Phylogenetic Analysis and Genetic Diversity of Colletotrichum falcatum Isolates Causing Sugarcane Red Rot Disease in Bangladesh
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Sampling, Isolation and Pure Culture Maintenance
2.2. Morphological and Colonies Variability
2.3. Pathogenicity Assay
2.4. Extraction and Purification of Genomic DNA
2.5. PCR Amplification and Genesequencing
2.6. Alignment of Genesequence and Phylogenetic Analysis
2.7. ISSR-PCR Amplification
| Primers | Sequences (5′–3′) | Primer Length (bp) | Annealing Temperature (°C) | References |
|---|---|---|---|---|
| ISSR-10 | CACCACCACCACCAC | 15 | 61.6 | Gupta et al. [40] |
| UBC-825 | ACACACACACACACACT | 17 | 51.4 | Arade et al. [41] |
| UBC-857 | ACACACACACACACACYG | 18 | 54.3 | Arade et al. [41] |
| UBC-873 | GACAGACAGACAGACA | 16 | 43.9 | Arade et al. [41] |
| ISSR845 | CTCTCTCTCTCTCTCTRG | 18 | 52.5 | Patel et al. [1] |
| UBC810 | GAGAGAGAGAGAGAGAT | 17 | 49.0 | Kaewchai et al. [42] |
| UBC828 | TGTGTGTGTGTGTGTGA | 17 | 53.0 | Kaewchaiet al. [42] |
| UBC850 | GTGTGTGTGTGTGTGTYC | 18 | 54.0 | Kaewchai et al. [42] |
| UBC860 | TGTGTGTGTGTGTGTGGA | 18 | 51.0 | Soytong & Kaewchai [43] |
| ISSR 7 | GGGCGAGAGAGAGAGAGAGA | 20 | 44.0 | Kaewchai et al. [42] |
2.8. ISSR Data Analysis
3. Results
3.1. Morphological and Colonies Variability
3.2. Pathogenic Variability
3.3. Molecular Identification of C. falcatum Isolates
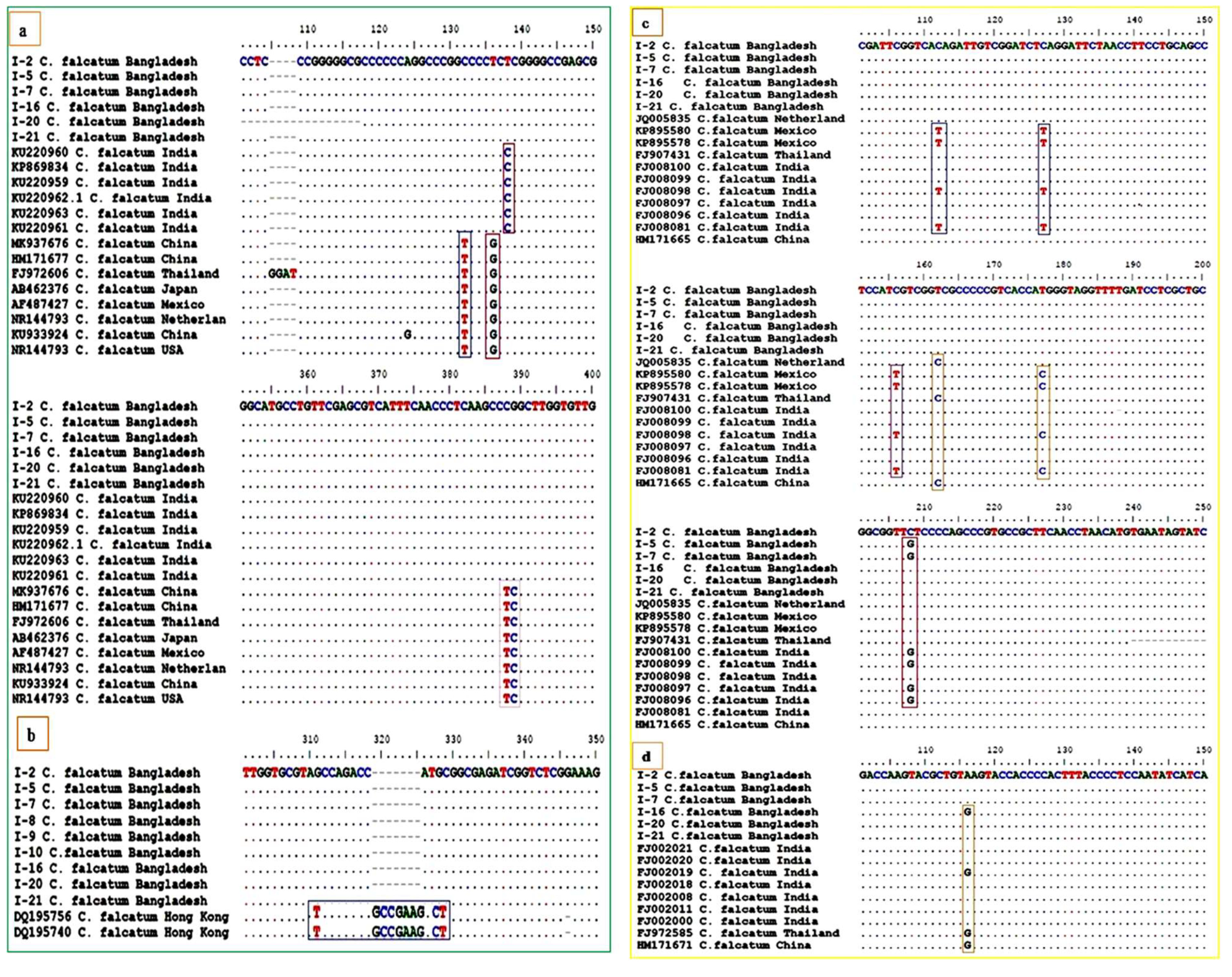
3.4. Phylogenetic Analysis of ITS-rDNA Region
3.5. Phylogenetic Analysis of β-Tubulin Gene Region
3.6. Phylogenetic Analysis of Actin Gene Region
3.7. Phylogenetic Analysis of GAPDH Gene Region
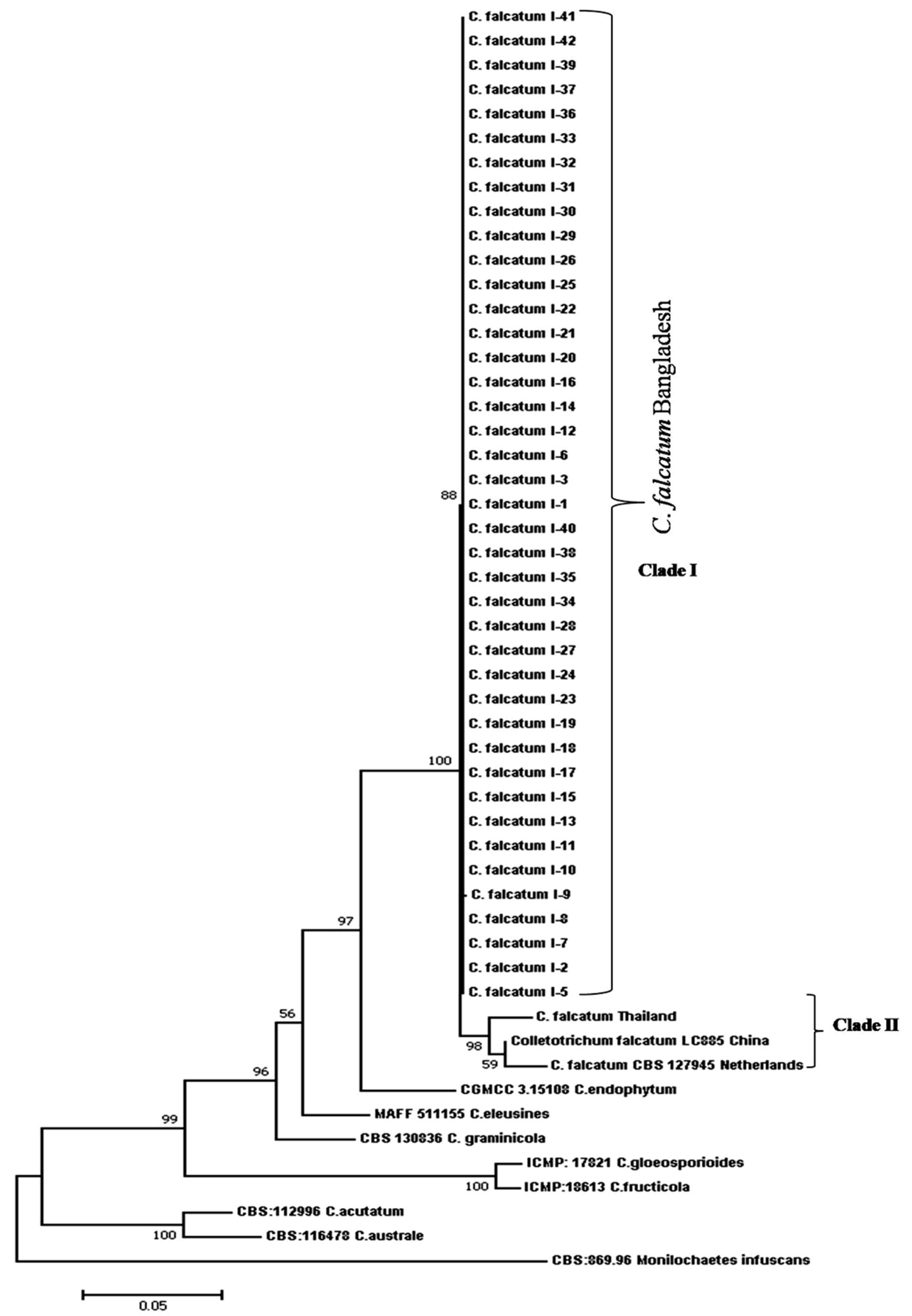
3.8. Phylogenetic Analysis of Combined Gene Regions
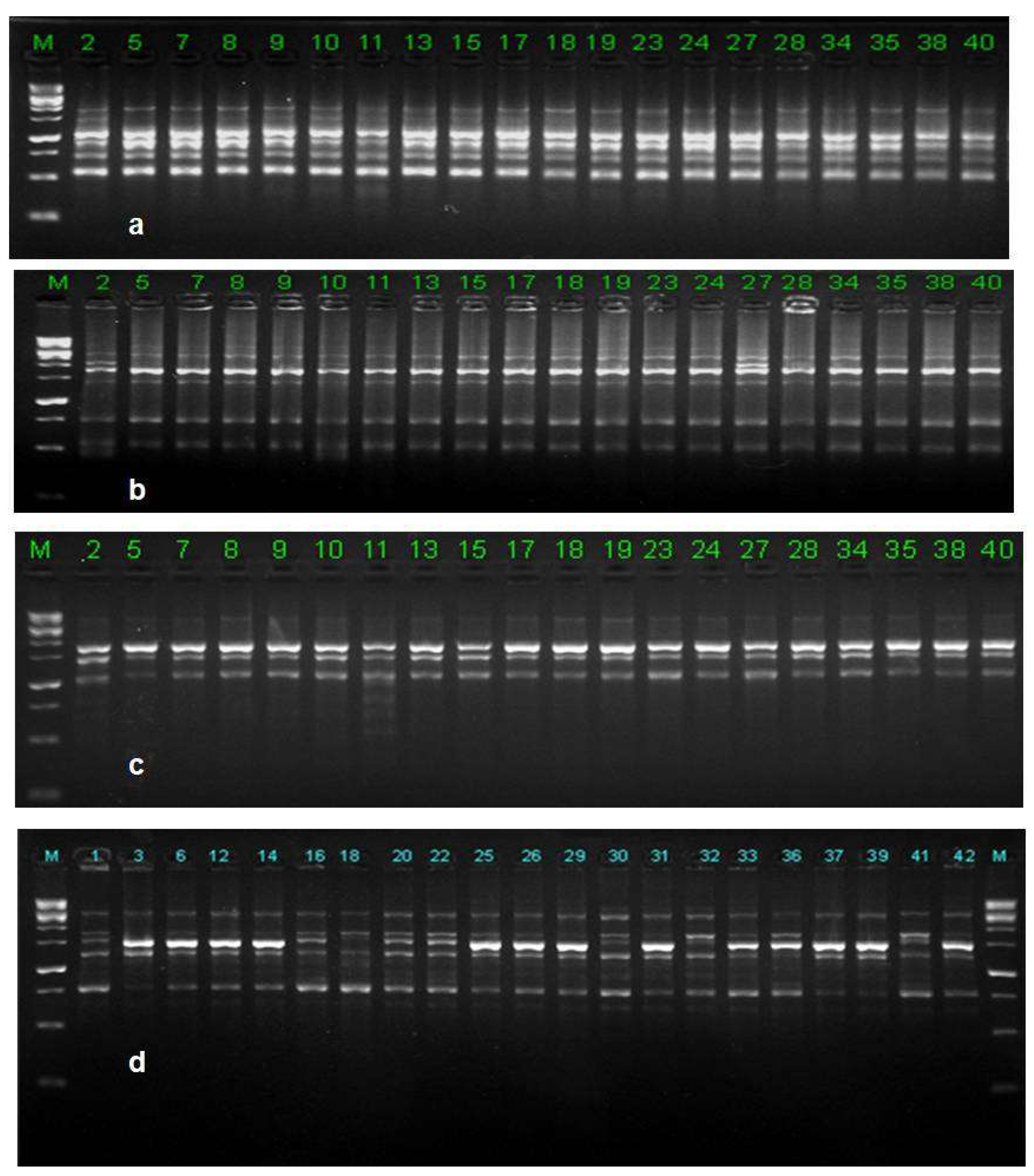
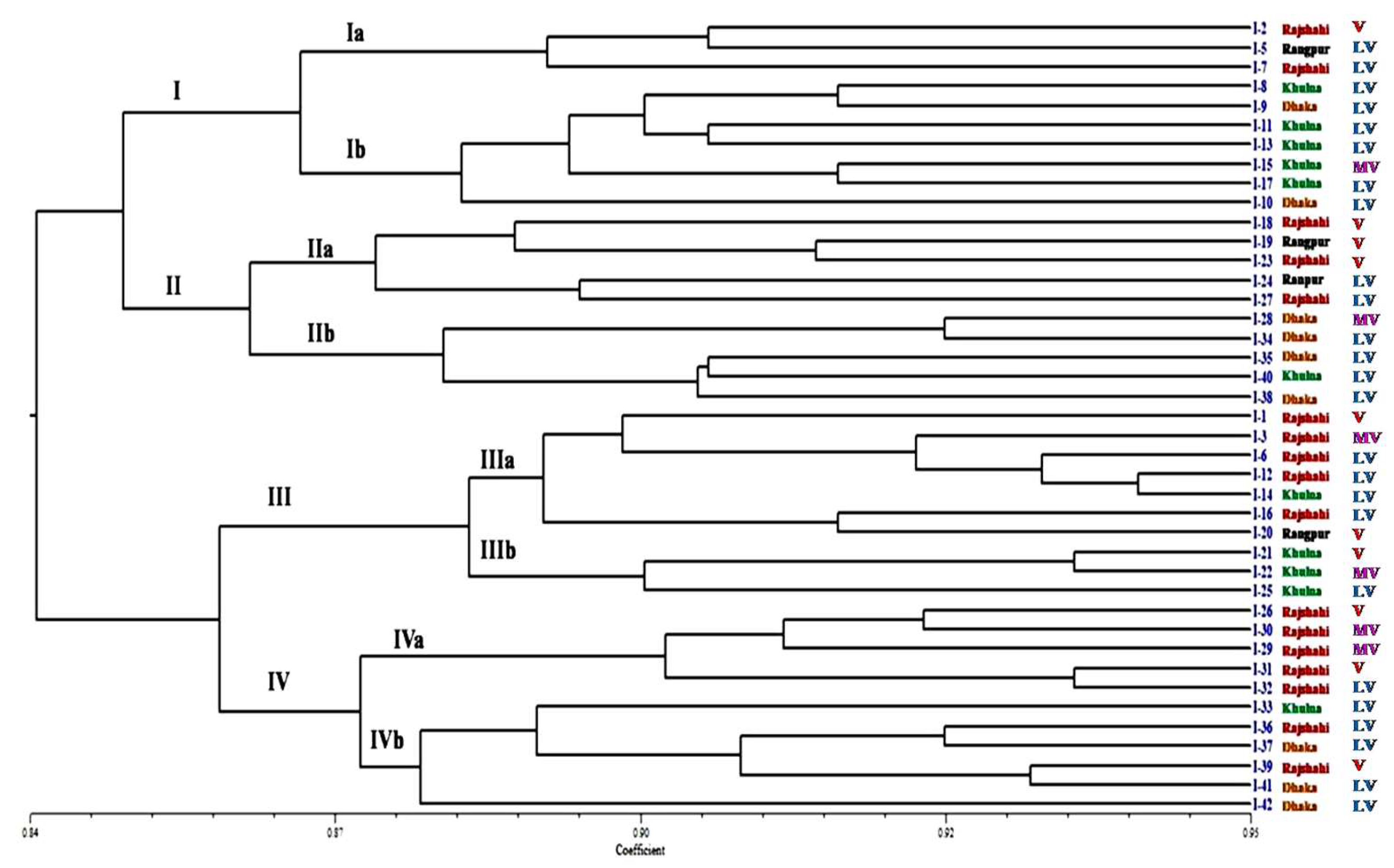
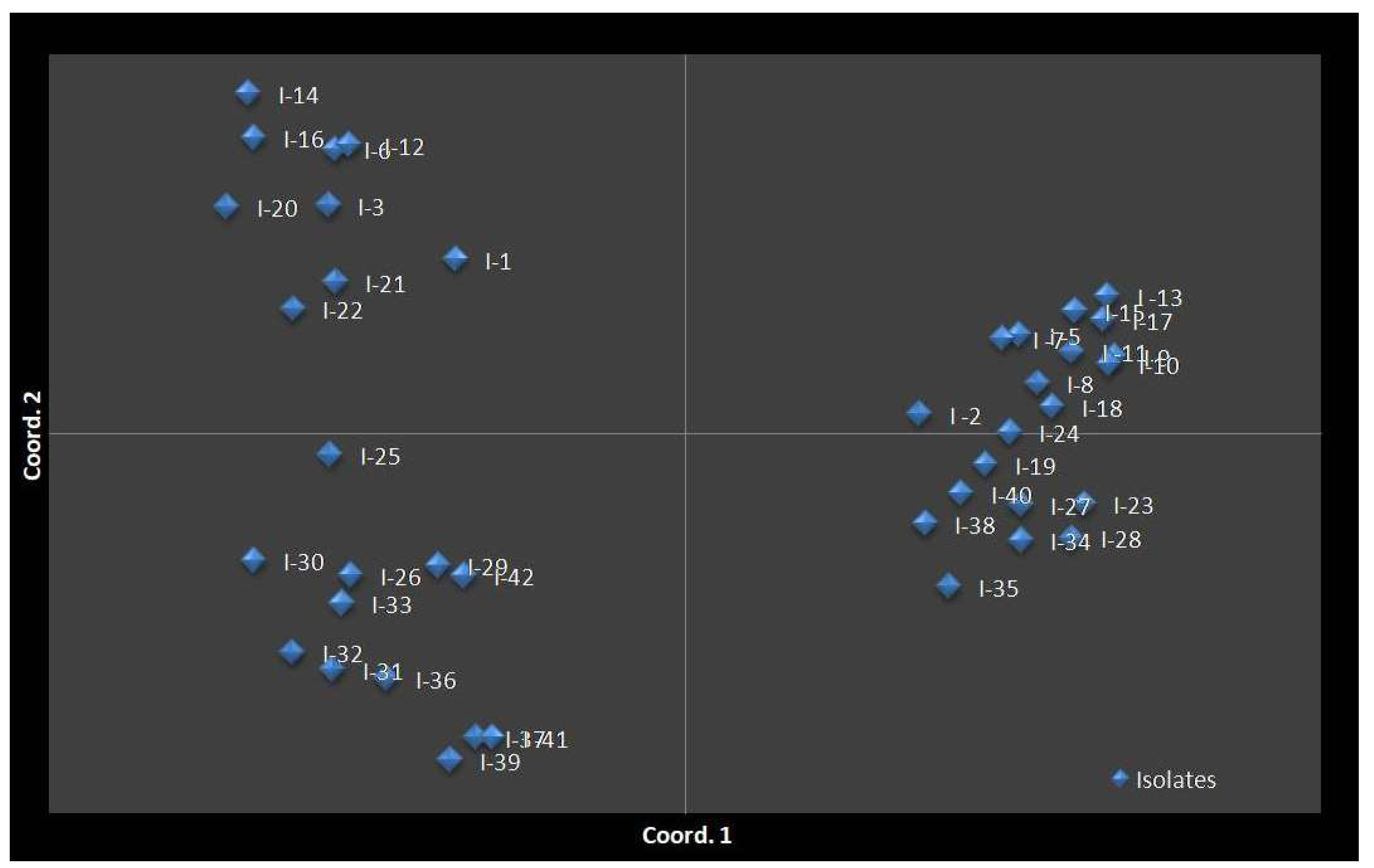
3.9. ISSR Analysis
3.9.1. Genetic Diversity
3.9.2. Populations Structure
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Patel, P.; Rajkumar, B.K.; Parmar, P.; Shah, R.; Krishnamurthy, R. Assessment of genetic diversity in Colletotrichum falcatum Went accessions based on RAPD and ISSR markers. J. Genet. Eng. Biotechnol. 2018, 16, 153–159. [Google Scholar] [CrossRef]
- Food and Agricultural Organization of United Nations; Economic and Social Department. The Statistical Division; FAO: Rome, Italy, 2018. [Google Scholar]
- Uddin, M.S.; Talukdar, M.I.; Rahman, M.S. Screening of red rot genotypes of sugarcane in Bangladesh. Bangladesh J. Sugarcane 2013, 33–34, 5–8. [Google Scholar]
- Rafay, S.A.; Singh, V.B. A new strain of Glomerella tucumanesis. Curr. Sci. 1957, 26, 19–20. [Google Scholar]
- Bailey, J.A.; Jeger, M.J. Colletotrichum: Biology, Pathology and Control; CAB International: Wallingford, UK, 1992.
- Talukder, M.I.; Kamal, M.M.; Iqbal, M.; Rahman, S. Bangladesh Akher Rog BalaiSomuho o Tar Pratikar; Publication No. 111; Bangladesh Sugarcane Research Institute: Ishurdi, Bangladesh, 2010.
- Ghazanfar, M.U.; Kamran, S. Evaluation of Different Plant Extracts against Colletotrichum falcatum Causing Red Rot in Sugarcane under Lab Conditions. J. Environ. Agric. 2016, 1, 68–73. [Google Scholar]
- Sengar, A.S.; Thind, K.S.; Kumar, B.; Pallavi, M.; Gosal, S.S. In vitro selection at cellular level for red rot resistance in sugarcane (Saccharum sp.). Plant Growth Regul. 2009, 58, 201–209. [Google Scholar] [CrossRef]
- Kumar, N.; Jhang, T.; Satyavir, N.; Sharma, T.R. Molecular and Pathological Characterization of Colletotrichum falcatum Infecting Subtropical Indian Sugarcane. J. Phytopathol. 2011, 159, 206–267. [Google Scholar] [CrossRef]
- Duttamajumdar, S.K. Red Rot of Sugarcane; Indian Institute of Sugarcane Research: Lucknow, India, 2008; p. 46.
- Freeman, S.; Shabi, E.; Katan, T. Characterization of Colletotrichum acutatum causing anthracnose of anemone (Anemone coronaria L.). Appl. Environ. Microbiol. 2000, 66, 5267–5272. [Google Scholar] [CrossRef]
- Hyde, K.D.; Cai, L.; McKenzie, E.H.C.; Yang, Y.L.; Zhang, J.Z.; Prihastuti, H. Colletotrichum: A catalogue of confusion. Fungal Divers. 2009, 39, 1–17. [Google Scholar]
- Bharti, Y.P.; Vishwakarma, S.K.; Kumar, A.; Singh, A.; Sharma, M.L.; Shukla, D.N. Physiological and Pathological Aspects of some new isolates of Colletotricum falcatum causing Red rot disease in Saccharum spp. complex. Acta Phytopathol. Entomol. Hung. 2012, 47, 35–50. [Google Scholar] [CrossRef]
- Cai, L.; Hyde, K.D.; Taylor, P.W.J.; Weir, B.; Waller, J.; Abang, M.M.; Prihastuti, H. A polyphasic approach for studying Colletotrichum. Fungal Divers. 2009, 39, 183–204. [Google Scholar]
- Martin, K.J.; Rygiewicz, P.T. Fungal-specific PCR primers developed for analysis of the ITS region of environmental DNA extracts. BMC Microbiol. 2005, 5, 1–11. [Google Scholar] [CrossRef]
- Talhinhas, P.; Sreenivasaprasad, S.; Neves-Martins, J.; Oliveira, H. Molecular and phenotypic analyses reveal association of diverse Colletotrichum acutatum groups and a low level of C. gloeosporioides with olive anthracnose. Appl. Environ. Microbiol. 2005, 71, 2987–2998. [Google Scholar] [CrossRef]
- McDonald, B.A.; Linde, C. The population genetics of plant pathogens and breeding strategies for durable resistance. Euphytica 2002, 124, 163–180. [Google Scholar] [CrossRef]
- Rangaswami, G. An agar blocks technique for isolating soil microorganisms with special reference to Pythiaceous fungi. Sci. Cult. 1958, 24, 85. [Google Scholar]
- Pandey, V.; Shukla, D.N. Morphological Studies on Red Rot of Sugarcane from Hardoi District of Uttar Pradesh. Int. J. Agric. Innov. Res. 2017, 6, 285–288. [Google Scholar]
- Abbas, H.; Anwar, S.A.; Javed, N.; Iqbal, M.A.; Abid, N. Morphological variability among isolates of Colletotrichum falcatum Went; infecting four cultivars of sugarcane. Pak. J. Phytopathol. 2010, 22, 101–104. [Google Scholar]
- Sangdit, P.; Leksomboon, C.; Lertsrutaiyotin, R. Cultural, morphological and pathological characterization of Colletotrichum falcatum causing red rot disease of sugarcane in Thailand. Agric. Nat. Resour. 2014, 48, 880–892. [Google Scholar]
- Prema, R.T.; Raguchander, T.; Kalaimani, T. Morphological characterization and reaction of partial purified toxin of sugarcane red rot pathogen Colletotrichum falcatum collected from Southern India. Int. J. Agric. Sci. 2013, 3, 59–76. [Google Scholar]
- Kaur, R.; Kumar, B.; Vikal, Y.; Sanghera, G.S. Genetic Diversity among Colletotrichum falcatum Isolates Causing Red Rot of Sugarcane in Subtropical Region of India. Not. Sci. Biol. 2014, 6, 308–315. [Google Scholar] [CrossRef][Green Version]
- Rahman, M.S.; Talukder, M.I.; Iqbal, M.; Begum, F.; Khatun, S. Reactions of some sugarcane genotypes against red rot (Colletotrichum falcatum Went.) disease. Indian Sugar 2009, 59, 35–40. [Google Scholar]
- Srinivasan, K.V.; Bhat, N.R. Red rot of sugarcane criteria for grading resistance. J. Indian Bot. Soc. 1961, 11, 566–577. [Google Scholar]
- Rahman, S.; Alam, S.M.; Joarder, O.I.; Alam, S. Red rot incidence of sugarcane in Bangladesh. Bangladesh J. Sugarcane 1998, 18–20, 106–114. [Google Scholar]
- Shukla, R.K.; Tripathi, D.N.P.; Pundey, A.K.; Singh, P.O.; Singh, S.B. A new mid late maturing and red rot resistant cultivar for high tonnage and recovery. Indian Sugar 2005, 55, 37–40. [Google Scholar]
- Viswanathan, R. Pathogen virulence in sugarcane red rot pathogen versus varieties in cultivation: Classical case of loss in virulence in the pathotype CF06 (Cf671). Sugar Tech. 2017, 19, 293–299. [Google Scholar] [CrossRef]
- Viswanathan, R.; Padmanaban, P.; Selvakumar, R. Emergence of new pathogenic variants in Colletotrichum falcatum, stalk infecting ascomycete in sugarcane: Role of host varieties. Sugar Tech. 2019, 22, 1–12. [Google Scholar] [CrossRef]
- Lin, Y.H.; Chang, J.Y.; Liu, E.T.; Chao, C.P.; Huang, J.W.; Chang, P.F.L. Development of a molecular marker for specific detection of Fusarium oxysporum f. sp. Cubenserace 4 Eur. J. Plant Pathol. 2009, 123, 353. [Google Scholar] [CrossRef]
- White, T.J.; Bruns, T.; Lee, S.; Taylor, J.W. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In PCR Protocols: A Guide to Methods and Applications; Innis, M.A., Gelfand, D.H., Sninsky, J.J., White, T.J., Eds.; Academic Press: New York, NY, USA, 1990; pp. 315–322. [Google Scholar]
- Gardes, M.; Bruns, T.D. ITS primers with enhanced specificity for Basidiomycetes application to the identification of mycorrhizae and rusts. Mol. Ecol. 1993, 2, 113–118. [Google Scholar] [CrossRef] [PubMed]
- Wang, R.Y.; Gao, B.; Li, X.H.; Ma, J.; Chen, S.L. First report of Fusarium solani causing Fusarium root rot and stem canker on storage roots of sweet potato in China. Plant Dis. 2014, 98, 160-160. [Google Scholar] [CrossRef]
- Carbone, I.; Kohn, L.M. A method for designing primer sets for speciation studies in filamentous ascomycetes. Mycologia 1999, 91, 553–556. [Google Scholar] [CrossRef]
- Templeton, M.D.; Rikkerink, E.H.A.; Solon, S.L.; Crowhurst, R.N. Cloning and molecular characterization of the glyceraldehyde-3-phosphate dehydrogenaseen coding gene and cDNA from the plant pathogenic fungus Glomerella cingulata. Gene 1992, 22, 225–230. [Google Scholar] [CrossRef]
- Hall, T.A. BioEdit: A user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp. Ser. 1999, 41, 95–98. [Google Scholar]
- Hall, T.; Biosciences, I.; Carlsbad, C. BioEdit: An important software for molecular biology. GERF Bull. Biosci. 2011, 2, 60–61. [Google Scholar]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular Evolutionary Genetics Analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef]
- Kimura, M. A simple method for estimating evolutionary rate of base substitutions through comparative studies of nucleotide sequences. J. Mol. Evol. 1980, 16, 111–120. [Google Scholar] [CrossRef]
- Gupta, M.; Chyi, Y.S.; Severson, R.J.; Owen, J.L. Amplification of DNA markers from evolutionary diverse genomes using single primers of simple sequence repeats. Theory Appl. Genet. 1994, 89, 998–1006. [Google Scholar] [CrossRef]
- Arade, P.C.; Singh, P.; Mahatma, M. Characterization of Colletotrichum falcatum Went. Causing Red Rot in Sugarcane Saccharum complex. Bioscan 2014, 9, 375–379. [Google Scholar]
- Kaewchai, S.; Wang, H.K.; Lin, F.C.; Hyde, K.D.; Soytong, K. Genetic variation among isolates of Rigidoporus microporus causing white root disease of rubber trees in southern Thailand revealed by ISSR markers and pathogenicity. Afr. J. Microbiol. Res. 2009, 3, 641–648. [Google Scholar]
- Soytong, K.; Kaewchai, S. Biological control of white root of rubber trees using Chaetomium cupreum. J. Agric. Technol. 2014, 10, 93–103. [Google Scholar]
- Rohlf, F.J. NTSYSpc Numerical Taxonomy and Multivariate Analysis System Version 2.0 User Guide; Applied Biostatistics Inc.: New York, NY, USA, 1998. [Google Scholar]
- Peakall, R.; Smouse, P.E. GenAlEx 6.5: Genetic Analysis in Excel. Population genetic software for teaching and research an update. Bioinformatics 2012, 28, 2537–2539. [Google Scholar] [CrossRef]
- Nei, M.; Li, W.H. Mathematical model for studying genetic variation in terms of restriction endonucleases. Proc. Natl. Acad. Sci. USA 1979, 76, 5269–5273. [Google Scholar] [CrossRef] [PubMed]
- Kimura, M.; Crow, J.F. The number of alleles that can be maintained in a finite population. Genetics 1964, 49, 725–738. [Google Scholar] [CrossRef]
- Yeh, F.C.; Boyle, T.J.B. Sample genetic analysis of co-dominant and dominant markers and quantitative traits. Belg. J. Bot. 1997, 130, 129–157. [Google Scholar]
- McDermott, J.M.; McDonald, B.A. Gene flow in plant pathosystems. Annu. Rev. Phytopathol. 1993, 31, 353–373. [Google Scholar] [CrossRef]
- Wright, S. The genetical structure of populations. Ann. Eugen. 1951, 15, 323–354. [Google Scholar] [CrossRef]
- Malathi, P.; Viswanathan, R.; Ramesh Sundar, A.; Padmanaban, P.; Prakasam, N.; Mohanraj, D.; Jothi, R. Phylogenetic analysis of Colletotrichum falcatum isolates causing red rot in sugarcane. J. Sugarcane Res. 2011, 1, 69–74. [Google Scholar]
- Rahman, M.S.; Khatun, K.; Rahman, K. Sugarcane and sugar industry in Bangladesh: An Overview. Sugar Tech. 2016, 18, 627–635. [Google Scholar] [CrossRef]
- Viswanathan, R.; Padmanaban, P.; Mohanraj, D.; Jothi, R. Indirect-ELISA technique for the detection of the red rot pathogen in sugarcane (Saccharum spp. hybrid) and resistance screening. Indian J. Agric. Sci. 2000, 70, 8–11. [Google Scholar]
- Mishra, M.K.; Behera, B. Pathogenic and molecular Variability of Colletotrichum falcatum Went. Isolates from sugarcane with red rot disease symptoms. J. Crop. Sci. Biotechnol. 2009, 12, 31–35. [Google Scholar] [CrossRef]
- Prittesh, P.; Amaresan, N.; Rushabh, S.; Krishnamurthy, R.; Bhasker, V.V. Isolation and pathogenic variability of Colletotrichum falcatum causing red rot in sugarcane. J. Plant Dis. Prot. 2016, 123, 273–277. [Google Scholar] [CrossRef]
- Malathi, P.; Viswanathan, R. Variations in Colletotrichum falcatum red rot pathogen of sugarcane in relation to host resistance. Sugar Tech. 2012, 14, 181–187. [Google Scholar] [CrossRef]
- Wang, L.; Liu, L.M.; Wang, Z.G.; Huang, S.W. Genetic Structure and Aggressiveness of Rhizoctonia solani AG 1-IA, the Cause of Sheath Blight of Rice in Southern China. J. Phytopathol. 2013, 161, 753–762. [Google Scholar] [CrossRef]
- Mahmodi, F.; Kadir, J.; Puteh, A. Genetic Diversity and Pathogenic Variability of Colletotrichum truncatum Causing Anthracnose of Pepper in Malaysia. J. Phytopathol. 2013, 162, 456–465. [Google Scholar] [CrossRef]
- Aktaruzzaman, M.; Afroz, T.; Lee, Y.G.; Kim, B.S. Post-harvest anthracnose of papaya caused by Colletotrichum truncatum in Korea. Eur. J. Plant Pathol. 2018, 150, 259–265. [Google Scholar] [CrossRef]
- Jayawardena, R.S.; Hyde, K.D.; Damm, U.; Cai, L.; Liu, M.; Li, X.H.; Yan, J.Y. Notes on currently accepted species of Colletotrichum. Mycosphere 2016, 7, 1192–1260. [Google Scholar] [CrossRef]
- Hyde, K.D.; Cai, L.; Cannon, P.F.; Crouch, J.A.; Crous, P.W.; Damm, U.; Goodwin, P.H. Colletotrichum names in current use. Fungal Divers. 2009, 39, 147–182. [Google Scholar]
- Cannon, P.F.; Damm, U.; Johnston, P.R.; Weir, B.S. Colletotrichum current status and future directions. Stud. Mycol. 2012, 73, 181–213. [Google Scholar] [CrossRef] [PubMed]
- Hossain, M.I.; Ahmad, K.; Siddiqui, Y.; Saad, N.; Rahman, Z.; Haruna, A.O.; Bejo, S.K. Current and Prospective Strategies on Detecting and Managing Colletotrichum falcatum Causing Red Rot of Sugarcane. Agronomy 2020, 10, 1253. [Google Scholar] [CrossRef]
- Shahnazi, S.; Meon, S.; Ebrahimi, M. Characterisation, dierentiation and biochemical diversity of Fusarium solani and Fusarium proliferatum based on cellular fatty acid profiles. Arch. Phytopathol. Plant Prot. 2013, 46, 1513–1522. [Google Scholar] [CrossRef]
- Oghenekaro, A.O.; Miettinen, O.; Omorusi, V.I.; Evueh, G.A.; Farid, M.A.; Gazis, R.; Asiegbu, F.O. Molecular phylogeny of Rigidoporus microporus isolates associated with white rot disease of rubber trees (Hevea brasiliensis). Fungal Biol. 2014, 118, 495–506. [Google Scholar] [CrossRef]
- Hyde, K.D.; Nilsson, R.H.; Alias, S.A.; Ariyawansa, H.A.; Blair, J.E.; Cai, L.; Gorczak, M. One stop shop: Backbones trees for important phytopathogenic genera: I. Fungal Divers. 2014, 67, 21–125. [Google Scholar] [CrossRef]
- Gruning, C.R.; Sieber, T.N.; Rogers, S.O.; Holdenrieder, O. Characterization of dark septate endophytic fungi (DSE) using inter simple sequence repeat anchored polymerase chain reaction (ISSR PCR) amplification. Mycol. Res. 2001, 105, 24–32. [Google Scholar] [CrossRef]
- Muller, M.M.; Valjakka, R.; Suokka, A.; Hantula, J. Diversity of endophytic fungi of single Norway spruce needles and their role as pioneer decomposers. Mol. Ecol. 2001, 10, 1801–1810. [Google Scholar] [CrossRef]
- Rodrigues, K.F.; Sieber, T.N.; Grünig, C.R.; Holdenrieder, O. Characterization of Guignardia mangiferae isolated from tropical plants based on morphology, ISSR-PCR amplifications and ITS1-5.8S-ITS2 sequences. Mycol. Res. 2004, 108, 45–52. [Google Scholar] [CrossRef] [PubMed]
- Rampersad, S.N. Genetic structure of Colletotrichum gloeosporioides sensual to isolates infecting papaya inferred by multilocus ISSR markers. Phytopathology 2013, 103, 182–189. [Google Scholar] [CrossRef] [PubMed]
- Zhou, S.; Smith, D.R.; Stanosz, G.R. Differentiation of Botryosphaeria species and related anamorphic fungi using inter simple or short sequence repeat (ISSR) fingerprinting. Mycol. Res. 2001, 105, 919–926. [Google Scholar] [CrossRef]
- McDonald, B.A. The population genetics of fungi: Tools and techniques. Phytopathology 1997, 87, 448–453. [Google Scholar] [CrossRef]
- Bennett, R.S.; Milgroom, M.G.; Bergstrom, G.C. Population structure of seed borne Phaeosphaerian odorum on New York wheat. Phytopathology 2005, 95, 300–305. [Google Scholar] [CrossRef][Green Version]
- Weeds, P.L.; Chakraborty, S.; Fernandes, C.D.; d’ACharchar, M.J.; Ramesh, C.R.; Kexian, Y.; Kelemu, S. Genetic diversity in Colletotrichum gloeosporioides from Stylosanthes spp. at centers of origin and utilization. Phytopathology 2003, 93, 176–185. [Google Scholar] [CrossRef][Green Version]
- Alsultan, W.; Vadamalai, G.; Khairulmazmi, A.; Saud, H.M.; Al-Sadi, A.M.; Rashed, O.; Jaaffar, A.K.M.; Nasehi, A. Isolation, identification and characterization of endophytic bacteria antagonistic to Phytophthora palmivora causing black pod of cocoa in Malaysia. Eur. J. Plant Pathol. 2019, 155, 1077–1091. [Google Scholar] [CrossRef]
- Taheri, P.; Gnanamanickam, S.; Höfte, M. Characterization, genetic structure, and pathogenicity of Rhizoctonia spp. associated with rice sheath diseases in India. Phytopathology 2007, 97, 373–383. [Google Scholar] [CrossRef] [PubMed]
- McDonald, B.A. Population genetics of plant pathogens. Plant Health Instr. 2004. [Google Scholar] [CrossRef]
- Milgroom, M.G.; Peever, P.T. Population biology of plant pathogens: The synthesis of plant disease epidemiology and population genetics. Plant Dis. 2003, 87, 608–617. [Google Scholar] [CrossRef] [PubMed]
- Rashed, O.; Abdullah, N.A.; Alsultan, W.; Misawa, T.; Ahmad, K.; Kutawa, A.B. Characterization of inter and intra anastomosis group of Rhizoctonia spp. isolated from different crops in Peninsular Malaysia. Trop. Plant Pathol. 2021. [Google Scholar] [CrossRef]

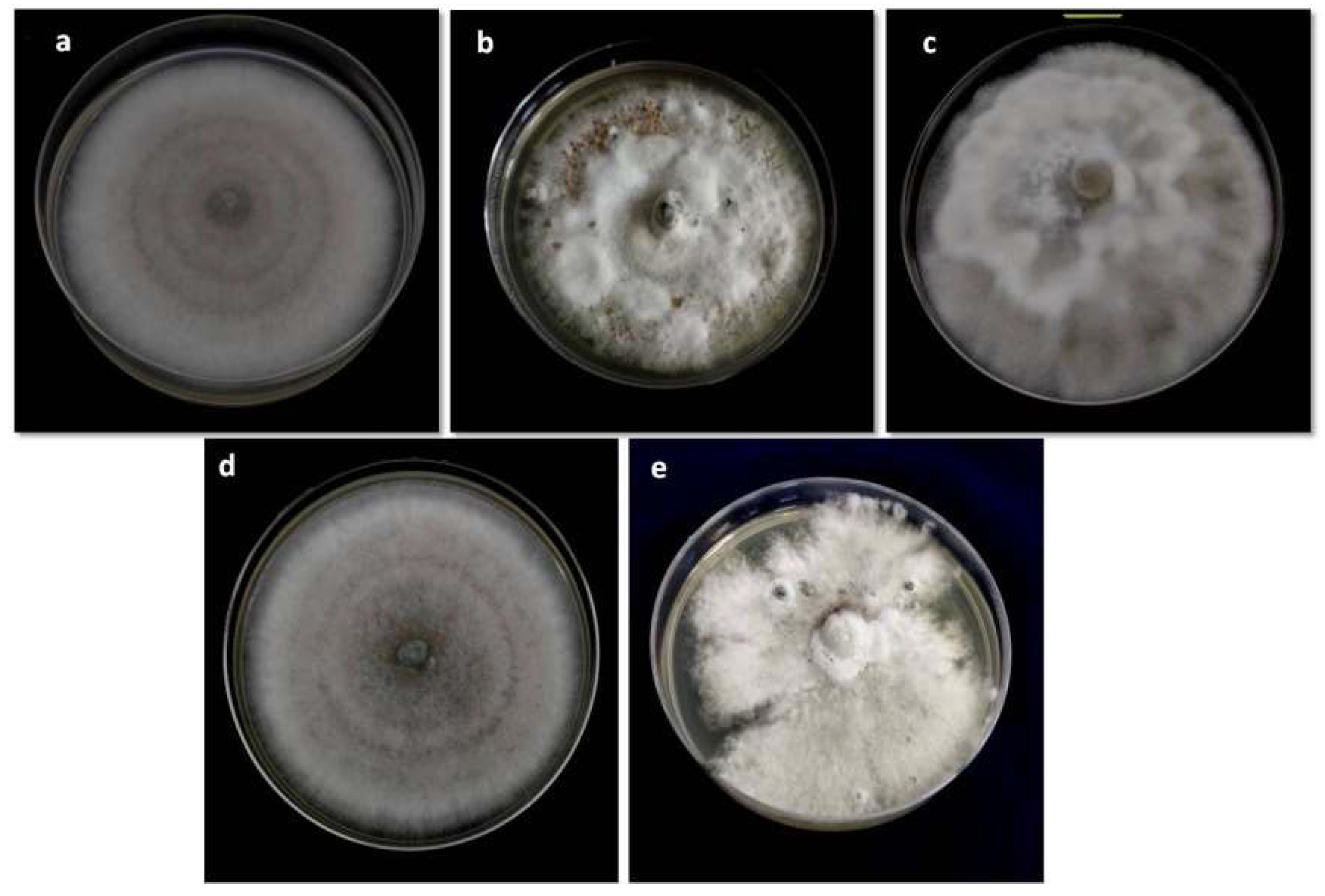
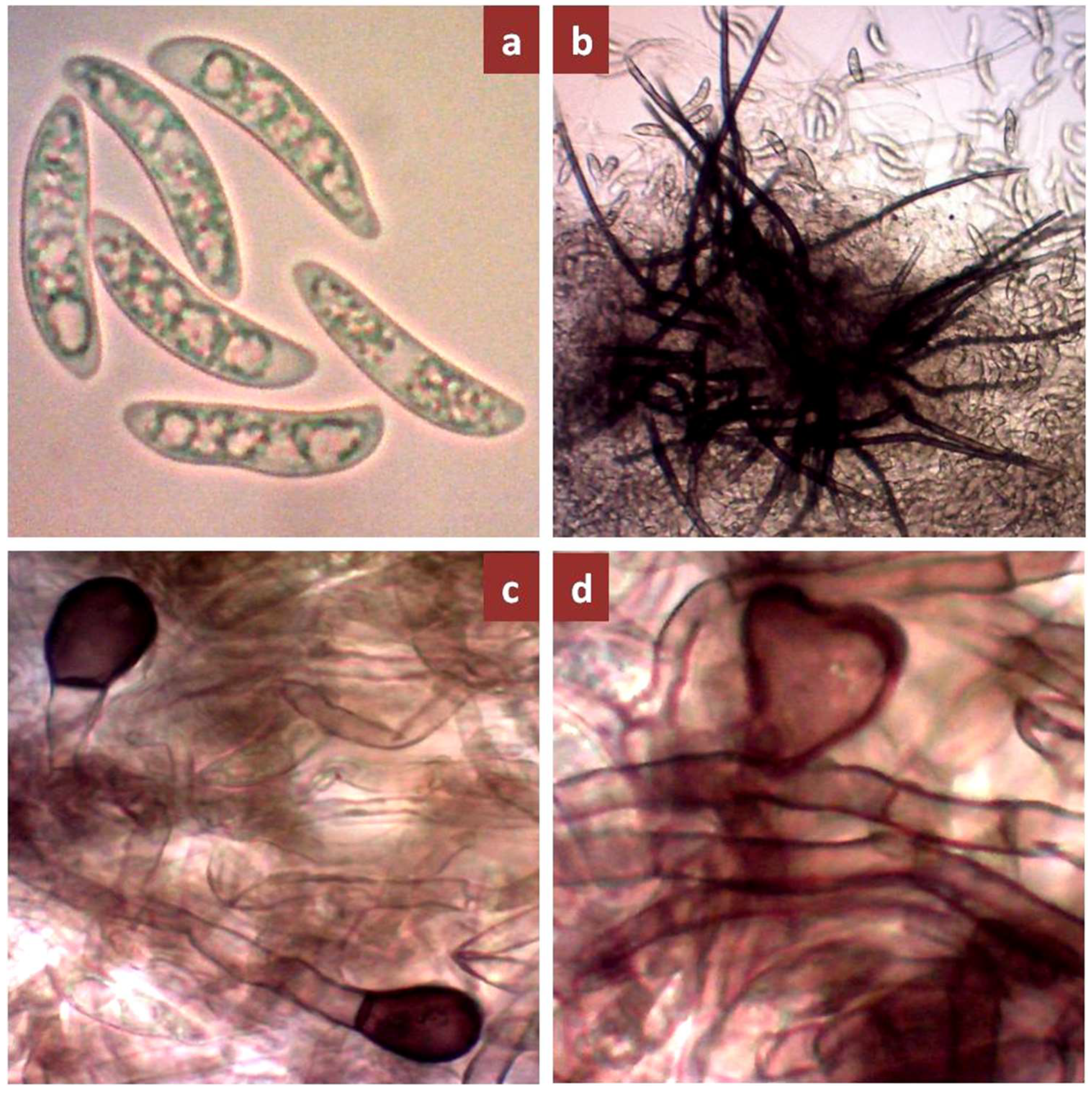

| SL No. | Isolates Code | Host Cultivar | Age of Plant (Month) | Site of Collection | Latitude (°N), Longitude (°E) | Name of Regions |
|---|---|---|---|---|---|---|
| 1 | I-1 | Isd 18 | 7 | BSRI, Ishurdi | 24.1153°, 89.0817° | Rajshahi Region |
| 2 | I-2 | Clone | 8 | BSRI, Ishurdi | 24.1153°, 89.0817° | |
| 3 | I-3 | Isd 18 | 7 | BSRI, Ishurdi | 24.1153°, 89.0817° | |
| 4 | I-6 | BSRI Akh 42 | 8 | NTSM, Natore | 24.4102°, 89.0076° | |
| 5 | I-7 | BSRI Akh 42 | 8 | Bonpara, Natore | 24.2942°, 89.0812° | |
| 6 | I-12 | I-291-87 | 8 | Chapainobabgonj | 23.1657°, 89.4990° | |
| 7 | I-16 | Q 69 | 7 | Ranihati | 24.6329°, 88.1929° | |
| 8 | I-18 | Q 69 | 8 | Ranihati | 24.6329°, 88.1929° | |
| 9 | I-23 | BSRI Akh 42 | 7 | NTSM, Natore | 24.4102°, 89.0076° | |
| 10 | I-26 | Isd 18 | 8 | BSRI, Ishurdi | 24.1153°, 89.0817° | |
| 11 | I-27 | Q 69 | 8 | Chapainawabgonj | 24.7413°, 88.2912° | |
| 12 | I-29 | Isd 18 | 6 | BSRI, Ishurdi | 24.1153°, 89.0817° | |
| 13 | I-30 | BSRI Akh 42 | 8 | Bonpara | 24.2942°, 89.0812° | |
| 14 | I-31 | BSRI Akh 42 | 8 | Bonbara | 24.2942°, 89.0812° | |
| 15 | I-32 | Isd 18 | 7 | BSRI, Ishurdi | 24.1153°, 89.0817° | |
| 16 | I-36 | Isd 18 | 8 | BSRI, Ishurdi | 24.1153°, 89.0817° | |
| 17 | I-39 | I 291-87 | 6 | BSRI, Ishurdi | 24.1153°, 89.0817° | |
| 18 | I-5 | BSRI Akh 42 | 8 | TSM | 26.0504°, 88.4219° | Rangpur Region |
| 19 | I-19 | Strains | 8 | Shampur | 25.6969°, 89.1451° | |
| 20 | I-20 | Clone | 7 | Shampur | 25.6969°, 89.1451° | |
| 21 | I-24 | Isd 16 | 11 | TSM | 26.0504°, 88.4219° | |
| 22 | I-8 | Co 975 | 8 | Patkelghata | 22.7652°, 89.1623° | Khulna Region |
| 23 | I-13 | Local Chewing | 8 | Chowgacha | 23.2632°, 89.0203° | |
| 24 | I-14 | Local Chewing | 8 | Sharsa (N) | 23.0693°, 88.9605° | |
| 25 | I-15 | Co-208 | 9 | Narail | 23.1657°, 89.4990° | |
| 26 | I-17 | Isd 18 | 7 | MKSM | 23.4139°, 89.1333° | |
| 27 | I-21 | Isd 18 | 8 | Magura | 23.4855°, 89.4198° | |
| 28 | I-22 | Co 975 | 9 | Patkelghata | 22.7652°, 89.1623° | |
| 29 | I-25 | Local Chewing | 6 | Jhikorgacha | 23.0999°, 89.0991° | |
| 30 | I-33 | Co 208 | 9 | Narail | 23.1657°, 89.4990° | |
| 31 | I-40 | GT 17 | 7 | Chuadanga | 23.6161°, 88.8263° | |
| 32 | I-11 | Isd 16 | 10 | Barisal | 22.7010°, 90.3535° | |
| 33 | I-9 | Isd 16 | 10 | Kalkini | 23.0724°, 90.2808° | Dhaka Region |
| 34 | I-10 | BSRI Akh 42 | 8 | V-Mirzapur | 24.0902°, 90.4073° | |
| 35 | I-28 | Isd 18 | 7 | Nokla | 24.9753°, 90.2057° | |
| 36 | I-35 | BSRI Akh 42 | 8 | Manikgonj | 23.8617°, 90.0003° | |
| 37 | I-37 | BSRI Akh 42 | 9 | Manikgonj | 23.8617°, 90.0003° | |
| 38 | I-38 | Local | 8 | Kalkini | 23.0724°, 90.2808° | |
| 39 | I-41 | BSRI Akh 42 | 9 | TungiPara | 22.9073°, 89.8985° | |
| 40 | I-42 | BSRI Akh 42 | 10 | Mirzapur | 24.1053°, 90.1051° | |
| 41 | I-34 | BSRI Akh 42 | 9 | Bandarban | 21.8311°, 92.3686° |
| Isolates | Colony Colour | Topography | Margin | Colony Radial Growth Rate/Day (mm) | Conidia Length (µm) * | Conidia Width (µm) * | Sporulation |
|---|---|---|---|---|---|---|---|
| I-1 | Whitish grey | Flat | Smooth | 8.40 g–k | 25.69 i–p | 6.02 bcde | +++ |
| I-2 | Greyish white | Flat | Smooth | 9.79 c–g | 25.52 k–p | 6.45 bcd | +++ |
| I-3 | Whitish grey | Less fluffy | Smooth | 10.57 a–f | 26.40 h–n | 6.28 bcd | +++ |
| I-5 | Greyish white | Flat | Smooth | 10.04 b–g | 24.66 m–p | 6.38 bcd | +++ |
| I-6 | Greyish white | Less fluffy | Irregular | 6.95 k–m | 25.17 l–p | 6.394 bcd | +++ |
| I-7 | Grey white | Flat | Smooth | 10.58 a–f | 29.29 c–g | 6.68 bcd | ++ |
| I-8 | Dark grey | Flat | Smooth | 10.63 a–e | 28.40 c–j | 6.67 bcd | ++ |
| I-9 | Grey | Flat | Smooth | 10.63 a–e | 28.38 c–k | 6.49 bcd | + |
| I-10 | Grey | Raised fluffy | Smooth | 10.63 a–e | 27.27 f–m | 6.43 bcd | ++ |
| I-11 | Greyish white | Less fluffy | Smooth | 9.02 e–i | 24.01 n–p | 6.71 bc | +++ |
| I-12 | Dark grey | Flat | Irregular | 6.04 m | 25.31 l–p | 5.94 cde | ++ |
| I-13 | Grey | Flat | Smooth | 10.63 a–e | 25.06 l–p | 6.05 bcde | ++ |
| I-14 | Greyish white | Raised fluffy | Smooth | 7.31 j–m | 25.55 j–p | 6.39 bcd | ++ |
| I-15 | Gray | Raised fluffy | Smooth | 10.63 a–e | 24.53 m–p | 5.33 e | ++ |
| I-16 | Greyish white | Flat | Irregular | 5.97 m | 27.57 e–k | 6.19 bcde | +++ |
| I-17 | Greyish white | Flat | Irregular | 9.71 c–h | 25.05 l–p | 6.77 bc | +++ |
| I-18 | Greyish white | Flat | Irregular | 8.09 i–m | 27.18 f–m | 5.99 cde | +++ |
| I-19 | Greyish white | Flat | Irregular | 9.42 c–h | 25.76 i–p | 6.40 bcd | +++ |
| I-20 | Greyish white | Raised fluffy | Irregular | 11.54 ab | 27.04 f–m | 6.45 bcd | +++ |
| I-21 | Whitish grey | Raised fluffy | Smooth | 10.95 a–d | 26.78 f–n | 5.94 cde | +++ |
| I-22 | Grey | Raised fluffy | Smooth | 10.12 b–f | 27.10 f–m | 6.36 bcd | ++ |
| I-23 | Dark grey | Flat | Smooth | 10.63 a–e | 27.73 d–l | 6.42 bcd | ++ |
| I-24 | Greyish white | Flat | Smooth | 10.21 b–f | 23.08 pq | 6.08 bcde | +++ |
| I-25 | Greyish white | Raised fluffy | Smooth | 8.95 f–j | 26.053 i–o | 6.46 bcd | +++ |
| I-26 | Greyish white | Fluffy | Irregular | 9.66 c–h | 26.15 i–o | 6.19 bcde | +++ |
| I-27 | Grey | Flat | Smooth | 10.10 b–f | 30.27 b–e | 6.53 bcd | ++ |
| I-28 | Grey | Flat | Smooth | 10.42 b–f | 33.51 a | 8.46 a | ++ |
| I-29 | Grey | Flat | Irregular | 8.09 h–l | 24.99 l–p | 6.25 bcd | ++ |
| I-30 | Grey | Raised fluffy | Irregular | 6.52 lm | 27.62 e–l | 6.293 bcd | + |
| I-31 | Whitish grey | Raised fluffy | Smooth | 10.64 a–e | 26.59 g–n | 6.19 bcde | +++ |
| I-32 | Grey | Raised fluffy | Smooth | 11.07 abc | 30.53 bcd | 6.52 bcd | + |
| I-33 | Grey | Flat | Smooth | 9.35 d–h | 30.76 abc | 6.63 bcd | + |
| I-34 | Grey | Flat | Irregular | 10.52 a–f | 27.37 f–m | 6.28 bcd | ++ |
| I-35 | Greyish white | Raised fluffy | Smooth | 10.63 a–e | 23.49 opq | 6.61 bcd | +++ |
| I-36 | Grey | Raised fluffy | Smooth | 10.35 b–f | 33.08 ab | 7.84 a | + |
| I-37 | Dark grey | Flat | Smooth | 9.52 c–h | 27.77 d–l | 6.49 bcd | ++ |
| I-38 | Grayish white | Raised fluffy | Smooth | 10.63 a–e | 20.86 q | 6.26 bcd | ++ |
| I-39 | Greyish white | Raised fluffy | Smooth | 10.43 b–f | 27.75 d–l | 5.83 ed | ++ |
| I-40 | Grey | Raised fluffy | Smooth | 10.63 a–e | 28.48 c–i | 6.89 b | + |
| I-41 | Greyish white | Raised fluffy | Smooth | 12.14 a | 29.03 c–h | 5.96 cde | ++ |
| I-42 | Greyish white | Flat | Irregular | 9.45 c–h | 29.53 cdef | 6.394 bcd | +++ |
| Mean | - | - | - | 9.70 ± 0.22 | 26.99 ± 0.49 | 6.41 ± 0.15 | - |
| Sl No. | Isolates | Disease Reactions & Score | Virulence Categories | Geographic Regions |
|---|---|---|---|---|
| 1 | I -1 | HS (9.0) | Virulent | Rajshahi |
| 2 | I-2 | HS (8.6) | Virulent | |
| 3 | I-3 | MS (5.9) | Moderately Virulent | |
| 4 | I-6 | MR (3.3) | Less Virulent | |
| 5 | I-7 | MR (3.3) | Less Virulent | |
| 6 | I-12 | MR (2.7) | Less Virulent | |
| 7 | I-16 | R (2.0) | Less Virulent | |
| 8 | I-18 | HS (8.7) | Virulent | |
| 9 | I-23 | HS (9.0) | Virulent | |
| 10 | I-26 | HS (8.6) | Virulent | |
| 11 | I-27 | MR (3.6) | Less Virulent | |
| 12 | I-29 | MS (5.3) | Moderately Virulent | |
| 13 | I-30 | MS (5.1) | Moderately Virulent | |
| 14 | I-31 | HS (9.0) | Virulent | |
| 15 | I-32 | MR (3.2) | Less Virulent | |
| 16 | I-36 | MR (2.4) | Less Virulent | |
| 17 | I-39 | S (6.3) | Virulent | |
| 18 | I-5 | MR (2.8) | Less Virulent | Rangpur |
| 19 | I-19 | HS (8.3) | Virulent | |
| 20 | I-20 | HS (8.5) | Virulent | |
| 21 | I-24 | MR (2.9) | Less Virulent | |
| 22 | I-8 | R (1.4) | Less Virulent | Khulna |
| 23 | I-11 | MR (3.7) | Less Virulent | |
| 24 | I-13 | MR (2.6) | Less Virulent | |
| 25 | I-14 | MR (3.1) | Less Virulent | |
| 26 | I-15 | MS (5.6) | Moderately Virulent | |
| 27 | I-17 | R (1.8) | Less Virulent | |
| 28 | I-21 | S (6.6) | Virulent | |
| 29 | I-22 | MS (4.7) | Moderately Virulent | |
| 30 | I-25 | MR (3.6) | Less Virulent | |
| 31 | I-33 | MR (2.8) | Less Virulent | |
| 32 | I-40 | MR (2.2) | Less Virulent | |
| 33 | I-9 | MS (4.1) | Moderately Virulent | Dhaka |
| 34 | I-10 | MR (2.7) | Less Virulent | |
| 35 | I-28 | MS (5.8) | Moderately Virulent | |
| 36 | I-34 | MR (2.6) | Less Virulent | |
| 37 | I-35 | MR (3.5) | Less Virulent | |
| 38 | I-37 | R (0.8) | Less Virulent | |
| 39 | I-38 | MR (2.1) | Less Virulent | |
| 40 | I-41 | R (1.7) | Less Virulent | |
| 41 | I-42 | R (0.9) | Less Virulent |
| SL No. | Marker | NAB | NMB | NPB | PPB (%) |
|---|---|---|---|---|---|
| 1 | UBC810 | 37 | 2 | 35 | 94.59 |
| 2 | UBC 850 | 36 | 0 | 36 | 100.00 |
| 3 | UBC 828 | 42 | 1 | 41 | 97.62 |
| 4 | ISSR 7 | 45 | 0 | 45 | 100.00 |
| 5 | UBC 860 | 42 | 0 | 42 | 100.00 |
| 6 | ISSR 10 | 43 | 0 | 43 | 100.00 |
| 7 | UBC 857 | 31 | 0 | 31 | 100.00 |
| 8 | UBC 825 | 42 | 0 | 42 | 100.00 |
| 9 | UBC-873 | 46 | 1 | 45 | 97.83 |
| 10 | ISSR 845 | 40 | 0 | 40 | 100.00 |
| Mean | 40.40 | 0.40 | 40.00 | 99.01 | |
| Populations | PPB (%) | Na | Ne | H | I |
|---|---|---|---|---|---|
| Raj | 68.81 | 1.6881 | 1.2388 | 0.1610 | 0.2640 |
| Ran | 34.16 | 1.3416 | 1.2356 | 0.1377 | 0.2021 |
| Khu | 55.45 | 1.5545 | 1.2324 | 0.1520 | 0.2422 |
| Dha | 55.20 | 1.5520 | 1.2413 | 0.1577 | 0.2500 |
| Mean Value | 53.40 | 1.5341 | 1.2370 | 0.1521 | 0.2396 |
| Mean at Species level | 99.01 | 1.9901 | 1.2425 | 0.1732 | 0.2386 |
| Source of Variation | Degree of Freedom | Sum of Square | Mean Square | Estimated Variance | Total Variance (%) | p Value |
|---|---|---|---|---|---|---|
| Among the populations | 3 | 326.373 | 108.791 | 8.260 | 22% | <0.001 |
| Within the Populations | 37 | 1108.164 | 29.950 | 29.950 | 78% | <0.001 |
| Total | 40 | 1434.537 | - | 38.211 | 100% | - |
| Populations | Raj | Ran | Khu | Dha |
|---|---|---|---|---|
| Raj | **** | 0.976 | 0.980 | 0.991 |
| Ran | 0.009 | **** | 0.971 | 0.977 |
| Khu | 0.023 | 0.020 | **** | 0.970 |
| Dha | 0.031 | 0.030 | 0.024 | **** |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hossain, M.I.; Ahmad, K.; Vadamalai, G.; Siddiqui, Y.; Saad, N.; Ahmed, O.H.; Hata, E.M.; Adzmi, F.; Rashed, O.; Rahman, M.Z.; et al. Phylogenetic Analysis and Genetic Diversity of Colletotrichum falcatum Isolates Causing Sugarcane Red Rot Disease in Bangladesh. Biology 2021, 10, 862. https://doi.org/10.3390/biology10090862
Hossain MI, Ahmad K, Vadamalai G, Siddiqui Y, Saad N, Ahmed OH, Hata EM, Adzmi F, Rashed O, Rahman MZ, et al. Phylogenetic Analysis and Genetic Diversity of Colletotrichum falcatum Isolates Causing Sugarcane Red Rot Disease in Bangladesh. Biology. 2021; 10(9):862. https://doi.org/10.3390/biology10090862
Chicago/Turabian StyleHossain, Md Imam, Khairulmazmi Ahmad, Ganesan Vadamalai, Yasmeen Siddiqui, Norsazilawati Saad, Osumanu Haruna Ahmed, Erneeza Mohd Hata, Fariz Adzmi, Osamah Rashed, Muhammad Ziaur Rahman, and et al. 2021. "Phylogenetic Analysis and Genetic Diversity of Colletotrichum falcatum Isolates Causing Sugarcane Red Rot Disease in Bangladesh" Biology 10, no. 9: 862. https://doi.org/10.3390/biology10090862
APA StyleHossain, M. I., Ahmad, K., Vadamalai, G., Siddiqui, Y., Saad, N., Ahmed, O. H., Hata, E. M., Adzmi, F., Rashed, O., Rahman, M. Z., & Kutawa, A. B. (2021). Phylogenetic Analysis and Genetic Diversity of Colletotrichum falcatum Isolates Causing Sugarcane Red Rot Disease in Bangladesh. Biology, 10(9), 862. https://doi.org/10.3390/biology10090862










