Phenotypic and Molecular Characteristics of the MDR Efflux Pump Gene-Carrying Stenotrophomonas maltophilia Strains Isolated in Warsaw, Poland
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Bacterial Strains
2.2. Antimicrobial Susceptibility Testing
2.3. Detection of MDR Efflux Pump Genes
2.4. Pulsed-Field Gel Electrophoresis (PFGE)
2.5. Multilocus Sequence Typing (MLST)
3. Results
3.1. Susceptibility Profiles of the Isolates
3.2. Occurrence of Genes Encoding the MDR Efflux Systems
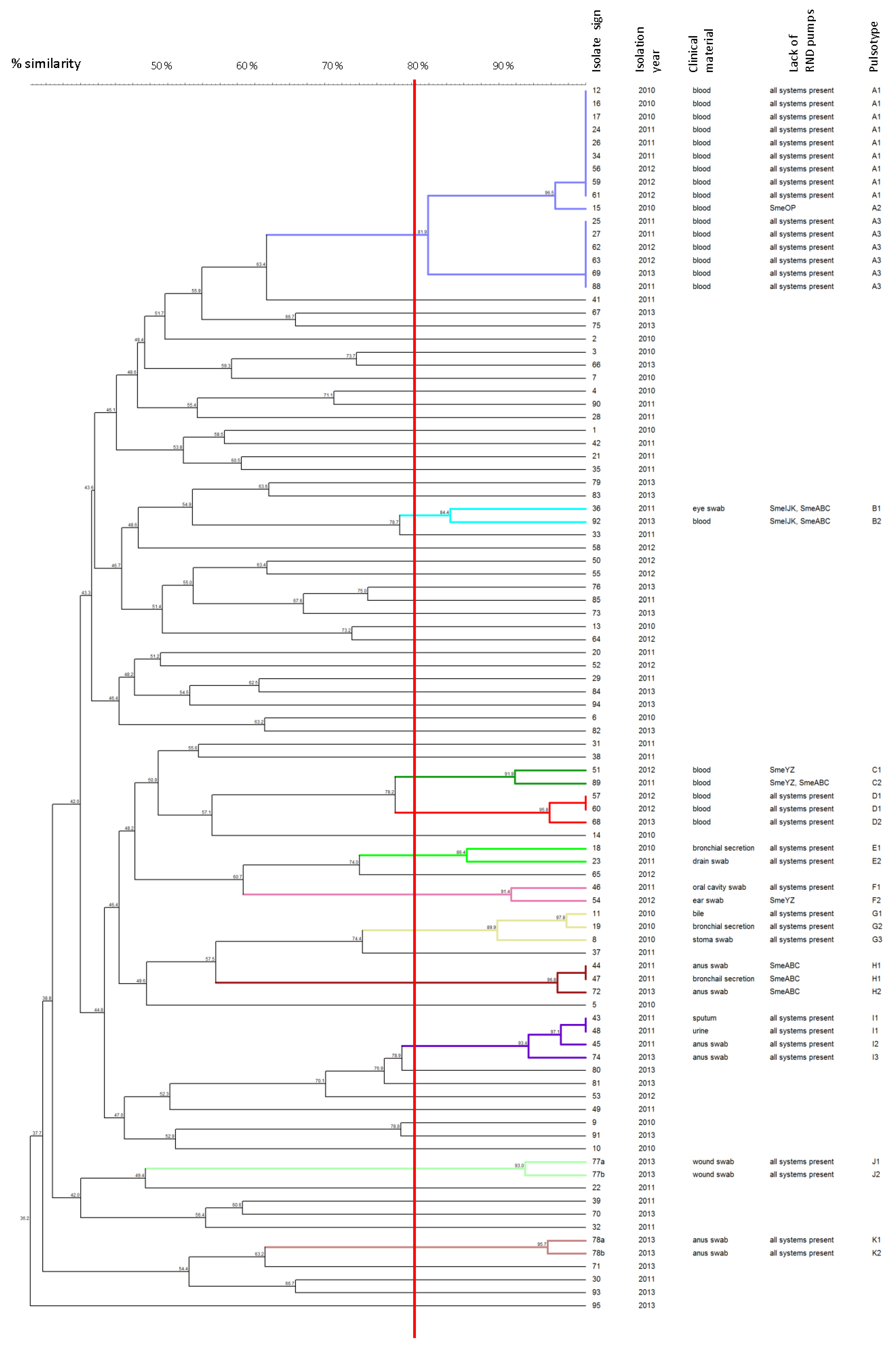
3.3. Molecular Typing of Isolates by Pulsed-Field Gel Electrophoresis (PFGE)
3.4. Multilocus Sequence Typing (MLST) Analysis
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Brooke, J.S. Advances in the microbiology of Stenotrophomonas maltophilia. Clin. Microbiol. Rev. 2021, 34, e0003019. [Google Scholar] [CrossRef]
- Brooke, J.S. Stenotrophomonas maltophilia: An emerging global opportunistic pathogen. Clin. Microbiol. Rev. 2012, 25, 2–41. [Google Scholar] [CrossRef]
- Paez, J.I.; Costa, S.F. Risk factors associated with mortality of infections caused by Stenotrophomonas maltophilia: A systematic review. J. Hosp. Infect. 2008, 70, 101–108. [Google Scholar] [CrossRef] [PubMed]
- Sánchez, M.B. Antibiotic resistance in the opportunistic pathogen Stenotrophomonas maltophilia. Front. Microbiol. 2015, 6, 658. [Google Scholar] [CrossRef] [PubMed]
- Alonso, A.; Martinez, J.L. Cloning and characterization of SmeDEF, a novel multidrug efflux pump from Stenotrophomonas maltophilia. Antimicrob. Agents Chemother. 2000, 44, 3079–3086. [Google Scholar] [CrossRef]
- Zhang, L.; Xian-Zhi, L.; Poole, K. SmeDEF multidrug efflux pump contributes to intrinsic multidrug resistance in Stenotrophomonas maltophilia. Antimicrob. Agents Chemother. 2001, 45, 3497–3503. [Google Scholar] [CrossRef]
- Garcia-Leon, G.; Hernandez, A.; Hernando-Amado, S.; Alavi, P.; Berg, G.; Martinez, J.L. A function of SmeDEF, the major quinolone resistance determinant of Stenotrophomonas maltophilia, is the colonization of plant roots. Appl. Environ. Microbiol. 2014, 80, 4559–4565. [Google Scholar] [CrossRef] [PubMed]
- Chang, L.L.; Chen, H.F.; Chang, C.Y.; Lee, T.M.; Wu, W.J. Contribution of integrons, and SmeABC and SmeDEF efflux pumps to multidrug resistance in clinical isolates of Stenotrophomonas maltophilia. J. Antimicrob. Chemother. 2004, 53, 518–521. [Google Scholar] [CrossRef]
- Gould, V.C.; Okazaki, A.; Avison, M.B. Coordinate hyperproduction of SmeZ and SmeJK efflux pumps extends drug resistance in Stenotophomonas maltophilia. Antimicrob. Agents Chemother. 2013, 57, 655–657. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.H.; Huang, C.C.; Chung, T.C.; Hu, R.M.; Huang, Y.W.; Yang, T.C. Contribution of resistance-nodulation-division efflux pump operon smeU1-V-W-U2-X to multidrug resistance of Stenotophomonas maltophilia. Antimicrob. Agents Chemother. 2011, 55, 5826–5833. [Google Scholar] [CrossRef]
- Lin, C.W.; Huang, Y.W.; Hu, R.M.; Yang, T.C. SmeOP-tolCsm efflux pump contributes to the multidrug resistance of Stenotrophomonas maltophilia. Antimicrob. Agents Chemother. 2014, 58, 2405–2408. [Google Scholar] [CrossRef]
- Crossman, L.C.; Gould, V.C.; Dow, J.M.; Vernikos, G.S.; Okazaki, A.; Sebaihia, M.; Saunders, D.; Arrowsmith, C.; Carver, T.; Peters, N.; et al. The complete genome, comparative and functional analysis of Stenotrophomonas maltophilia reveals an organism heavily shielded by drug resistance determinants. Genome Biol. 2008, 9, R74. [Google Scholar] [CrossRef] [PubMed]
- Al-Hamad, A.; Upton, M.; Burnie, J. Molecular cloning and characterization of SmrA, a novel ABC multidrug efflux pump from Stenotrophomonas maltophilia. J. Antimicrob. Chemother. 2009, 64, 731–734. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Lin, Y.T.; Huang, Y.W.; Liou, R.S.; Chang, Y.C.; Yang, T.C. MacABCsm, an ABC-type tripartite efflux pump of Stenotrophomonas maltophilia involved in drug resistance, oxidative and envelope stress tolerances and biofilm formation. J. Antimicrob. Chemother. 2014, 69, 3221–3226. [Google Scholar] [CrossRef]
- Huang, Y.W.; Hu, R.M.; Chu, F.Y.; Lin, H.R.; Yang, T.C. Characterization of a major facilitator superfamily (MFS) tripartite efflux pump EmrCABsm from Stenotrophomonas maltophilia. J. Antimicrob. Chemother. 2013, 68, 2498–2505. [Google Scholar] [CrossRef]
- Hu, R.M.; Liao, S.T.; Huang, C.C.; Huang, Y.W.; Yang, T.C. An inducible fusaric acid tripartite efflux pump contributes to the fusaric acid resistance in Stenotrophomonas maltophilia. PLoS ONE 2012, 7, e51053. [Google Scholar] [CrossRef] [PubMed]
- Dulyayangkul, P.; Calvopiña, K.; Kate, J.; Heesom, K.J.; Avison, M.B. Novel mechanisms of efflux-mediated levofloxacin resistance and reduced amikacin susceptibility in Stenotrophomonas maltophilia. Antimicrob. Agents Chemother. 2021, 65, e01284-20. [Google Scholar] [CrossRef]
- Gherardi, G.; Creti, R.; Pompilio, A.; Di Bonaventura, G. An overview of various typing methods for clinical epidemiology of the emerging pathogen Stenothophomonas maltophilia. Diagn. Microbiol. Infect. Dis. 2015, 81, 219–226. [Google Scholar] [CrossRef]
- Pompilio, A.; Crocetta, V.; Ghosh, D.; Chakrabarti, M.; Gherardi, G.; Vitali, L.A.; Fiscarelli, E.; Di Bonaventura, G. Stenotrophomonas maltophilia phenotypic and genotypic diversity during a 10-year colonization in the lungs of a cystic fibrosis patient. Front. Microbiol. 2016, 7, 1551. [Google Scholar] [CrossRef]
- Rizek, C.F.; Jonas, D.; Paez, J.I.G.; Rosa, J.F.; Neto, L.V.P.; Martins, R.R.; Moreno, L.Z.; Rossi, A., Jr.; Levin, A.S.; Costa, S.F. Multidrug-resistant Stenotrophomonas maltophilia: Description of new MLST profiles and resistance and virulence genes using whole-genome sequencing. J. Glob. Antimicrob. Resist. 2018, 15, 212–214. [Google Scholar] [CrossRef]
- Clinical and Laboratory Standards Institute (CLSI). M02: Performance Standards for Antimicrobial Disk Susceptibility Tests, 13th ed.; CLSI: Wayne, PA, USA, 2018. [Google Scholar]
- Liofilchem Inc. Liofilchem MTS—MIC Test Strip Guide. 2020. Available online: https://www.liofilchem.com/solutions/clinical/arm/mts-mic-test-strip.html (accessed on 18 September 2021).
- Clinical and Laboratory Standards Institute (CLSI). M100: Performance Standards for Antimicrobial Susceptibility Testing, 31st ed.; CLSI: Wayne, PA, USA, 2021. [Google Scholar]
- Jumaa, P.A.; Sonnevend, A.; Pàl, T.; El Hag, M.; Amith, R.; Trad, O. The molecular epidemiology of Stenotrophomonas maltophilia bacteraemia in a tertiary referral hospital in the United Arab Emirates 2000–2004. Ann. Clin. Microb. Antimicrob. 2006, 5, 32. [Google Scholar] [CrossRef][Green Version]
- Tenover, F.C.; Arbeit, R.D.; Goering, R.V.; Mickelsen, P.A.; Murray, B.E.; Persing, D.H.; Swaminathan, B. Interpreting chromosomal DNA restriction patterns produced by pulsed-field gel electrophoresis: Criteria for bacterial strain typing. J. Clin. Microbiol. 1995, 33, 2233–2239. [Google Scholar] [CrossRef]
- Kaiser, S.; Biehler, K.; Jonas, D. A Stenotrophomonas maltophilia multilocus sequence typing scheme for inferring population structure. J. Bacteriol. 2009, 191, 2934–2943. [Google Scholar] [CrossRef]
- Chang, Y.T.; Lin, C.Y.; Chen, Y.H.; Hsueh, P.R. Update of infections caused by Stenotrophomonas maltophilia with particular attention to resistance mechanisms and therapeutic options. Front. Microbiol. 2015, 6, 893. [Google Scholar] [CrossRef]
- Cruz-Cordova, A.; Mancilla-Rojano, J.; Luna-Pineda, V.M.; Escalona-Venegas, G.; Cázares-Domínguez, V.; Ormsby, C.; Franco-Hernández, I.; Zavala-Vega, S.; Andrés-Hernández, M.; Medina-Pelcastre, M.; et al. Molecular epidemiology, antibiotic resistance, and virulence traits of Stenotrophomonas maltophilia strains associated with an outbreak in a Mexican tertiary care hospital. Front. Cell. Infect. Microbiol. 2020, 10, 50. [Google Scholar] [CrossRef]
- Insuwanno, W.; Kiratisin, P.; Jitmuang, A. Stenotrophomonas maltophilia infections: Clinical characteristics and factors associated with mortality of hospitalized patients. Infect. Drug Resist. 2020, 3, 1559–1566. [Google Scholar] [CrossRef]
- Anđelković, M.V.; Janković, S.M.; Kostić, M.J.; Živković Zarić, R.S.; Opančina, V.D.; Živić, M.Ž.; Milosavljević, M.J.; Pejčić, A.V. Antimicrobial treatment of Stenotrophomonas maltophilia invasive infections: Systematic review. J. Chemother. 2019, 31, 297–306. [Google Scholar]
- Sader, H.S.; Farrell, D.J.; Flamm, R.K.; Jones, R.N. Antimicrobial susceptibility of Gram-negative organisms isolated from patients hospitalized with pneumonia in US and European hospitals: Results from the SENTRY Antimicrobial Surveillance Program, 2009–2012. Int. J. Antimicrob. Agents 2014, 43, 328–334. [Google Scholar] [CrossRef]
- Wang, C.-H.; Lin, J.-C.; Lin, H.-A.; Chang, F.-Y.; Wang, N.-C.; Chiu, S.-K.; Lin, T.-Y.; Yang, Y.-S.; Kan, L.-P.; Yang, C.-H.; et al. Comparisons between patients with trimethoprim-sulfamethoxazole-susceptible and trimethoprim-sulfamethoxazole-resistant Stenotrophomonas maltophilia monomicrobial bacteraemia: A 10 year retrospective study. J. Microbiol. Immunol. Infect. 2016, 49, 378–386. [Google Scholar] [CrossRef]
- Nys, C.; Cherabuddi, K.; Venugopalan, V.; Klinker, K.P. Clinical and microbiologic outcomes in patients with monomicrobial Stenotrophomonas maltophilia infections. Antimicrob. Agents Chemother. 2019, 63, e00788-19. [Google Scholar] [CrossRef]
- Azimi, A.; Rezaei, F.; Yaseri, M.; Jafari, S.; Rahbar, M.; Douraghi, M. Emergence of fuoroquinolone resistance and possible mechanisms in clinical isolates of Stenotrophomonas maltophilia from Iran. Sci. Rep. 2021, 11, 9582. [Google Scholar] [CrossRef]
- Hotta, G.; Matsumura, Y.; Kato, K.; Nakano, S.; Yunoki, T.; Yamamoto, M.; Nagao, M.; Ito, Y.; Takakura, S.; Ichiyama, S. Risk factors and outcomes of Stenotrophomonas maltophilia bacteraemia: A comparison with bacteraemia caused by Pseudomonas aeruginosa and Acinetobacter species. PLoS ONE 2014, 9, e112208. [Google Scholar] [CrossRef]
- Nakamura, R.; Oota, M.; Matsumoto, S.; Sato, T.; Yamano, Y. In Vitro activity and In Vivo efficacy of cefiderocol against Stenotrophomonas maltophilia. Antimicrob. Agents Chemother. 2021, 65, e01436-20. [Google Scholar] [CrossRef]
- Cho, H.H.; Sung, J.Y.; Kwon, K.C.; Koo, S.H. Expression of Sme efflux pumps and multilocus sequence typing in clinical isolates of Stenotrophomonas maltophilia. Ann. Lab. Med. 2012, 32, 38–43. [Google Scholar] [CrossRef]
- Mercier-Darty, M.; Royer, G.; Lamy, B.; Charron, C.; Lemenand, O.; Gomart, C.; Fourreau, F.; Madec, J.-Y.; Jumas-Bilak, E.; Decousser, J.-W.; et al. Comparative whole-genome phylogeny of animal, environmental, and human strains confirms the genogroup organization and diversity of the Stenotrophomonas maltophilia complex. Appl. Environ. Microbiol. 2020, 86, e02919-19. [Google Scholar] [CrossRef]
- Blanco, P.; Corona, F.; Sánchez, M.B.; Martínez, J.L. Vitamin K3 induces the expression of the Stenotrophomonas maltophilia SmeVWX multidrug efflux pump. Antimicrob. Agents Chemother. 2017, 61, e02453-16. [Google Scholar] [CrossRef]
- Roscetto, E.; Rocco, F.; Carlomagno, M.S.; Casalino, M.; Colonna, B.; Zarrilli, R.; Di Nocera, P.P. PCR-based rapid genotyping of Stenotrophomonas maltophilia isolates. BMC Microbiol. 2008, 8, 202. [Google Scholar] [CrossRef]
- Kardan-Yamchi, J.; Hajihasani, A.; Talebi, M.; Khodaparast, S.; Azimi, A.; Rahbar, M.; Fallah, F.; Douraghi, M. Intra-hospital dissemination of clinical and environmental isolates of Stenotrophomonas maltophilia from Tehran. Lett. Appl. Microbiol. 2021, 72, 325–331. [Google Scholar] [CrossRef]
- Nasr, P. Genetics, epidemiology, and clinical manifestations of multidrug-resistant Acinetobacter baumannii. J. Hosp. Infect. 2020, 104, 4–11. [Google Scholar] [CrossRef]
- Pirzadian, J.; Harteveld, S.P.; Ramdutt, S.N.; van Wamel, W.J.B.; Klaassen, C.H.W.; Vos, M.C.; Severin, J.A. Novel use of culturomics to identify the microbiota in hospital sink drains with and without persistent VIM-positive Pseudomonas aeruginosa. Sci. Rep. 2020, 10, 17052. [Google Scholar] [CrossRef]
- Kac, G.; Podglajen, I.; Vaupré, S.; Colardelle, N.; Buu-Hof, A.; Gutmann, L. Molecular epidemiology of extended-spectrum beta-lactamase-producing Enterobacteriaceae isolated from environmental and clinical specimens in a cardiac surgery intensive care unit. Infect. Control Hosp. Epidemiol. 2004, 25, 852–855. [Google Scholar] [CrossRef]
| Efflux System | Target Gene | Primer | Sequence (5′–3′) | Product Length (bp) | Annealing Temperature (°C) | Reference |
|---|---|---|---|---|---|---|
| SmeABC | smeB | B-F | GGGCCGGAAAGCTACGA | 200 | 59 | Chang et al. [8] |
| B-R | AGCGAAATGGTCACGAATGG | |||||
| smeA | A-F | AAGGCCATCGATGGCAAGGC | 146 | 59 | This study | |
| A-R | TCCGGGTTCGGAATGACCG | |||||
| SmeDEF | smeD | D-F | CCAAGAGCCTTTCCGTCAT | 150 | 59 | Zhang et al. [6] |
| D-R | TCTCGGACTTCAGCGTGAC | |||||
| RT-D-F | CGGTCAGCATCCTGATGGA | 73 | 59 | Garcia-Leon et al. [7] | ||
| RT-D-R | ACGCTGACTTCGGAGAACTC | |||||
| SmeYZ | smeZ | Z-F | AGTGGACCAGCCAGTCGCT | 508 | 59 | This study |
| Z-R | ACTACATAGAAGACCGGCACG | |||||
| SmeIJK | smeK | K-F | GACCTCGCAGACGCAGTCG | 505 | 59 | Gould et al. [9] modified |
| K-R | CAGGTAGTCGCGCAGGGTC | |||||
| smeI | I-F | TTCCGCGAAGGCCAGGAAGT | 107 | 59 | This study | |
| I-R | TCGTTCTGGCGCTTGGCTG | |||||
| SmeOP | smeP | P-F | GGTGCTGGCGATGACCTTC | 372 | 58 | This study |
| P-R | TCCGGCAGCA TCTTGTCGC | |||||
| SmeMN | smeN | N-F | GGTCTCCTCG ACCATGGAC | 314 | 58 | This study |
| N-R | CCTTGCCCAGCGGGATG | |||||
| SmeVWX | smeW | W-F | TTCGGCGACATCGTGCTCAA | 843 | 58 | This study |
| W-R | CTTGAAGAAGCGGTTGAACGG | |||||
| SmeGH | smeH | H-F | GTGGATGATCGGCTTCACGAT | 556 | 58 | This study |
| H-R | CGCATAGCCCTGGTCTTCTT | |||||
| MacABCsm | macB | MacB-F | GTGATCGACGAGAACACCCA | 589 | 58 | This study |
| MacB-R | GGCCGATCATCGAGCCCA | |||||
| SmrA | smrA | SmrA-F | GGTGTGGCCGGTGCTGCT | 677 | 63 | This study |
| SmrA-R | CGCGGTGCTTGACCGCCA |
| Agent | Disc-Diffusion Method | Etest Method | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| No. of Isolates (%) | GIZ Range a (mm) | No. of Isolates (%) | MIC Range (mg/L) | MIC50 (mg/L) | MIC90 (mg/L) | |||||
| S | I | R | S | I | R | |||||
| Minocycline | 94 (100) | 0 | 0 | 21–36 | 94 (100) | 0 | 0 | 0.19–3 | 0.75 | 1.5 |
| Levofloxacin | 86 (91) | 6 (6) | 2 (2) | 9–34 | 50 (53) | 37 (39.5) | 7 (7.5) | 1–16 | 2 | 6 |
| Trimethoprim- sulfamethoxazole | 94 (100) | 0 | 0 | 18–37 | 94 (100) | 0 | 0 | 0.047–0.75 | 0.125 | 0.25 |
| Efflux System Family | Efflux System | No. of Isolates (%) |
|---|---|---|
| RND | SmeABC | 72 (76.6) |
| SmeDEF | 94 (100) | |
| SmeIJK | 86 (91.5) | |
| SmeYZ | 89 (94.7) | |
| SmeMN | 94 (100) | |
| SmeOP | 90 (95.7) | |
| SmeVWX | 93 (98.9) | |
| SmeGH | 94 (100) | |
| ABC | SmrA | 93 (98.9) |
| MacABCsm | 94 (100) |
| Strain | Alleles | ST | ||||||
|---|---|---|---|---|---|---|---|---|
| recA | gapA | guaA | atpD | nuoD | ppsA | mutM | ||
| 56/2012 | 74 | 157 a | 273 a | 130 a | 139 | 186 a | 148 a | 498 b |
| 57/2012 | 150 a | 18 | 274 a | 6 | 142 a | 185 a | 148 a | 499 b |
| 62/2012 | 74 | 157 a | 273 a | 130 a | 139 | 186 a | 148 a | 498 b |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zając, O.M.; Tyski, S.; Laudy, A.E. Phenotypic and Molecular Characteristics of the MDR Efflux Pump Gene-Carrying Stenotrophomonas maltophilia Strains Isolated in Warsaw, Poland. Biology 2022, 11, 105. https://doi.org/10.3390/biology11010105
Zając OM, Tyski S, Laudy AE. Phenotypic and Molecular Characteristics of the MDR Efflux Pump Gene-Carrying Stenotrophomonas maltophilia Strains Isolated in Warsaw, Poland. Biology. 2022; 11(1):105. https://doi.org/10.3390/biology11010105
Chicago/Turabian StyleZając, Olga M., Stefan Tyski, and Agnieszka E. Laudy. 2022. "Phenotypic and Molecular Characteristics of the MDR Efflux Pump Gene-Carrying Stenotrophomonas maltophilia Strains Isolated in Warsaw, Poland" Biology 11, no. 1: 105. https://doi.org/10.3390/biology11010105
APA StyleZając, O. M., Tyski, S., & Laudy, A. E. (2022). Phenotypic and Molecular Characteristics of the MDR Efflux Pump Gene-Carrying Stenotrophomonas maltophilia Strains Isolated in Warsaw, Poland. Biology, 11(1), 105. https://doi.org/10.3390/biology11010105






