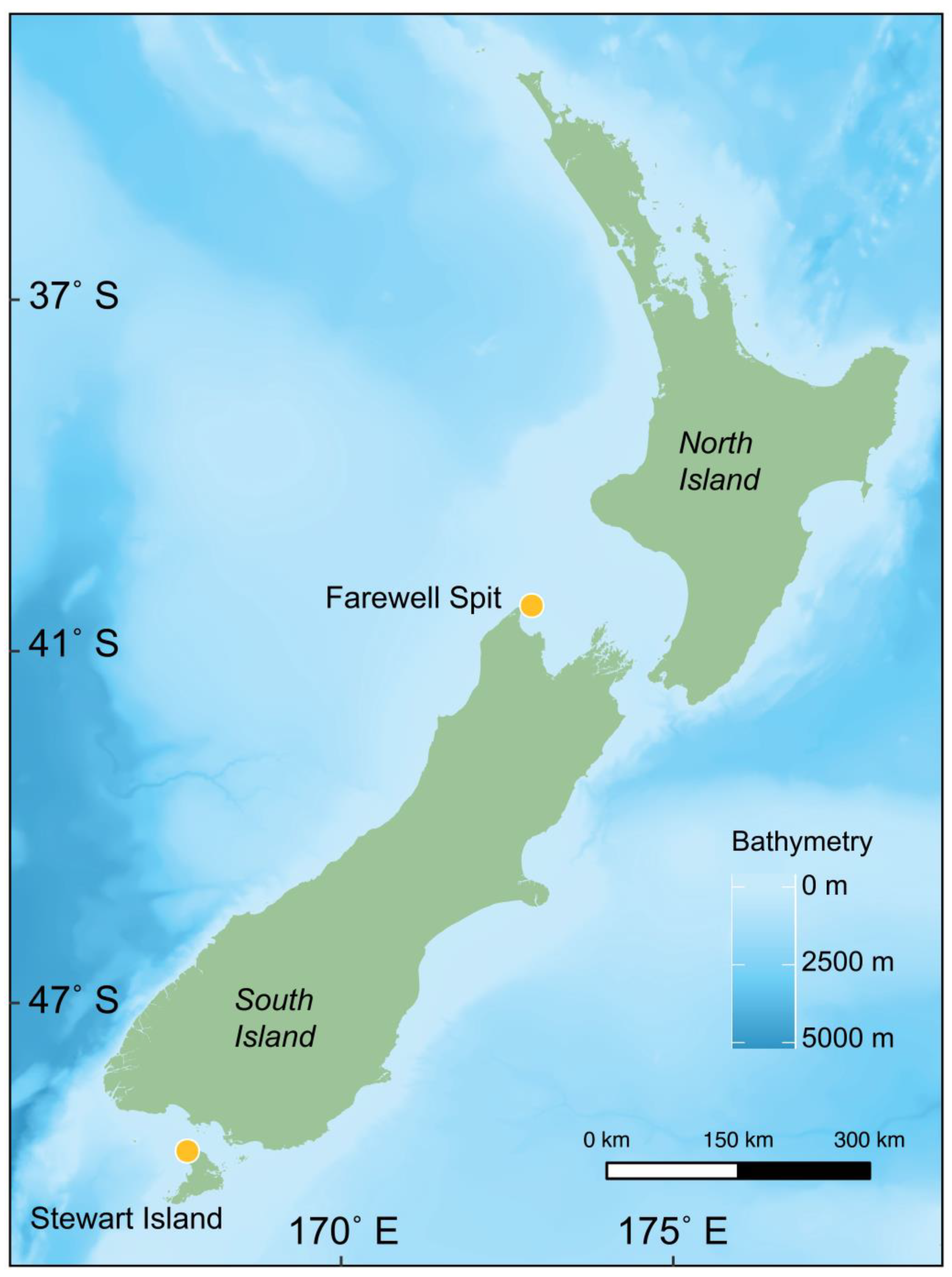
2.1. Sampling
2.2. Sample Preparation
2.3. Carbon and Nitrogen Isotope Analysis
2.4. Sulphur Analysis
2.5. Correction Equations
2.6. Statistical Analysis
3. Results
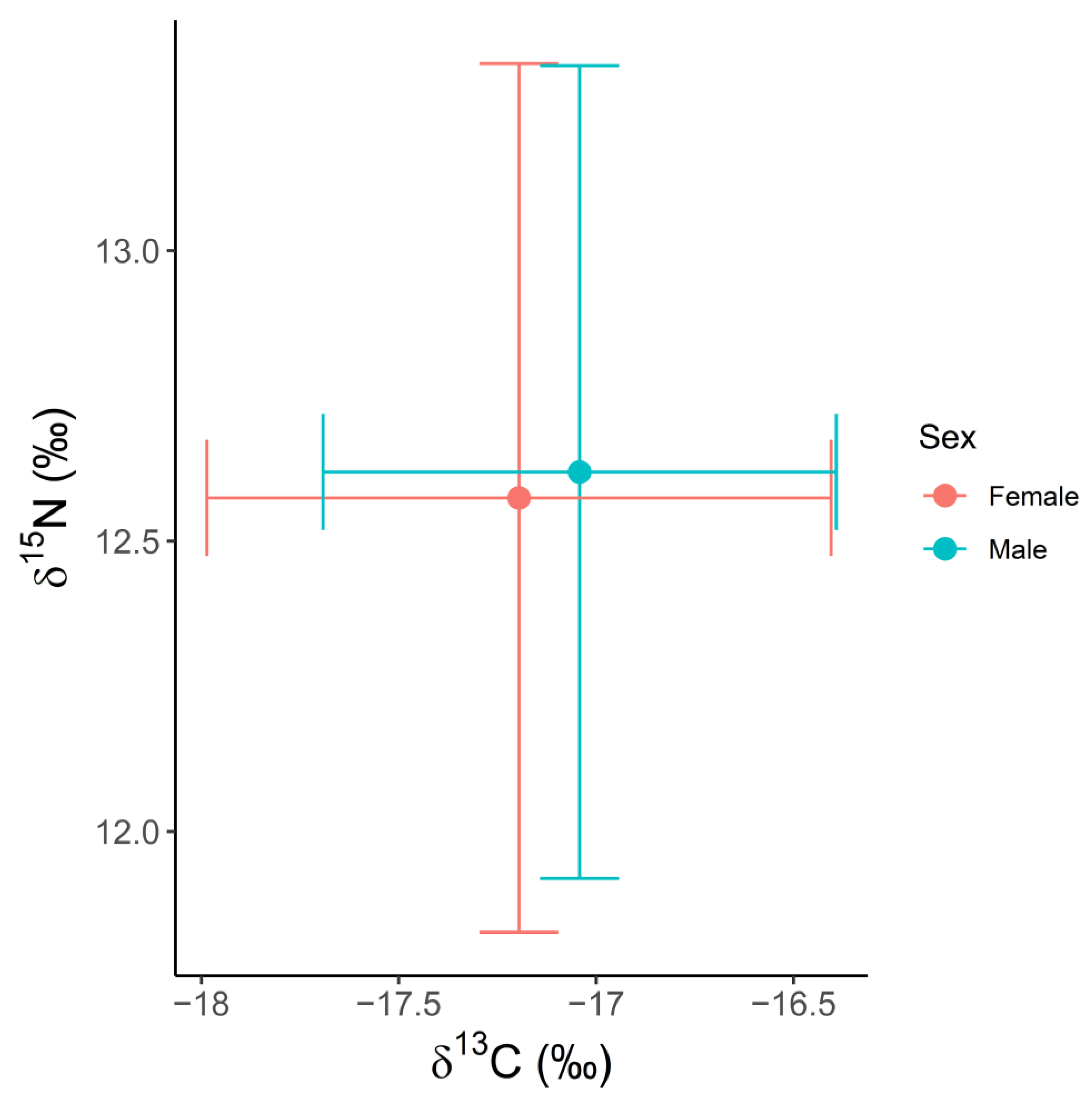
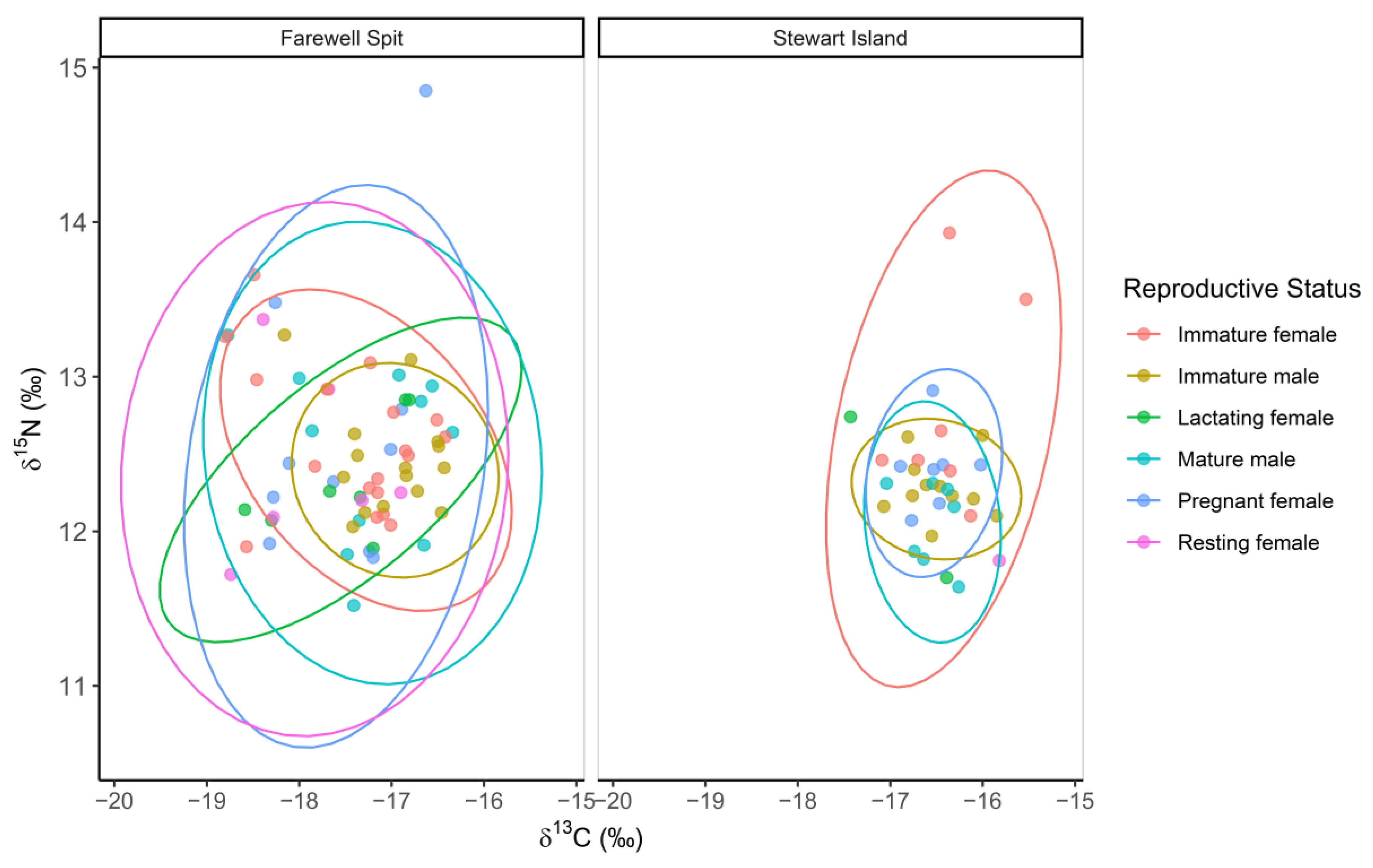
3.1. Ontogenetic Variation in δ13C, δ15N and δ34S Values
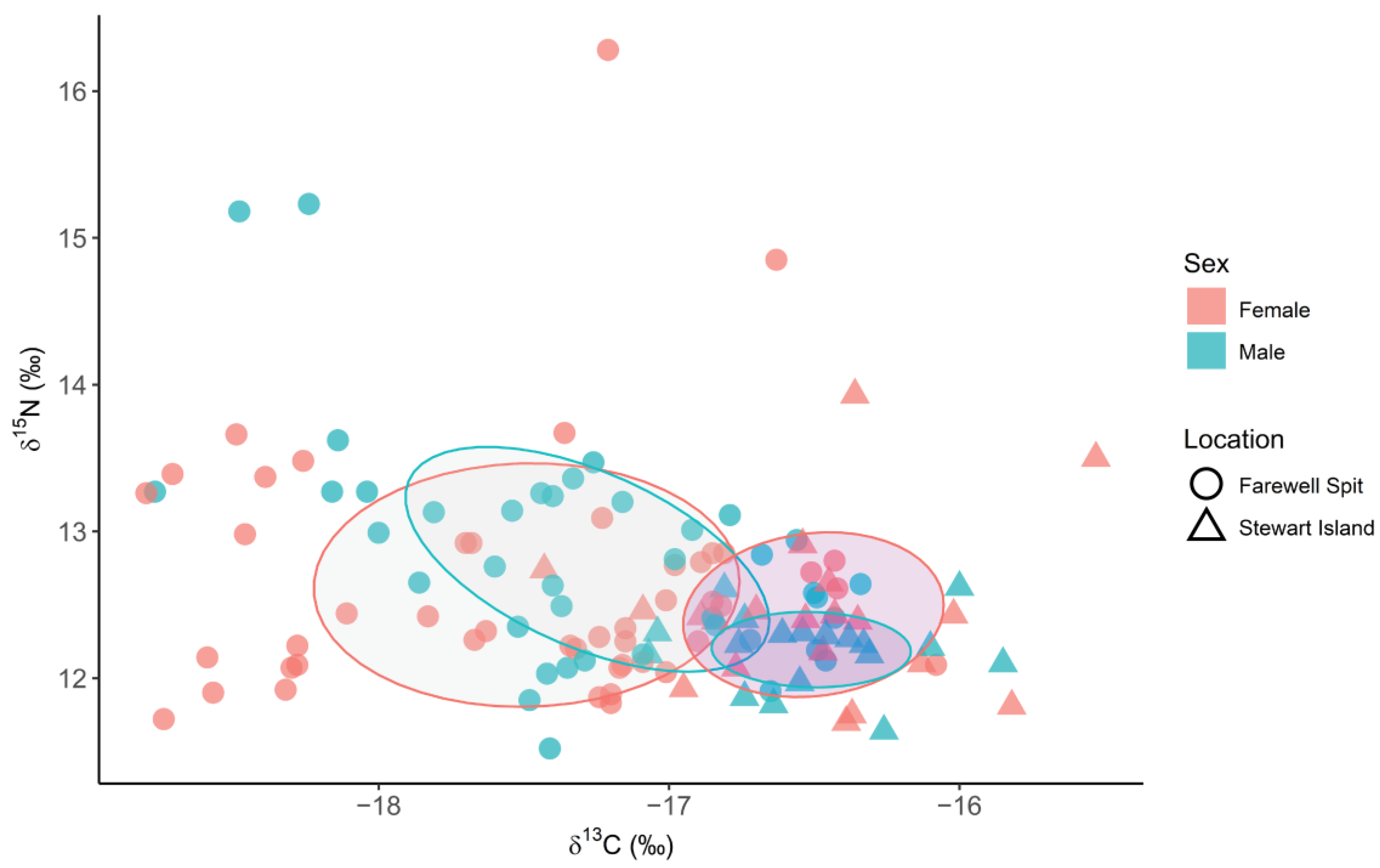
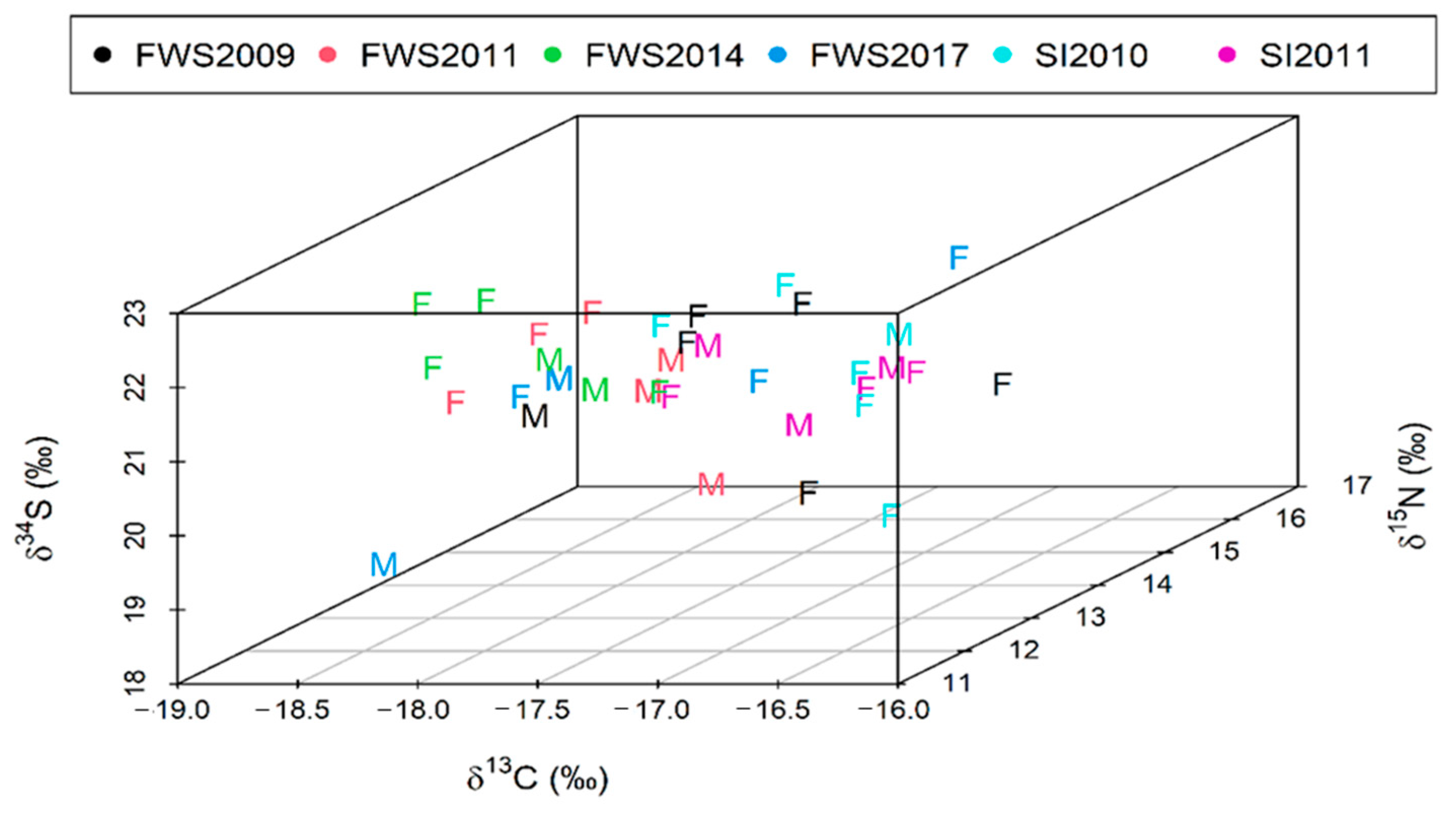
3.2. Spatial and Temporal Variation in δ13C, δ15N and δ34S Values
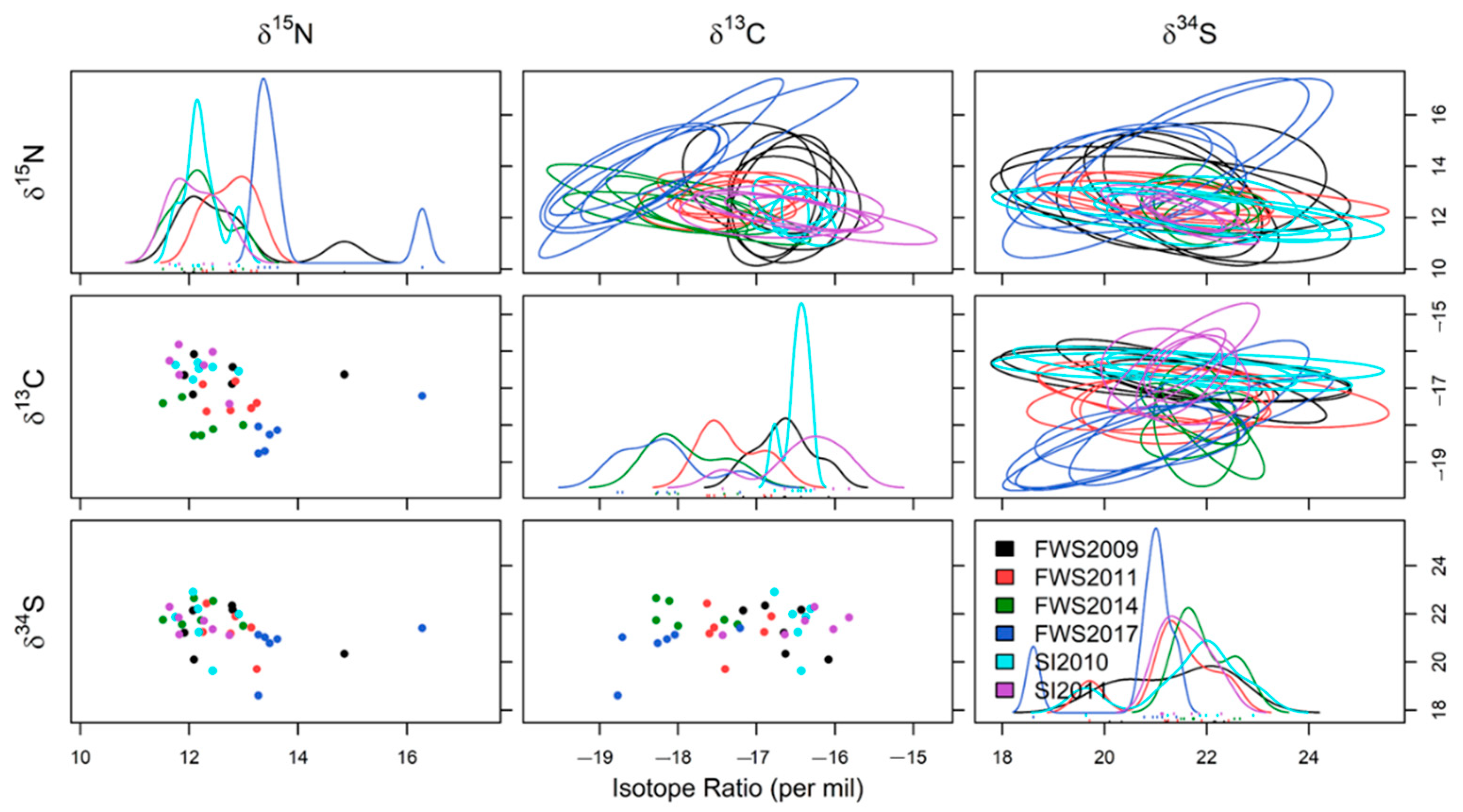
3.3. Triple Isotope Niche Regions
4. Discussion
4.1. Ontogenetic Variation in Isotope Values
4.2. Spatial and Temporal Variation in Stable Isotope Values
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Newsome, S.D.; Clementz, M.T.; Koch, P.L. Using stable isotope biogeochemistry to study marine mammal ecology. Mar. Mammal Sci. 2010, 26, 509–572. [Google Scholar] [CrossRef]
- Bearhop, S.; Adams, C.E.; Waldron, S.; Fuller, R.A.; Macleod, H. Determining trophic niche width: A novel approach using stable isotope analysis. J. Anim. Ecol. 2004, 73, 1007–1012. [Google Scholar] [CrossRef]
- Crawford, K.; McDonald, R.A.; Bearhop, S. Applications of stable isotope techniques to the ecology of mammals. Mammal Rev. 2008, 38, 87–107. [Google Scholar]
- Boecklen, W.J.; Yarnes, C.T.; Cook, B.A.; James, A.C. On the use of stable isotopes in trophic ecology. Annu. Rev. Ecol. Evol. Syst. 2011, 42, 411–440. [Google Scholar] [CrossRef]
- Gillespie, J.H. Application of stable isotope analysis to study temporal changes in foraging ecology in a highly endangered amphibian. PLoS ONE 2013, 8, e53041. [Google Scholar] [CrossRef]
- Jackson, M.C.; Britton, J.R. Divergence in the trophic niche of sympatric freshwater invaders. Biol. Invasions 2014, 16, 1095–1103. [Google Scholar] [CrossRef]
- Navarro, J.; Coll, M.; Somes, C.J.; Olson, R.J. Trophic niche of squids: Insights from isotopic data in marine systems worldwide. Deep. Sea Res. Part II: Top. Stud. Oceanogr. 2013, 95, 93–102. [Google Scholar] [CrossRef]
- Newsome, S.D.; Martinez del Rio, C.; Bearhop, S.; Phillips, D.L. A niche for isotopic ecology. Front. Ecol. Environ. 2007, 5, 429–436. [Google Scholar] [CrossRef]
- Mendes, S.; Newton, J.; Reid, R.J.; Zuur, A.F.; Pierce, G.J. Stable carbon and nitrogen isotope ratio profiling of sperm whale teeth reveals ontogenetic movements and trophic ecology. Oecologia 2007, 151, 605–615. [Google Scholar] [CrossRef]
- Borrell, A.; Gazo, M.; Aguilar, A.; Raga, J.A.; Degollada, E.; Gozalbes, P.; García-Vernet, R. Niche partitioning amongst northwestern Mediterranean cetaceans using stable isotopes. Prog. Oceanogr. 2021, 193, 102559. [Google Scholar] [CrossRef]
- Cherel, Y.; Hobson, K.A. Geographical variation in carbon stable isotope signatures of marine predators: A tool to investigate their foraging areas in the Southern Ocean. Mar. Ecol. Prog. Ser. 2007, 329, 281–287. [Google Scholar] [CrossRef]
- McCutchan, J.H., Jr.; Lewis, W.M., Jr.; Kendall, C.; McGrath, C.C. Variation in trophic shift for stable isotope ratios of carbon, nitrogen, and sulfur. Oikos 2003, 102, 378–390. [Google Scholar] [CrossRef]
- Matthews, C.J.D.; Ferguson, S.H. Spatial segregation and similar trophic-level diet among eastern Canadian Arctic/north-west Atlantic killer whales inferred from bulk and compound specific isotopic analysis. J. Mar. Biol. Assoc. U. K. 2014, 94, 1343–1355. [Google Scholar] [CrossRef]
- Balasse, M.; Tresset, A.; Dobney, K.; Ambrose, S.H. The use of isotope ratios to test for seaweed eating in sheep. J. Zool. 2005, 266, 283–291. [Google Scholar] [CrossRef]
- Crowley, B.E.; Slater, P.A.; Arrigo-Nelson, S.J.; Baden, A.L.; Karpanty, S.M. Strontium isotopes are consistent with low-elevation foraging limits for Henst’s goshawk. Wildl. Soc. Bull. 2017, 41, 743–751. [Google Scholar] [CrossRef]
- Hobson, K.A. Stable isotope analysis of marbled murrelets: Evidence for freshwater feeding and determination of trophic level. Condor 1990, 92, 897–903. [Google Scholar] [CrossRef]
- Kiszka, J.; Simon-Bouhet, B.; Martinez, L.; Pusineri, C.; Richard, P.; Ridoux, V. Ecological niche segregation within a community of sympatric dolphins around a tropical island. Mar. Ecol. Prog. Ser. 2011, 433, 273–288. [Google Scholar] [CrossRef]
- Oelbermann, K.; Scheu, S. Stable isotope enrichment (δ15N and δ13C) in a generalist predator (Pardosa lugubris, Araneae: Lycosidae): Effects of prey quality. Oecologia 2002, 130, 337–344. [Google Scholar] [CrossRef]
- Peterson, B.J.; Fry, B. Stable isotopes in ecosystem studies. Annu. Rev. Ecol. Syst. 1987, 18, 293–320. [Google Scholar] [CrossRef]
- Connolly, R.; Guest, M.; Melville, A.; Oakes, J. Sulfur stable isotopes separate producers in marine food-web analysis. Oecologia 2004, 138, 161–167. [Google Scholar] [CrossRef]
- Telsnig, J.I.D.; Jennings, S.; Mill, A.C.; Walker, N.D.; Parnell, A.C.; Polunin, N.V.C. Estimating contributions of pelagic and benthic pathways to consumer production in coupled marine food webs. J. Anim. Ecol. 2019, 88, 405–415. [Google Scholar]
- Matthews, C.; Ferguson, S. Seasonal foraging behaviour of Eastern Canada-West Greenland bowhead whales: An assessment of isotopic cycles along baleen. Mar. Ecol. Prog. Ser. 2015, 522, 269–286. [Google Scholar] [CrossRef]
- Wilson, R.M.; Tyson, R.B.; Nelson, J.A.; Balmer, B.C.; Chanton, J.P.; Nowacek, D.P. Niche differentiation and prey selectivity among common bottlenose dolphins (Tursiops truncatus) sighted in St. George Sound, Gulf of Mexico. Front. Mar. Sci. 2017, 4, 235. [Google Scholar] [CrossRef]
- Valenzuela, L.O.; Rowntree, V.J.; Sironi, M.; Seger, J. Stable isotopes (δ15N, δ13C, δ34S) in skin reveal diverse food sources used by Southern right whales Eubalaena australis. Mar. Ecol. Prog. Ser. 2018, 603, 243–255. [Google Scholar]
- Hette-Tronquart, N. Isotopic niche is not equal to trophic niche. Ecol. Lett. 2019, 22, 1987–1989. [Google Scholar] [CrossRef] [PubMed]
- Shipley, O.N.; Matich, P. Studying animal niches using bulk stable isotope ratios: An updated synthesis. Oecologia 2020, 193, 27–51. [Google Scholar] [CrossRef]
- Jackson, A.; Inger, R.; Parnell, A.C.; Bearhop, S. Comparing isotopic niche widths among and within communities: SIBER—Stable Isotope Bayesian Ellipses in R. J. Anim. Ecol. 2011, 80, 595–602. [Google Scholar] [CrossRef]
- Marshall, H.H.; Inger, R.; Jackson, A.L.; McDonald, R.A.; Thompson, F.J.; Cant, M.A. Stable isotopes are quantitative indicators of trophic niche. Ecol. Lett. 2019, 22, 1990–1992. [Google Scholar] [CrossRef]
- Gause, G.F. The Struggle for Existence: A Classic of Mathematical Biology and Ecology; Dover Publications, Inc.: Mineola, NY, USA, 2019. [Google Scholar]
- Hutchinson, G.E. A Treatise on Limnology; John Wiley and Sons, Inc.: Hoboken, NJ, USA, 1957; Volume 1, p. 243. [Google Scholar]
- Durante, C.A.; Crespo, E.A.; Loizaga, R. Isotopic niche partitioning between two small cetacean species. Mar. Ecol. Prog. Ser. 2021, 659, 247–259. [Google Scholar] [CrossRef]
- Costa, A.F.; Botta, S.; Siciliano, S.; Giarrizzo, T. Resource partitioning among stranded aquatic mammals from Amazon and Northeastern coast of Brazil revealed through carbon and nitrogen stable isotopes. Sci. Rep. 2020, 10, 12897. [Google Scholar] [CrossRef]
- Gibbs, S.E.; Harcourt, R.G.; Kemper, C.M. Niche differentiation of bottlenose dolphin species in South Australia revealed by stable isotopes and stomach contents. Wildl. Res. 2011, 38, 261–270. [Google Scholar] [CrossRef]
- Giménez, J.; Cañadas, A.; Ramírez, F.; Afán, I.; García-Tiscar, S.; Fernández-Maldonado, C.; Castillo, J.J.; de Stephanis, R. Living apart together: Niche partitioning among Alboran Sea cetaceans. Ecol. Indic. 2018, 95, 32–40. [Google Scholar] [CrossRef]
- Praca, E.; Laran, S.; Lepoint, G.; Thomé, J.P.; Quetglas, A.; Belcari, P.; Sartor, P.; Dhermain, F.; Ody, D.; Tapie, N.; et al. Toothed whales in the northwestern Mediterranean: Insight into their feeding ecology using chemical tracers. Mar. Pollut. Bull. 2011, 62, 1058–1065. [Google Scholar] [CrossRef]
- Nicholson, K.; Bejder, L.; Loneragan, N. Niche partitioning among social clusters of a resident estuarine apex predator. Behav. Ecol. Sociobiol. 2021, 75, 160. [Google Scholar] [CrossRef]
- De Stephanis, R.; García-Tíscar, S.; Verborgh, P.; Esteban-Pavo, R.; Pérez, S.; Minvielle-Sebastia, L.; Guinet, C. Diet of the social groups of long-finned pilot whales (Globicephala melas) in the Strait of Gibraltar. Mar. Biol. 2008, 154, 603–612. [Google Scholar] [CrossRef]
- Rossman, S.; Ostrom, P.H.; Stolen, M.; Barros, N.B.; Gandhi, H.; Stricker, C.A.; Wells, R.S. Individual specialization in the foraging habits of female bottlenose dolphins living in a trophically diverse and habitat rich estuary. Oecologia 2015, 178, 415–425. [Google Scholar] [CrossRef] [PubMed]
- Marcoux, M.; McMeans, B.C.; Fisk, A.T.; Ferguson, S.H. Composition and temporal variation in the diet of beluga whales, derived from stable isotopes. Mar. Ecol. Prog. Ser. 2012, 471, 283–291. [Google Scholar] [CrossRef]
- Reisinger, R.R.; Gröcke, D.R.; Lübcker, N.; McClymont, E.L.; Hoelzel, A.R.; de Bruyn, P.J.N. Variation in the diet of killer whales Orcinus orca at Marion Island, Southern Ocean. Mar. Ecol. Prog. Ser. 2016, 549, 263–274. [Google Scholar] [CrossRef]
- Riccialdelli, L.; Goodall, N. Intra-specific trophic variation in false killer whales (Pseudorca crassidens) from the southwestern South Atlantic Ocean through stable isotopes analysis. Mamm. Biol. 2015, 80, 298–302. [Google Scholar] [CrossRef]
- Meissner, A.M.; MacLeod, C.D.; Richard, P.; Ridoux, V.; Pierce, G. Feeding ecology of striped dolphins, Stenella coeruleoalba, in the north-western Mediterranean Sea based on stable isotope analyses. J. Mar. Biol. Assoc. U. K. 2012, 92, 1677–1687. [Google Scholar] [CrossRef]
- Jackson-Ricketts, J.; Ruiz-Cooley, R.I.; Junchompoo, C.; Thongsukdee, S.; Intongkham, A.; Ninwat, S.; Kittiwattanawong, K.; Hines, E.M.; Costa, D.P. Ontogenetic variation in diet and habitat of Irrawaddy dolphins (Orcaella brevirostris) in the Gulf of Thailand and the Andaman Sea. Mar. Mammal Sci. 2019, 35, 492–521. [Google Scholar] [CrossRef]
- Riccialdelli, L.; Newsome, S.D.; Dellabianca, N.A.; Bastida, R.; Fogel, M.L.; Goodall, R.N.P. Ontogenetic diet shift in Commerson’s dolphin (Cephalorhynchus commersonii commersonii) off Tierra del Fuego. Polar Biol. 2013, 36, 617–627. [Google Scholar] [CrossRef]
- Borrell, A.; Abad-Oliva, N.; Gómez-Campos, E.; Giménez, J.; Aguilar, A. Discrimination of stable isotopes in fin whale tissues and application to diet assessment in cetaceans. Rapid Commun. Mass Spectrom. 2012, 26, 1596–1602. [Google Scholar]
- Gannon, D.P.; Waples, D.M. Diets of coastal bottlenose dolphins from the US mid-Atlantic coast differ by habitat. Mar. Mammal Sci. 2004, 20, 527–545. [Google Scholar] [CrossRef]
- Abend, A.G.; Smith, T.D. Differences in ratios of stable isotopes of nitrogen in long-finned pilot whales (Globicephala melas) in the western and eastern North Atlantic. ICES J. Mar. Sci. 1995, 52, 837–841. [Google Scholar] [CrossRef]
- Santos, M.B.; Monteiro, S.S.; Vingada, J.V.; Ferreira, M.; López, A.; Martínez Cedeira, J.A.; Reid, R.J.; Brownlow, A.; Pierce, G.J. Patterns and trends in the diet of long-finned pilot whales (Globicephala melas) in the northeast Atlantic. Mar. Mammal Sci. 2014, 30, 1–19. [Google Scholar] [CrossRef]
- Desportes, G.; Mouritsen, R. Diet of the pilot whale, Globicephala melas, around the Faroe Islands; ICES: Toronto, ON, Canada, 1988; 15p. [Google Scholar]
- Betty, E.L.; Bollard, B.; Murphy, S.; Ogle, M.; Hendriks, H.; Orams, M.B.; Stockin, K.A. Using emerging hot spot analysis of stranding records to inform conservation management of a data-poor cetacean species. Biodivers. Conserv. 2020, 29, 643–665. [Google Scholar] [CrossRef]
- Beatson, E.; O’Shea, S.; Ogle, M. First report on the stomach contents of long-finned pilot whales, Globicephala melas, stranded in New Zealand. N. Z. J. Zool. 2007, 34, 51–56. [Google Scholar] [CrossRef]
- Beatson, E.; O’Shea, S.; Stone, C.; Shortland, T. Notes on New Zealand mammals 6. Second report on the stomach contents of long-finned pilot whales, Globicephala melas. N. Z. J. Zool. 2007, 34, 359–362. [Google Scholar] [CrossRef][Green Version]
- Beatson, E.L.; O’Shea, S. Stomach contents of long-finned pilot whales, Globicephala melas, mass-stranded on Farewell Spit, Golden Bay in 2005 and 2008. N. Z. J. Zool. 2009, 36, 47–58. [Google Scholar] [CrossRef]
- Sekiguchi, K.; Best, P.B. In vitro digestibility of some prey species of dolphins. Fish. Bull. Natl. Ocean. Atmos. Adm. 1997, 95, 386–393. [Google Scholar]
- Hyslop, E.J. Stomach contents analysis—A review of methods and their application. J. Fish Biol. 1980, 17, 411–429. [Google Scholar] [CrossRef]
- Post, D.M. Using stable isotopes to estimate trophic position: Models, methods, and assumptions. Ecology 2002, 83, 703–718. [Google Scholar] [CrossRef]
- CANZ. New Zealand Region Bathymetry, 1:4,000,000, 2nd ed.; NIWA Chart Miscellaneous Series No. 85; National Institute of Water & Atmospheric Research Ltd.: Auckland, New Zealand, 2008. [Google Scholar]
- Geraci, J.R.; Lounsbury, V.J. Marine Mammals Ashore: A Field Guide for Strandings; National Aquarium in Baltimore: Baltimore, MD, USA, 2005. [Google Scholar]
- Betty, E.L.; Stockin, K.A.; Hinton, B.; Bollard, B.A.; Smith, A.N.H.; Orams, M.B.; Murphy, S. Age, growth, and sexual dimorphism of the Southern Hemisphere long-finned pilot whale (Globicephala melas edwardii). J. Mammal. 2022, 103, 560–575. [Google Scholar] [CrossRef]
- Betty, E. Life History of the long-finned pilot whale (Globicephala melas edwardii); Insights from strandings on the New Zealand coast. Ph.D. Thesis, Auckland University of Technology, Auckland, New Zealand, 2019. [Google Scholar]
- Betty, E.L.; Stockin, K.A.; Smith, A.N.H.; Bollard, B.; Orams, M.B.; Murphy, S. Sexual maturation in male long-finned pilot whales (Globicephala melas edwardii): Defining indicators of sexual maturity. J. Mammal. 2019, 100, 1387–1402. [Google Scholar] [CrossRef]
- Olin, J.A.; Poulakis, G.R.; Stevens, P.W.; DeAngelo, J.A.; Fisk, A.T. Preservation effects on stable isotope values of archived elasmobranch fin tissue: Comparisons between frozen and ethanol-stored samples. Trans. Am. Fish. Soc. 2014, 143, 1569–1576. [Google Scholar]
- Wild, L.A.; Chenoweth, E.M.; Mueter, F.J.; Straley, J.M. Evidence for dietary time series in layers of cetacean skin using stable carbon and nitrogen isotope ratios. Rapid Commun. Mass Spectrom. 2018, 32, 1425–1438. [Google Scholar] [CrossRef]
- Paul, D.; Skrzypek, G.; Fórizs, I. Normalization of measured stable isotopic compositions to isotope reference scales—A review. Rapid Commun. Mass Spectrom. 2007, 21, 3006–3014. [Google Scholar]
- Focken, U.; Becker, K. Metabolic fractionation of stable carbon isotopes: Implications of different proximate compositions for studies of the aquatic food webs using δ13C data. Oecologia 1998, 115, 337–343. [Google Scholar] [CrossRef]
- Choy, E.S.; Roth, J.D.; Loseto, L.L. Lipid removal and acidification affect nitrogen and carbon stable isotope ratios of beluga whales (Delphinapterus leucas) and their potential prey species in the Beaufort Sea ecosystem. Mar. Biol. 2016, 163, 220. [Google Scholar] [CrossRef]
- Groß, J.; Fry, B.; Burford, M.A.; Nash, S.B. Assessing the effects of lipid extraction and lipid correction on stable isotope values (δ13C and δ15N) of blubber and skin from southern hemisphere humpback whales. Rapid Commun. Mass Spectrom. 2021, 35, e9140. [Google Scholar] [CrossRef] [PubMed]
- Viola, M.N.P.; Riccialdelli, L.; Negri, M.F.; Panebianco, M.V.; Panarello, H.O.; Cappozzo, H.L. Intra-specific isotope variations of franciscana dolphin Pontoporia blainvillei regarding biological parameters and distinct environment. Mamm. Biol. 2017, 85, 47–54. [Google Scholar] [CrossRef]
- Logan, J.M.; Jardine, T.D.; Miller, T.J.; Bunn, S.E.; Cunjak, R.A.; Lutcavage, M.E. Lipid corrections in carbon and nitrogen stable isotope analyses: Comparison of chemical extraction and modelling methods. J. Anim. Ecol. 2008, 77, 838–846. [Google Scholar] [CrossRef] [PubMed]
- Wilson, R.M.; Chanton, J.P.; Balmer, B.C.; Nowacek, D.P. An evaluation of lipid extraction techniques for interpretation of carbon and nitrogen isotope values in bottlenose dolphin (Tursiops truncatus) skin tissue. Mar. Mammal Sci. 2014, 30, 85–103. [Google Scholar] [CrossRef]
- Giménez, J.; Ramírez, F.; Forero, M.G.; Almunia, J.; de Stephanis, R.; Navarro, J. Lipid effects on isotopic values in bottlenose dolphins (Tursiops truncatus) and their prey with implications for diet assessment. Mar. Biol. 2017, 164, 122. [Google Scholar] [CrossRef]
- Arostegui, M.C.; Schindler, D.E.; Holtgrieve, G.W. Does lipid-correction introduce biases into isotopic mixing models? Implications for diet reconstruction studies. Oecologia 2019, 191, 745–755. [Google Scholar] [CrossRef]
- Fry, B. Stable isotopic indicators of habitat use by Mississippi River fish. J. N. Am. Benthol. Soc. 2002, 21, 676–685. [Google Scholar] [CrossRef]
- Post, D.M.; Layman, C.A.; Arrington, D.A.; Takimoto, G.; Quattrochi, J.; Montana, C.G. Getting to the fat of the matter: Models, methods and assumptions for dealing with lipids in stable isotope analyses. Oecologia 2007, 152, 179–189. [Google Scholar] [CrossRef]
- Peters, K.J.; Bury, S.J.; Hinton, B.; Betty, E.L.; Casano-Bally, D.; Parra, G.J.; Stockin, K.A. Too close for comfort? Isotopic niche segregation in New Zealand’s odontocetes. Biology 2022, 11, 1179. [Google Scholar] [CrossRef]
- Elliott, K.H.; Davis, M.; Elliott, J.E. Equations for Lipid Normalization of Carbon Stable Isotope Ratios in Aquatic Bird Eggs. PLoS ONE 2014, 9, e83597. [Google Scholar] [CrossRef] [PubMed]
- Körtzinger, A.; Quay, P.D.; Sonnerup, R.E. Relationship between anthropogenic CO2 and the 13C Suess effect in the North Atlantic Ocean. Glob. Biogeochem. Cycles 2003, 17, 5-1–5-20. [Google Scholar] [CrossRef]
- Quay, P.; Sonnerup, R.; Westby, T.; Stutsman, J.; McNichol, A. Changes in the 13C/12C of dissolved inorganic carbon in the ocean as a tracer of anthropogenic CO2 uptake. Glob. Biogeochem. Cycles 2003, 17, 4-1–4-20. [Google Scholar] [CrossRef]
- Kassambara, A. Rstatix: Pipe-Friendly Framework for Basic Statistical Tests, Version 0.7.0;; 2020. Available online: https://CRAN.R-project.org/package=rstatix (accessed on 8 March 2022).
- Fernández, R.; García-Tiscar, S.; Begoña Santos, M.; López, A.; Martínez-Cedeira, J.A.; Newton, J.; Pierce, G.J. Stable isotope analysis in two sympatric populations of bottlenose dolphins Tursiops truncatus: Evidence of resource partitioning? Mar. Biol. 2011, 158, 1043–1055. [Google Scholar] [CrossRef]
- Hastie, T.; Tibshirani, R. Generalized Additive Models; Monographs on Statistics & Applied Probability 43; Chapman and Hall/CRC: Boca Raton, FL, USA, 1990; p. 1. [Google Scholar]
- Wood, S.; Wood, M.S. Package ‘mgcv’. R Package Version 2015, 1, 729. [Google Scholar]
- Wood, S.N. Introducing GAMs, in Generalized Additive Models; Chapman and Hall/CRC: Boca Raton, FL, USA, 2017; pp. 161–194. [Google Scholar]
- Burnham, K.P.; Anderson, D.R.; Huyvaert, K.P. AIC model selection and multimodel inference in behavioral ecology: Some background, observations, and comparisons. Behav. Ecol. Sociobiol. 2011, 65, 23–35. [Google Scholar] [CrossRef]
- Ritz, C.; Spiess, A.N. qpcR: An R package for sigmoidal model selection in quantitative real-time polymerase chain reaction analysis. Bioinformatics 2008, 24, 1549–1551. [Google Scholar] [CrossRef]
- Wickham, H. ggplot2. WIREs Comput. Stat. 2011, 3, 180–185. [Google Scholar] [CrossRef]
- Ligges, U.; Maechler, M.; Schnackenberg, S.; Ligges, M.U. Package ‘Scatterplot3d’. 2018. Available online: https://cran.r-project.org/web/packages/scatterplot3d/index.html (accessed on 10 March 2022).
- Lysy, M.; Stasko, A.D.; Swanson, H.K. Package ‘nicheROVER’:(Niche)(R)egion and Niche (Over) Lap Metrics for Multidimensional Ecological Niches (Version 1.0). 2014. Available online: https://cran.r-project.org/web/packages/nicheROVER/index.html (accessed on 11 March 2022).
- Swanson, H.K.; Lysy, M.; Power, M.; Stasko, A.D.; Johnson, J.D.; Reist, J.D. A new probabilistic method for quantifying n-dimensional ecological niches and niche overlap. Ecology 2015, 96, 318–324. [Google Scholar] [CrossRef]
- Graham, H.V.; Patzkowsky, M.E.; Wing, S.L.; Parker, G.G.; Fogel, M.L.; Freeman, K.H. Isotopic characteristics of canopies in simulated leaf assemblages. Geochim. Cosmochim. Acta 2014, 144, 82–95. [Google Scholar] [CrossRef]
- R Core Team. R: A Language and Environment for Statistical Computing, Version 4.0.5; R Foundation for Statistical Computing: Vienna, Austria, 2021; Available online: https://www.R-project.org/:2021 (accessed on 31 March 2021).
- Peters, K.J.; Bury, S.J.; Betty, E.L.; Parra, G.J.; Tezanos-Pinto, G.; Stockin, K.A. Foraging ecology of the common dolphin Delphinus delphis revealed by stable isotope analysis. Mar. Ecol. Prog. Ser. 2020, 652, 173–186. [Google Scholar] [CrossRef]
- Guerra, M.; Wing, L.; Dawson, S.; Rayment, W. Stable isotope analyses reveal seasonal and inter-individual variation in the foraging ecology of sperm whales. Mar. Ecol. Prog. Ser. 2020, 638, 207–219. [Google Scholar] [CrossRef]
- Torres, L.G.; Gill, P.C.; Graham, B.; Steel, D.; Hamner, R.M.; Baker, S.; Constantine, R.; Escobar-Flores, P.; Sutton, P.; Bury, S. Population, Habitat and Prey Characteristics of blue Whales Foraging in the South Taranaki Bight, New Zealand; Report SC/66a/SH/6 to the Scientific Committee of the International Whaling Commission: Cambridge, UK, 2015. [Google Scholar]
- Torres, L.G.; Rayment, W.; Olavarría, C.; Thompson, D.R.; Graham, B.; Baker, C.S.; Patenaude, N.; Bury, S.J.; Boren, L.; Parker, G.; et al. Demography and ecology of southern right whales Eubalaena australis wintering at sub-Antarctic Campbell Island, New Zealand. Polar Biol. 2017, 40, 95–106. [Google Scholar] [CrossRef]
- Abrantes, K.G.; Barnett, A. Intrapopulation variations in diet and habitat use in a marine apex predator, the broadnose sevengill shark Notorynchus cepedianus. Mar. Ecol. Prog. Ser. 2011, 442, 133–148. [Google Scholar] [CrossRef]
- Troina, G.C.; Dehairs, F.; Botta, S.; Tullio, J.C.D.; Elskens, M.; Secchi, E.R. Zooplankton-based δ13C and δ15N isoscapes from the outer continental shelf and slope in the subtropical western South Atlantic. Deep. Sea Res. Part I Oceanogr. Res. Pap. 2020, 159, 103235. [Google Scholar] [CrossRef]
- Ward-Paige, C.A.; Risk, M.J.; Sherwood, O.A.; Jaap, W.C. Clionid sponge surveys on the Florida Reef Tract suggest land-based nutrient inputs. Mar. Pollut. Bull. 2005, 51, 570–579. [Google Scholar] [CrossRef]
- Fontaine, M.; Carravieri, A.; Simon-Bouhet, B.; Bustamante, P.; Gasco, N.; Bailleul, F.; Guinet, C.; Cherel, Y. Ecological tracers and at-sea observations document the foraging ecology of southern long-finned pilot whales (Globicephala melas edwardii) in Kerguelen waters. Mar. Biol. 2015, 162, 207–219. [Google Scholar] [CrossRef]
- Monteiro, S.; Ferreira, M.; Vingada, J.V.; López, A.; Brownlow, A.; Méndez-Fernandez, P. Application of stable isotopes to assess the feeding ecology of long-finned pilot whale (Globicephala melas) in the Northeast Atlantic Ocean. J. Exp. Mar. Biol. Ecol. 2015, 465, 56–63. [Google Scholar] [CrossRef]
- Becker, Y.A.; Fioramonti, N.E.; Dellabianca, N.A.; Riccialdelli, L. Feeding ecology of the long finned pilot whale, Globicephala melas edwardii, in the southwestern Atlantic Ocean, determined by stable isotopes analysis. Polar Biol. 2021, 44, 1655–1667. [Google Scholar]
- Pinzone, M.; Damseaux, F.; Michel, L.N.; Das, K. Stable isotope ratios of carbon, nitrogen and sulphur and mercury concentrations as descriptors of trophic ecology and contamination sources of Mediterranean whales. Chemosphere 2019, 237, 124448. [Google Scholar] [CrossRef]
- Abend, A.G.; Smith, T.D. Differences in stable isotope ratios of carbon and nitrogen between long-finned pilot whales (Globicephala melas) and their primary prey in the western north Atlantic. ICES J. Mar. Sci. 1997, 54, 500–503. [Google Scholar] [CrossRef][Green Version]
- MacAvoy, S.E.; Cortese, N.; Cybulski, J.; Hohn, A.A.; Macko, S.A. Sources of stable isotope variation among stranded Western Atlantic dolphins (Tursiops truncatus) in North Carolina. Mar. Mammal Sci. 2017, 33, 1224–1234. [Google Scholar] [CrossRef]
- Olin, J.A.; Fair, P.A.; Recks, M.A.; Zolman, E.; Adams, J.; Fisk, A.T. Unique seasonal forage bases within a local population of bottlenose dolphin (Tursiops truncatus). Mar. Mammal Sci. 2012, 28, E28–E40. [Google Scholar] [CrossRef]
- Cardona, L.; Aguilar, A.; Pazos, L. Delayed ontogenic dietary shift and high levels of omnivory in green turtles (Chelonia mydas) from the NW coast of Africa. Mar. Biol. 2009, 156, 1487–1495. [Google Scholar] [CrossRef]
- Abend, A.G.; Smith, T.D. Review of Distribution of the Long-Finned Pilot Whale (Globicephala melas) in the North Atlantic and Mediterranean; NOAA technical memorandum NMFS-NE Series 117; Northeast Fisheries Science Center: Woods Hole, MA, USA, 1999. [Google Scholar]
- Sekiguchi, K.; Klages, N.T.W.; Best, P.B. Comparative analysis of the diets of smaller odontocete cetaceans along the coast of Southern Africa. S. Afr. J. Mar. Sci. 1992, 12, 843–861. [Google Scholar] [CrossRef]
- Rossman, S.; Berens McCabe, E.; Barros, N.B.; Gandhi, H.; Ostrom, P.H.; Stricker, C.A.; Wells, R.S. Foraging habits in a generalist predator: Sex and age influence habitat selection and resource use among bottlenose dolphins (Tursiops truncatus). Mar. Mammal Sci. 2015, 31, 155–168. [Google Scholar] [CrossRef]
- Lischka, A.; Betty, E.L.; Braid, H.E.; Pook, C.J.; Gaw, S.; Bolstad, K.S.R. Trace element concentrations, including Cd and Hg, in long-finned pilot whales (Globicephala melas edwardii) mass stranded on the New Zealand coast. Mar. Pollut. Bull. 2021, 165, 112084. [Google Scholar] [CrossRef]
- Méndez-Fernandez, P.; Pierce, G.J.; Bustamante, P.; Chouvelon, T.; Ferreira, M.; González, A.F.; López, A.; Read, F.L.; Santos, M.B.; Spitz, J.; et al. Ecological niche segregation among five toothed whale species off the NW Iberian Peninsula using ecological tracers as multi-approach. Mar. Biol. 2013, 160, 2825–2840. [Google Scholar] [CrossRef]
- Bustamante, P.; Caurant, F.; Fowler, S.W.; Miramand, P. Cephalopods as a vector for the transfer of cadmium to top marine predators in the north-east Atlantic Ocean. Sci. Total Environ. 1998, 220, 71–80. [Google Scholar] [CrossRef]
- Jefferson, T.A.; Webber, M.A.; Pitman, R. Marine Mammals of the World: A Comprehensive Guide to Their Identification; Elsevier: Amsterdam, The Netherlands, 2011. [Google Scholar]
- Palmer, E.; Alexander, A.; Liggins, L.; Guerra, M.; Bury, S.J.; Hendriks, H.; Stockin, K.A.; Peters, K.J. A piece of the puzzle: Analyses of recent strandings and historical records reveal new genetic and ecological insights on New Zealand sperm whales. Mar. Ecol. Prog. Ser. 2022, 690, 201–217. [Google Scholar] [CrossRef]
- Giménez, J.; Cañadas, A.; Ramírez, F.; Afán, I.; García-Tiscar, S.; Fernández-Maldonado, C.; Castillo, J.J.; de Stephanis, R. Intra-and interspecific niche partitioning in striped and common dolphins inhabiting the southwestern Mediterranean Sea. Mar. Ecol. Prog. Ser. 2017, 567, 199–210. [Google Scholar] [CrossRef]
- Niño Torres, C.; Gallo-Reynoso, J.P.; Galván-Magaña, F.; Escobar-Briones, E.; Macko, S. Isotopic analysis of δ13C, δ15N, and δ34S “A feeding tale” in teeth of the longbeaked common dolphin, Delphinus capensis. Mar. Mammal Sci. 2006, 22, 831–846. [Google Scholar] [CrossRef]
- Gelippi, M.; Popp, B.; Gauger, M.F.W.; Caraveo-Patiño, J. Tracing gestation and lactation in free ranging gray whales using the stable isotopic composition of epidermis layers. PLoS ONE 2020, 15, e0240171. [Google Scholar] [CrossRef] [PubMed]
- Knoff, A.; Hohn, A.; Macko, S. Ontogenetic diet changes in bottlenose dolphins (Tursiops truncatus) reflected through stable isotopes. Mar. Mammal Sci. 2008, 24, 128–137. [Google Scholar] [CrossRef]
- Bernard, H.J.; Hohn, A.A. Differences in feeding habits between pregnant and lactating spotted dolphins (Stenella attenuata). J. Mammal. 1989, 70, 211–215. [Google Scholar] [CrossRef]
- Rechsteiner, E.U.; Rosen, D.A.S.; Trites, A.W. Energy requirements of Pacific white-sided dolphins (Lagenorhynchus obliquidens) as predicted by a bioenergetic model. J. Mammal. 2013, 94, 820–832. [Google Scholar] [CrossRef]
- Malinowski, C.R.; Herzing, D.L. Prey use and nutritional differences between reproductive states and age classes in Atlantic spotted dolphins (Stenella frontalis) in the Bahamas. Mar. Mammal Sci. 2015, 31, 1471–1493. [Google Scholar] [CrossRef]
- Clark, C.T.; Fleming, A.H.; Calambokidis, J.; Kellar, N.M.; Allen, C.D.; Catelani, K.N.; Robbins, M.; Beaulieu, N.E.; Steel, D.; Harvey, J.T. Heavy with child? Pregnancy status and stable isotope ratios as determined from biopsies of humpback whales. Conserv. Physiol. 2016, 4, cow050. [Google Scholar] [CrossRef]
- Engen, S.; Stenseth, N.C. Age-specific optimal diets and optimal foraging tactics: A life-historic approach. Theor. Popul. Biol. 1989, 36, 281–295. [Google Scholar] [CrossRef]
- Takahashi, M.; Tamura, T.; Bando, T.; Konishi, K. Feeding habits of Bryde’s and sei whales in the western North Pacific inferred from stomach contents and skin stable isotope ratios. J. Sea Res. 2022, 184, 102204. [Google Scholar] [CrossRef]
- Browning, N.E.; McCulloch, S.D.; Bossart, G.D.; Worthy, G.A.J. Fine-scale population structure of estuarine bottlenose dolphins (Tursiops truncatus) assessed using stable isotope ratios and fatty acid signature analyses. Mar. Biol. 2014, 161, 1307–1317. [Google Scholar] [CrossRef]
- Rau, G.H.; Sweeney, R.E.; Kaplan, I.R. Plankton 13C: 12C ratio changes with latitude: Differences between northern and southern oceans. Deep. Sea Res. Part A Oceanogr. Res. Pap. 1982, 29, 1035–1039. [Google Scholar]
- Chikaraishi, Y.; Steffan, S.A.; Ogawa, N.O.; Ishikawa, N.F.; Sasaki, Y.; Tsuchiya, M.; Ohkouchi, N. High-resolution food webs based on nitrogen isotopic composition of amino acids. Ecol. Evol. 2014, 4, 2423–2449. [Google Scholar] [CrossRef]
- McClelland, J.W.; Montoya, J.P. Trophic relationships and the nitrogen isotopic composition of amino acids in plankton. Ecology 2002, 83, 2173–2180. [Google Scholar] [CrossRef]
- Hannides, C.C.S.; Popp, B.N.; Landry, M.R.; Graham, B.S. Quantification of zooplankton trophic position in the North Pacific subtropical gyre using stable nitrogen isotopes. Limnol. Oceanogr. 2009, 54, 50–61. [Google Scholar]
- Chikaraishi, Y.; Ogawa, N.O.; Kashiyama, Y.; Takano, Y.; Suga, H.; Tomitani, A.; Miyashita, H.; Kitazato, H.; Ohkouchi, N. Determination of aquatic food-web structure based on compound-specific nitrogen isotopic composition of amino acids. Limnol. Oceanogr. Methods 2009, 7, 740–750. [Google Scholar]
- Foote, A.D. Mortality rate acceleration and post-reproductive lifespan in matrilineal whale species. Biol. Lett. 2008, 4, 189–191. [Google Scholar] [CrossRef]
- Whitehead, H.; Vachon, F.; Frasier, T.R. Cultural hitchhiking in the matrilineal whales. Behav. Genet. 2017, 47, 324–334. [Google Scholar] [CrossRef]
- Oremus, M.; Gales, R.; Kettles, H.; Baker, C.S. Genetic evidence of multiple matrilines and spatial disruption of kinship bonds in mass strandings of long-finned pilot whales, Globicephala melas. J. Hered. 2013, 104, 301–311. [Google Scholar]
- Dammhahn, M.; Randriamoria, T.M.; Goodman, S.M. Broad and flexible stable isotope niches in invasive non-native Rattus spp. in anthropogenic and natural habitats of central eastern Madagascar. BMC Ecol. 2017, 17, 16. [Google Scholar]
- Scholz, C.; Firozpoor, J.; Kramer-Schadt, S.; Gras, P.; Schulze, C.; Kimmig, S.E.; Voigt, C.C.; Ortmann, S. Individual dietary specialization in a generalist predator: A stable isotope analysis of urban and rural red foxes. Ecol. Evol. 2020, 10, 8855–8870. [Google Scholar] [CrossRef] [PubMed]
- Källberg Normark, L.; Liénart, C.; Pillay, D.; Garbaras, A.; Savage, C.; Karlson, A.M.L. Isotopic niche size variability in an ecosystem engineer along a disturbance gradient in a South African lagoon. Mar. Environ. Res. 2022, 173, 105541. [Google Scholar] [CrossRef] [PubMed]
- Vander Zanden, H.B.; Bjorndal, K.A.; Reich, K.J.; Bolten, A.B. Individual specialists in a generalist population: Results from a long-term stable isotope series. Biol. Lett. 2010, 6, 711–714. [Google Scholar] [CrossRef]
- Jourdain, E.; Andvik, C.; Karoliussen, R.; Ruus, A.; Vongraven, D.; Borgå, K. Isotopic niche differs between seal and fish-eating killer whales (Orcinus orca) in northern Norway. Ecol. Evol. 2020, 10, 4115–4127. [Google Scholar] [CrossRef] [PubMed]
- Barros, N.B.; Ostrom, P.H.; Stricker, C.A.; Wells, R.S. Stable isotopes differentiate bottlenose dolphins off west-central Florida. Mar. Mammal Sci. 2010, 26, 324–336. [Google Scholar] [CrossRef]
- Cañadas, A.; Sagarminaga, R.; García-Tiscar, S. Cetacean distribution related with depth and slope in the Mediterranean waters off southern Spain. Deep. Sea Res. Part I Oceanogr. Res. Pap. 2002, 49, 2053–2073. [Google Scholar] [CrossRef]
- Kritzler, H. Observations on the Pilot Whale in Captivity. J. Mammal. 1952, 33, 321–334. [Google Scholar] [CrossRef]
- Lorrain, A.; Pethybridge, H.; Cassar, N.; Receveur, A.; Allain, V.; Bodin, N.; Bopp, L.; Choy, C.A.; Duffy, L.; Fry, B.; et al. Trends in tuna carbon isotopes suggest global changes in pelagic phytoplankton communities. Glob. Change Biol. 2020, 26, 458–470. [Google Scholar] [CrossRef]
- Nicholson, K.; Loneragan, N.; Finn, H.; Bejder, L. Social, spatial and isotopic niche partitioning identify an estuarine community of bottlenose dolphins as a discrete management unit. Aquat. Conserv. Mar. Freshw. Ecosyst. 2021, 31, 3526–3542. [Google Scholar] [CrossRef]
- Gannon, D.P.; Read, A.J.; Craddock, J.E.; Fristrup, K.M.; Nicolas, J.R. Feeding ecology of long-finned pilot whales Globicephala melas in the western North Atlantic. Mar. Ecol. Prog. Ser. 1997, 148, 1–10. [Google Scholar] [CrossRef]
- Ruiz-Cooley, R.I.; Gerrodette, T.; Chivers, S.J.; Danil, K. Cooperative feeding in common dolphins as suggested by ontogenetic patterns in δ15N bulk and amino acids. J. Anim. Ecol. 2021, 90, 1583–1595. [Google Scholar] [CrossRef] [PubMed]
- Isojunno, S.; Sadykova, D.; Deruiter, S.; Cure, C.; Visser, F.; Thomas, L.; Miller, P.J.O.; Harris, C.M. Individual, ecological, and anthropogenic influences on activity budgets of long-finned pilot whales. Ecosphere 2017, 8, e02044. [Google Scholar] [CrossRef]
- Sivle, L.; Kvadsheim, P.; Fahlman, A.; Lam, F.-P.; Tyack, P.; Miller, P. Changes in dive behavior during naval sonar exposure in killer whales, long-finned pilot whales, and sperm whales. Front. Physiol. 2012, 3, 400. [Google Scholar] [CrossRef] [PubMed]
- Baird, R.W.; Borsani, J.F.; Hanson, M.B.; Tyack, P.L. Diving and night-time behavior of long-finned pilot whales in the Ligurian Sea. Mar. Ecol. Prog. Ser. 2002, 237, 301–305. [Google Scholar] [CrossRef]
- Mengual, R.; García, M.; Segovia, Y.; Pertusa, J. Ocular morphology, topography of ganglion cell distribution and visual resolution of the pilot whale (Globicephala melas). Zoomorphology 2015, 134, 339–349. [Google Scholar] [CrossRef]
- Werth, A. A kinematic study of suction feeding and associated behavior in the long-finned pilot whale, Globicephala melas (Traill). Mar. Mammal Sci. 2000, 16, 299–314. [Google Scholar] [CrossRef]
- Quick, N.J.; Isojunno, S.; Sadykova, D.; Bowers, M.; Nowacek, D.P.; Read, A.J. Hidden Markov models reveal complexity in the diving behaviour of short-finned pilot whales. Sci. Rep. 2017, 7, 45765. [Google Scholar] [CrossRef]
| Ontogenetic Status | n | Body Length Range (cm) | Age Range (Years) |
|---|---|---|---|
| Maturity status | |||
| Immature | 56 | 168–482 | 0–13 |
| Mature | 69 | 364–595 | 6–33 |
| Reproductive group | |||
| Immature male | 26 | 255–482 | 1–13 |
| Mature male | 18 | 467–581 | 14–31 |
| Immature female | 25 | 168–375 | 0–8 |
| Pregnant female | 17 | 364–461 | 6–33 |
| Lactating female | 9 | 380–446 | 7–30 |
| Resting female | 7 | 397–453 | 11–30 |
| Unknown | 23 | 194–595 | 5–32 |
| δ13C (‰) | δ15N (‰) | ||||||
|---|---|---|---|---|---|---|---|
| n | Range | Mean | SD | Range | Mean | SD | |
| All | 125 | −18.80 to −15.53 | −17.12 | 0.73 | 11.52 to 16.28 | 12.59 | 0.72 |
| Maturity status | |||||||
| Immature | 56 | −18.80 to −15.53 | −17.00 | 0.70 | 11.90 to 15.23 | 12.59 | 0.58 |
| Mature | 69 | −18.77 to −15.82 | −17.23 | 0.74 | 11.52 to 16.28 | 12.60 | 0.82 |
| Reproductive group | |||||||
| Immature male | 26 | −18.16 to −16.26 | −16.79 | 0.52 | 11.97 to 13.27 | 12.38 | 0.30 |
| Mature male | 18 | −18.77 to −16.26 | −17.00 | 0.69 | 11.52 to 13.27 | 12.34 | 0.53 |
| Immature female | 25 | −18.80 to −15.53 | −17.12 | 0.81 | 11.90 to 13.93 | 12.64 | 0.53 |
| Pregnant female | 17 | −18.32 to −16.02 | −17.13 | 0.73 | 11.83 to 14.85 | 12.53 | 0.72 |
| Lactating female | 9 | −18.59 to −16.39 | −17.40 | 0.71 | 11.70 to 12.85 | 12.30 | 0.42 |
| Resting female | 7 | −18.74 to −15.82 | −17.59 | 1.01 | 11.72 to 13.37 | 12.34 | 0.60 |
| Unknown | 23 | −18.62 to −15.99 | −17.26 | 0.68 | 11.75 to 16.28 | 13.23 | 1.10 |
| δ13C (‰) | δ15N (‰) | δ34S (‰) | |||||
|---|---|---|---|---|---|---|---|
| n | Mean | SD | Mean | SD | Mean | SD | |
| Male | 13 | −17.32 | 0.81 | 12.59 | 0.72 | 21.14 | 0.99 |
| Female | 23 | −17.06 | 0.82 | 12.70 | 1.04 | 21.58 | 0.83 |
| Pregnant/Lactating female | 14 | −17.12 | 0.73 | 12.68 | 0.75 | 21.56 | 0.90 |
| All | 36 | −17.14 | 0.78 | 12.66 | 0.93 | 21.42 | 0.91 |
| Model | R2 | LL | % DE | ∆AICc | wAICc |
|---|---|---|---|---|---|
| δ15N | |||||
| Stranding event | 0.431 | 1.000 | 45.40 | - | 0.145 |
| Location + Stranding event | 0.431 | 0.885 | 45.40 | 0.250 | 0.128 |
| Year + Location | 0.425 | 0.553 | 44.90 | 1.190 | 0.080 |
| δ13C | |||||
| Maturity + Stranding event | 0.679 | 1.000 | 69.40 | - | 0.119 |
| Maturity + Year + Location | 0.679 | 1.000 | 69.40 | - | 0.119 |
| Sex + Maturity + Year + Location | 0.680 | 0.87 | 69.80 | 0.284 | 0.103 |
| δ34S | |||||
| Sex | 0.030 | 1.000 | 5.59 | - | 0.211 |
| Year | 0.020 | 0.885 | 4.77 | 0.250 | 0.186 |
| Location | 0.030 | 0.486 | 8.85 | 1.440 | 0.102 |
| (a) | |||||||
| Mature Male | Mature Female | Pregnant/Lactating Female | |||||
| Male | 82.14 | 75.73 | |||||
| Female | 48.41 | 74.90 | |||||
| Pregnant/Lactating female | 57.30 | 91.45 | |||||
| (b) | |||||||
| FWS2009 | FWS2011 | FWS2014 | FWS2017 | SI2010 | SI2011 | ||
| FWS2009 | 21.05 | 2.65 | 0.86 | 25.02 | 21.11 | ||
| FWS2011 | 36.13 | 17.57 | 4.07 | 5.81 | 27.04 | ||
| FWS2014 | 7.94 | 26.35 | 9.61 | 0.38 | 8.16 | ||
| FWS2017 | 1.49 | 3.51 | 2.48 | 0.00 | 0.75 | ||
| SI2010 | 74.78 | 20.34 | 1.09 | 0.01 | 41.57 | ||
| SI2011 | 58.80 | 29.13 | 7.26 | 0.76 | 37.41 | ||
| Isotope Range (‰) | Overall | FWS2009 | FWS2011 | FWS2014 | FWS2017 | SI2010 | SI2011 |
|---|---|---|---|---|---|---|---|
| n | 125 | 20 | 20 | 27 | 20 | 19 | 19 |
| δ13C | 3.27 | 1.15 | 0.96 | 1.64 | 1.58 | 0.76 | 1.90 |
| δ 15N | 4.76 | 2.95 | 1.96 | 1.47 | 3.85 | 2.18 | 1.86 |
| n | 36 | 6 | 6 | 6 | 6 | 6 | 6 |
| δ34S | 4.30 | 2.24 | 2.72 | 1.15 | 2.80 | 3.27 | 1.18 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hinton, B.; Stockin, K.A.; Bury, S.J.; Peters, K.J.; Betty, E.L. Isotopic Niche Analysis of Long-Finned Pilot Whales (Globicephala melas edwardii) in Aotearoa New Zealand Waters. Biology 2022, 11, 1414. https://doi.org/10.3390/biology11101414
Hinton B, Stockin KA, Bury SJ, Peters KJ, Betty EL. Isotopic Niche Analysis of Long-Finned Pilot Whales (Globicephala melas edwardii) in Aotearoa New Zealand Waters. Biology. 2022; 11(10):1414. https://doi.org/10.3390/biology11101414
Chicago/Turabian StyleHinton, Bethany, Karen A. Stockin, Sarah J. Bury, Katharina J. Peters, and Emma L. Betty. 2022. "Isotopic Niche Analysis of Long-Finned Pilot Whales (Globicephala melas edwardii) in Aotearoa New Zealand Waters" Biology 11, no. 10: 1414. https://doi.org/10.3390/biology11101414
APA StyleHinton, B., Stockin, K. A., Bury, S. J., Peters, K. J., & Betty, E. L. (2022). Isotopic Niche Analysis of Long-Finned Pilot Whales (Globicephala melas edwardii) in Aotearoa New Zealand Waters. Biology, 11(10), 1414. https://doi.org/10.3390/biology11101414






