Uncovering Mechanisms of Zanthoxylum piperitum Fruits for the Alleviation of Rheumatoid Arthritis Based on Network Pharmacology
Abstract
Simple Summary
Abstract
1. Introduction
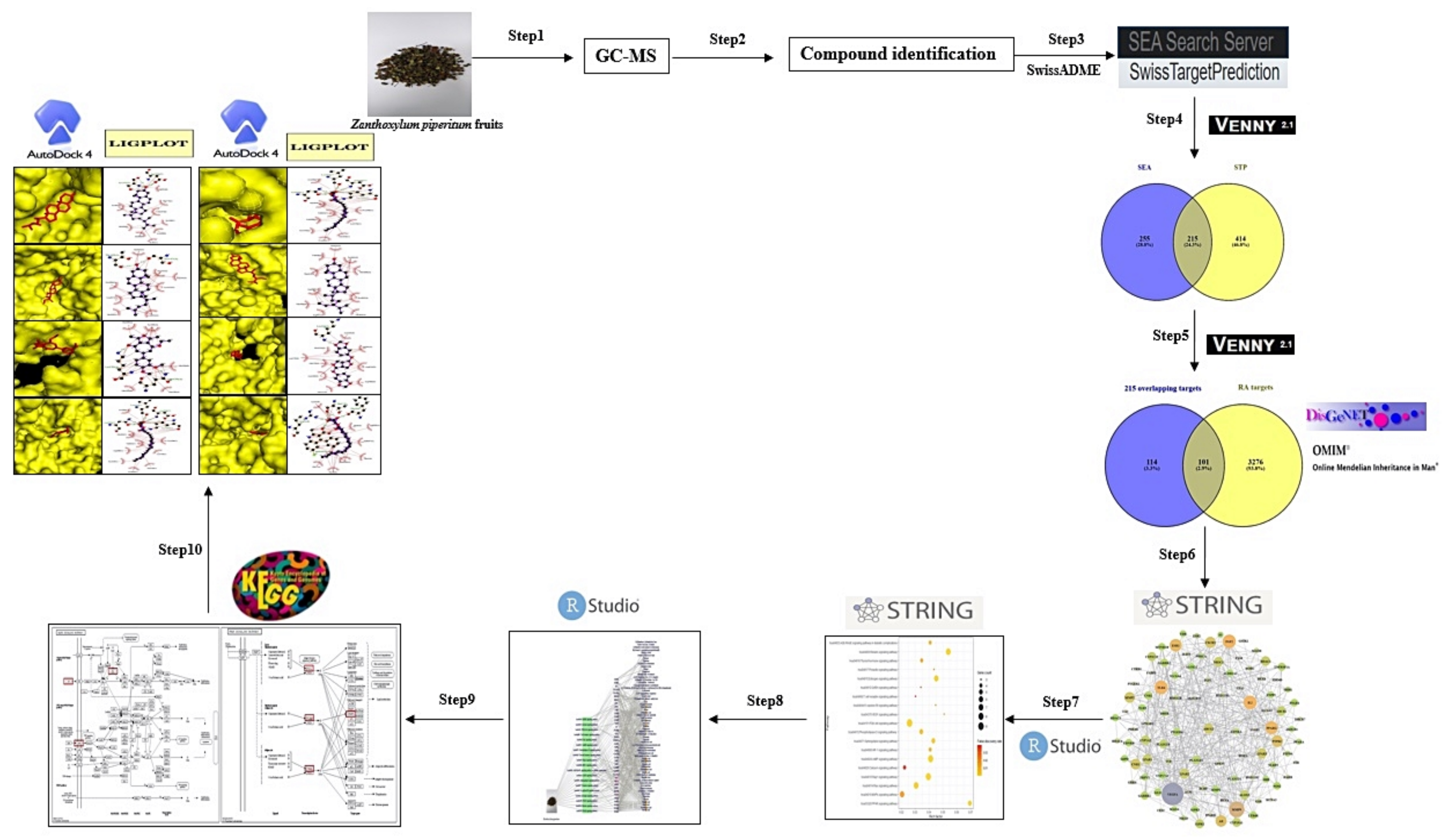
2. Materials and Methods
2.1. Plant Material Collection and Classification
2.2. Plant Preparation, Extraction
2.3. GC-MS Analysis Condition
2.4. Chemical Compounds Identification and Drug-Likeness Screening
2.5. Targets Associated with Compounds from ZPFs or Rheumatoid Arthritis
2.6. PPI Networks and Bubble Chart
2.7. Construction of STC Networks
2.8. Preparation of Targets for MDT
2.9. Preparation of Compounds from ZPFs for MDT
2.10. Preparation of Positive Standard Ligands for MDT
2.11. Ligand-Protein Docking
2.12. Toxicological Properties Prediction by AdmetSAR
3. Results
3.1. Chemical Compounds from ZPFs
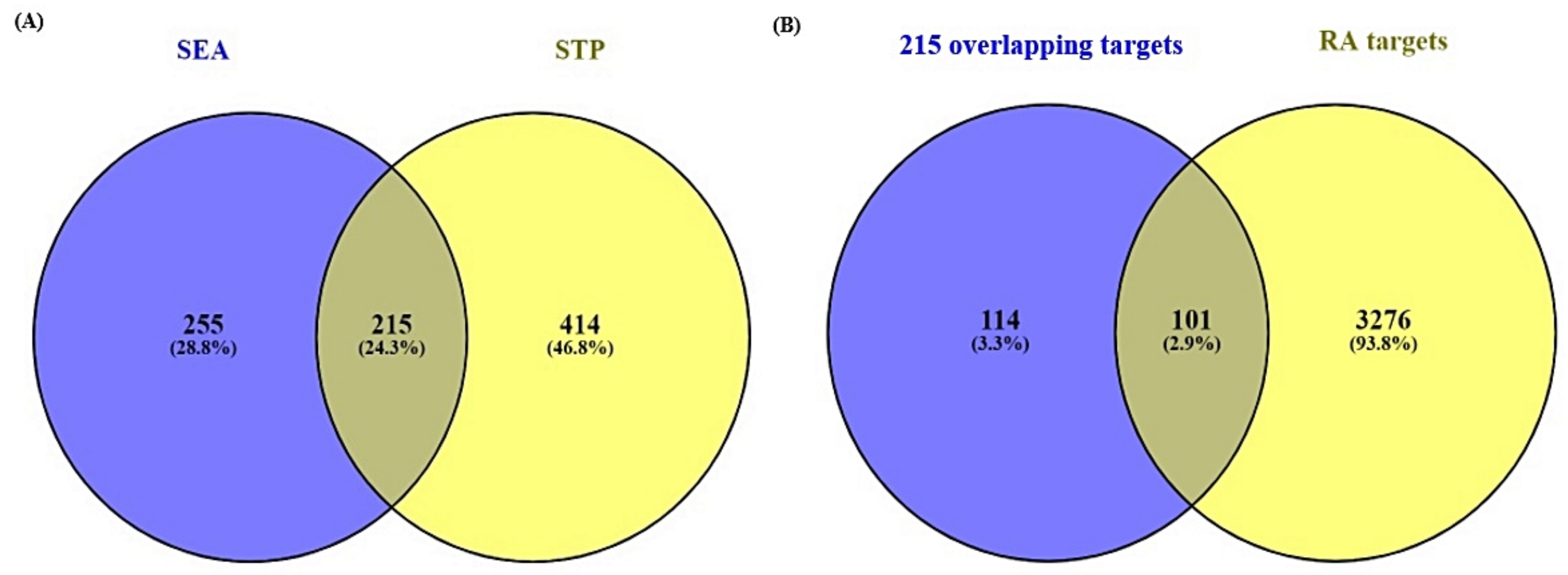
3.2. Overlapping Targets between SEA and STP Related to Chemical Compounds
3.3. Overlapping Targets between RA-Related Genes and the Final 101 Overlapping Targets
3.4. Acquisition of a Key Target from PPI Networks
3.5. Identification of Two Key Signaling Pathways from a Bubble Chart
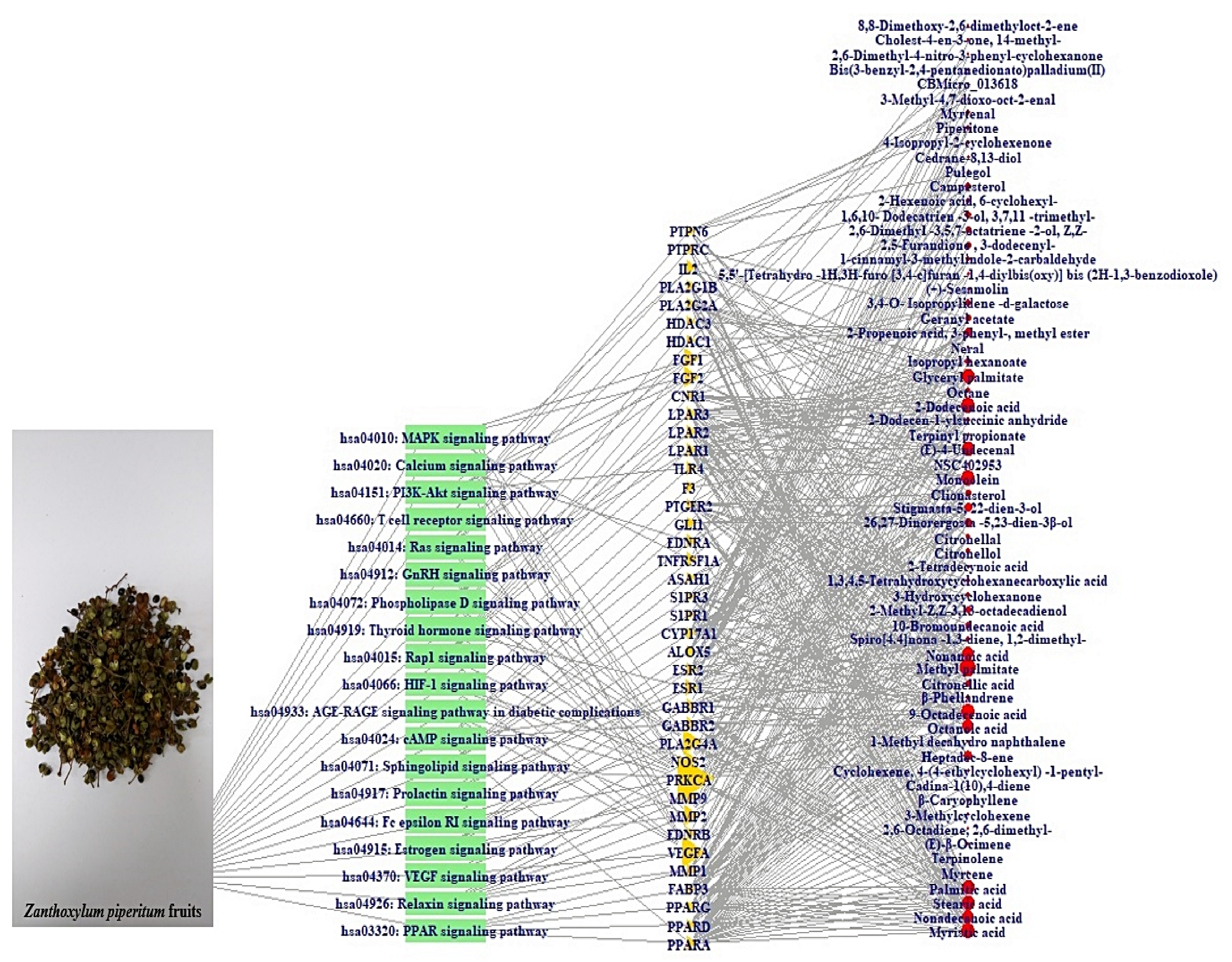
3.6. Construction of Signaling Pathway-Target-Compound Network
3.7. KEGG Pathway Enrichment Analysis
3.8. MDT of 6 Targets and 23 Chemical Compounds Associated with MAPK Signaling Pathway
3.9. MDT of 5 Targets and 43 Chemical Compounds Associated with PPAR Signaling Pathway
3.10. Identification of the Uppermost Seven Targets and Eight Compounds from Two Key Signaling Pathways against RA
3.11. Toxicological Properties of 8 Compounds
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| AGE | Advanced Glycation End-product; |
| AMPK | AMP-activated protein kinase; |
| cAMP | Cyclic AMP; |
| DMARDs | Disease-Modifying Arthritis Drugs; |
| FLSs | Fibroblast-Like Synoviocytes; |
| GC-MS | Gas Chromatography Mass Spectrum; |
| GnRH | Gonadotropin releasing hormone; |
| HIF-1 | Hypoxia Inducible Factor-1; |
| KEGG | Kyoto Encyclopedia of Genes and Genomes; |
| LPS | Lipopolysaccharide; |
| MAPK | Mitogen Activated Protein Kinase; |
| MDT | Molecular Docking Test; |
| NF-κB | Nuclear Factor Kappa B; |
| NSAIDs | Non-Steroidal Anti-Inflammatories Drugs; |
| OMIM | Online Mendelian Inheritance in Man; |
| PI3K-Akt | Phosphoinositide 3-kinase—Akt; |
| PLD | Phospholipase D; |
| PLD1 | Phospholipase D1; |
| PPAR | Peroxisome Proliferator Activated Receptor; |
| PPARA | Peroxisome Proliferator Activated Receptor Alpha; |
| PPARD | Peroxisome Proliferator Activated Receptor Delta; |
| PPARG | Peroxisome Proliferator Activated Receptor Gamma; |
| PPI | Protein-protein interaction; |
| PRKCA | Protein Kinase C Alpha; |
| RA | Rheumatoid Arthritis; |
| RAGE | The Receptor for Advanced Glycation End Products; |
| Rap1 | Ras-associated protein-1; |
| SEA | Similarity Ensemble Approach; |
| SMILES | Simplified Molecular Input Line Entry System; |
| STP | SwissTargetPrediction; |
| VEGF | Vascular Endothelial Growth Factor; |
| VEGFA | Vascular Endothelial Growth Factor A; |
| ZP | Zanthoxylum piperitum; |
| ZPFs | Zanthoxylum piperitum fruits |
References
- Guo, Q.; Wang, Y.; Xu, D.; Nossent, J.; Pavlos, N.J.; Xu, J. Rheumatoid arthritis: Pathological mechanisms and modern pharmacologic therapies. Bone Res. 2018, 6, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Heidari, B. Rheumatoid arthritis: Early diagnosis and treatment outcomes. Casp. J. Intern. Med. 2011, 2, 161–170. [Google Scholar]
- El Miedany, Y.; Youssef, S.; Mehanna, A.N.; El Gaafary, M. Development of a scoring system for assessment of outcome of early undifferentiated inflammatory synovitis. Jt. Bone Spine 2008, 75, 155–162. [Google Scholar] [CrossRef]
- McInnes, I.B.; Schett, G. Pathogenetic insights from the treatment of rheumatoid arthritis. Lancet 2017, 389, 2328–2337. [Google Scholar] [CrossRef]
- Noack, M.; Miossec, P. Selected cytokine pathways in rheumatoid arthritis. Semin. Immunopathol. 2017, 39, 365–383. [Google Scholar] [CrossRef] [PubMed]
- Scherer, H.U.; Häupl, T.; Burmester, G.R. The etiology of rheumatoid arthritis. J. Autoimmun. 2020, 110, 102400. [Google Scholar] [CrossRef]
- Kumar, P.; Banik, S. Pharmacotherapy options in rheumatoid arthritis. Clin. Med. Insights: Arthritis Musculoskelet. Disord. 2013, 6, 35–43. [Google Scholar] [CrossRef]
- Grove, M.L.; Hassell, A.B.; Hay, E.M.; Shadforth, M.F. Adverse reactions to disease-modifying anti-rheumatic drugs in clinical practice. QJM Mon. J. Assoc. Physicians 2001, 94, 309–319. [Google Scholar] [CrossRef] [PubMed]
- Wongrakpanich, S.; Wongrakpanich, A.; Melhado, K.; Rangaswami, J. A comprehensive review of non-steroidal anti-inflammatory drug use in the elderly. Aging Dis. 2018, 9, 143–150. [Google Scholar] [CrossRef]
- Hajja, G.; Bahlouli, A. Medicinal plants in the prevention and treatment of rheumatoid arthritis. MOJ Bioequivalence Bioavailab. 2018, 5. [Google Scholar] [CrossRef]
- Farzaei, M.H.; Farzaei, F.; Abdollahi, M.; Abbasabadi, Z.; Abdolghaffari, A.H.; Mehraban, B. A mechanistic review on medicinal plants used for rheumatoid arthritis in traditional Persian medicine. J. Pharm. Pharmacol. 2016, 68, 1233–1248. [Google Scholar] [CrossRef]
- Chung, M.-S. Volatile Compounds of Zanthoxylum piperitum A.P. DC. Food Sci. Biotechnol. 2005, 14, 529–532. [Google Scholar]
- Lee, J.-H.; Chang, K.-M.; Kim, G.-H. Anti-inflammatory Activities of Chopi (Zanthoxylum piperitum A.P. DC) Essential Oil: Suppression of the Inducible Nitric Oxide Synthase and Cellular Adhesion. Food Sci. Biotechnol. 2009, 18, 1371–1378. [Google Scholar]
- Hee, C.Y.; Yil, M.N. The Anti-inflammatory Mechanism of the Peel of Zanthoxylum piperitum D.C. is by Suppressing NF-κB/Caspase-1 Activation in LPS-Induced RAW264.7 Cells. Korean J. Plant Reources. 2019, 32, 669–676. [Google Scholar]
- Oh, K.K.; Adnan, M.; Cho, D.H. Network pharmacology of bioactives from Sorghum bicolor with targets related to diabetes mellitus. PLoS ONE 2020, 15, e0240873. [Google Scholar] [CrossRef]
- Oh, K.K.; Adnan, M.; Cho, D.H. Network pharmacology approach to decipher signaling pathways associated with target proteins of NSAIDs against COVID-19. Sci. Rep. 2021, 11, 9606. [Google Scholar] [CrossRef]
- Oh, K.-K.; Adnan, M.; Cho, D.-H. Network Pharmacology Study to Interpret Signaling Pathways of Ilex cornuta Leaves against Obesity. Processes 2021, 9, 1106. [Google Scholar] [CrossRef]
- Chandran, U.; Mehendale, N.; Patil, S.; Chaguturu, R.; Patwardhan, B. Network Pharmacology. In Innovative Approaches in Drug Discovery: Ethnopharmacology, Systems Biology and Holistic Targeting; Elsevier Inc.: Amsterdam, The Netherlands, 2017; pp. 127–164. ISBN 9780128018224. [Google Scholar]
- Lai, X.; Wang, X.; Hu, Y.; Su, S.; Li, W.; Li, S. Editorial: Network Pharmacology and Traditional Medicine. Front. Pharmacol. 2020, 11, 1194. [Google Scholar] [CrossRef]
- Oh, K.K.; Adnan, M.; Ju, I.; Cho, D.H. A network pharmacology study on main chemical compounds fromHibiscus cannabinusL. leaves. RSC Adv. 2021, 11, 11062–11082. [Google Scholar] [CrossRef]
- Keiser, M.J.; Roth, B.L.; Armbruster, B.N.; Ernsberger, P.; Irwin, J.J.; Shoichet, B.K. Relating protein pharmacology by ligand chemistry. Nat. Biotechnol. 2007, 25, 197–206. [Google Scholar] [CrossRef]
- Daina, A.; Michielin, O.; Zoete, V. SwissTargetPrediction: Updated data and new features for efficient prediction of protein targets of small molecules. Nucleic Acids Res. 2019, 47, W357–W3664. [Google Scholar] [CrossRef] [PubMed]
- Szklarczyk, D.; Gable, A.L.; Lyon, D.; Junge, A.; Wyder, S.; Huerta-Cepas, J.; Simonovic, M.; Doncheva, N.T.; Morris, J.H.; Bork, P.; et al. STRING v11: Protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 2019, 47, D607–D613. [Google Scholar] [CrossRef]
- Khanal, P.; Patil, B.M.; Chand, J.; Naaz, Y. Anthraquinone Derivatives as an Immune Booster and their Therapeutic Option Against COVID-19. Nat. Prod. Bioprospecting 2020, 10, 325–335. [Google Scholar] [CrossRef] [PubMed]
- Kanehisa, M.; Furumichi, M.; Sato, Y.; Ishiguro-Watanabe, M.; Tanabe, M. KEGG: Integrating viruses and cellular organisms. Nucleic Acids Res. 2021, 49, D545–D551. [Google Scholar] [CrossRef]
- Barton, A.; Woolmore, J.A.; Ward, D.; Eyre, S.; Hinks, A.; Ollier, W.E.R.; Strange, R.C.; Fryer, A.A.; John, S.; Hawkins, C.P.; et al. Association of protein kinase C alpha (PRKCA) gene with multiple sclerosis in a UK population. Brain 2004, 127, 1717–1722. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Makino, H.; Seki, S.; Yahara, Y.; Shiozawa, S.; Aikawa, Y.; Motomura, H.; Nogami, M.; Watanabe, K.; Sainoh, T.; Ito, H.; et al. A selective inhibition of c-Fos/activator protein-1 as a potential therapeutic target for intervertebral disc degeneration and associated pain. Sci. Rep. 2017, 7, 1–14. [Google Scholar] [CrossRef]
- Arias de la Rosa, I.; Escudero-Contreras, A.; Rodríguez-Cuenca, S.; Ruiz-Ponce, M.; Jiménez-Gómez, Y.; Ruiz-Limón, P.; Pérez-Sánchez, C.; Ábalos-Aguilera, M.C.; Cecchi, I.; Ortega, R.; et al. Defective glucose and lipid metabolism in rheumatoid arthritis is determined by chronic inflammation in metabolic tissues. J. Intern. Med. 2018, 284, 61–77. [Google Scholar] [CrossRef]
- Shityakov, S.; Förster, C. In silico predictive model to determine vector-mediated transport properties for the blood-brain barrier choline transporter. Adv. Appl. Bioinform. Chem. 2014, 7, 23–36. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Wang, J.; Luo, S.; Zhan, Y.; Lu, Q. The roles of PPARγ and its agonists in autoimmune diseases: A comprehensive review. J. Autoimmun. 2020, 113, 102510. [Google Scholar] [CrossRef] [PubMed]
- Yamasaki, S.; Nakashima, T.; Kawakami, A.; Miyashita, T.; Ida, H.; Migita, K.; Nakata, K.; Eguchi, K. Functional changes in rheumatoid fibroblast-like synovial cells through activation of peroxisome proliferator-activated receptor γ-mediated signalling pathway. Clin. Exp. Immunol. 2002, 129, 379–384. [Google Scholar] [CrossRef]
- Szekanecz, Z.; Koch, A.E. VEGF as an activity marker in rheumatoid arthritis. Int. J. Clin. Rheumatol. 2010, 5, 287–289. [Google Scholar] [CrossRef]
- Fernanda Romo-García, M.; Zapata-Zuñiga, M.; Antonio Enciso-Moreno, J.; Enrique Castañeda-Delgado, J. The Role of Estrogens in Rheumatoid Arthritis Physiopathology. In Rheumatoid Arthritis—Other Perspectives towards a Better Practice; IntechOpen: London, UK, 2020. [Google Scholar]
- Huang, L.; Lv, Q.; Xie, D.; Shi, T.; Wen, C. Deciphering the Potential Pharmaceutical Mechanism of Chinese Traditional Medicine (Gui-Zhi-Shao-Yao-Zhi-Mu) on Rheumatoid Arthritis. Sci. Rep. 2016, 6, 1–12. [Google Scholar] [CrossRef]
- Tang, M.W.; Reedquist, K.A.; Garcia, S.; Fernandez, B.M.; Codullo, V.; Vieira-Sousa, E.; Goffin, V.; Reuwer, A.Q.; Twickler, M.T.; Gerlag, D.M.; et al. The prolactin receptor is expressed in rheumatoid arthritis and psoriatic arthritis synovial tissue and contributes to macrophage activation. Rheumatology 2016, 55, 2248–2259. [Google Scholar] [CrossRef]
- Miltenberger-Miltenyi, G.; Cruz-Machado, A.R.; Saville, J.; Conceição, V.A.; Calado, Â.; Lopes, I.; Fuller, M.; Fonseca, J.E. Increased monohexosylceramide levels in the serum of established rheumatoid arthritis patients. Rheumatology 2020, 59, 2085–2089. [Google Scholar] [CrossRef]
- Lorenowicz, M.J.; Fernandez-Borja, M.; Hordijk, P.L. cAMP signaling in leukocyte transendothelial migration. Arterioscler. Thromb. Vasc. Biol. 2007, 27, 1014–1022. [Google Scholar] [CrossRef]
- Quiñonez-Flores, C.M.; González-Chávez, S.A.; Pacheco-Tena, C. Hypoxia and its implications in rheumatoid arthritis. J. Biomed. Sci. 2016, 23, 1–9. [Google Scholar] [CrossRef]
- Remans, P.; Reedquist, K.; Bos, J.; Verweij, C.; Breedveld, F.; van Laar, J.; Gringhuis, S. Deregulated Ras and Rap1 signaling in rheumatoid arthritis T cells leads to persistent production of free radicals. Arthritis Res. Ther. 2002, 4, 1–38. [Google Scholar] [CrossRef]
- Silman, A.J.; Ollier, W.E.R.; Bubel, M.A. Autoimmune thyroid disease and thyroid autoantibodies in rheumatoid arthritis patients and their families. Rheumatology 1989, 28, 18–21. [Google Scholar] [CrossRef]
- Mancini, A.; Di Segni, C.; Raimondo, S.; Olivieri, G.; Silvestrini, A.; Meucci, E.; Currò, D. Thyroid Hormones, Oxidative Stress, and Inflammation. Mediat. Inflamm. 2016, 2016, 1–12. [Google Scholar] [CrossRef]
- Yoo, H.J.; Hwang, W.C.; Min, D.S. Targeting of phospholipase d1 ameliorates collagen-induced arthritis via modulation of treg and Th17 cell imbalance and suppression of osteoclastogenesis. Int. J. Mol. Sci. 2020, 21, 3230. [Google Scholar] [CrossRef]
- Kåss, A.; Hollan, I.; Fagerland, M.W.; Gulseth, H.C.; Torjesen, P.A.; Førre, Ø.T. Rapid anti-inflammatory effects of gonadotropin-releasing hormone antagonism in rheumatoid arthritis patients with high gonadotropin levels in the Agra trial. PLoS ONE 2015, 10. [Google Scholar] [CrossRef] [PubMed]
- Zayoud, M.; Marcu-Malina, V.; Vax, E.; Jacob-Hirsch, J.; Elad-Sfadia, G.; Barshack, I.; Kloog, Y.; Goldstein, I. Ras signaling inhibitors attenuate disease in adjuvant-induced arthritis via targeting pathogenic antigen-specific Th17-type cells. Front. Immunol. 2017, 8, 799. [Google Scholar] [CrossRef]
- Mellado, M.; Martínez-Muñoz, L.; Cascio, G.; Lucas, P.; Pablos, J.L.; Rodríguez-Frade, J.M. T cell migration in rheumatoid arthritis. Front. Immunol. 2015, 6, 384. [Google Scholar] [CrossRef]
- Ito, K.; Caramori, G.; Adcock, I.M. Therapeutic potential of phosphatidylinositol 3-kinase inhibitors in inflammatory respiratory disease. J. Pharmacol. Exp. Ther. 2007, 321, 1–8. [Google Scholar] [CrossRef]
- Smith, M.D.; Weedon, H.; Papangelis, V.; Walker, J.; Roberts-Thomson, P.J.; Ahern, M.J. Apoptosis in the rheumatoid arthritis synovial membrane: Modulation by disease-modifying anti-rheumatic drug treatment. Rheumatology 2010, 49, 862–875. [Google Scholar] [CrossRef]
- Ye, Z.; Shen, Y.; Jin, K.; Qiu, J.; Hu, B.; Jadhav, R.R.; Sheth, K.; Weyand, C.M.; Goronzy, J.J. Arachidonic acid-regulated calcium signaling in T cells from patients with rheumatoid arthritis promotes synovial inflammation. Nat. Commun. 2021, 12, 1–17. [Google Scholar] [CrossRef]
- Li, Z.Z.; Tan, J.P.; Wang, L.L.; Li, Q.H. Andrographolide Benefits Rheumatoid Arthritis via Inhibiting MAPK Pathways. Inflammation 2017, 40, 1599–1605. [Google Scholar] [CrossRef]
- Ali, I.; Manzoor, Z.; Koo, J.E.; Moon, S.R.; Byeon, S.H.; Yoo, E.S.; Kang, H.K.; Hyun, J.W.; Lee, N.H.; Koh, Y.S. Monoolein, isolated from Ishige sinicola, inhibits lipopolysaccharide-induced inflammatory response by attenuating mitogen-activated protein kinase and NF-κB pathways. Food Sci. Biotechnol. 2017, 26, 507–511. [Google Scholar] [CrossRef]
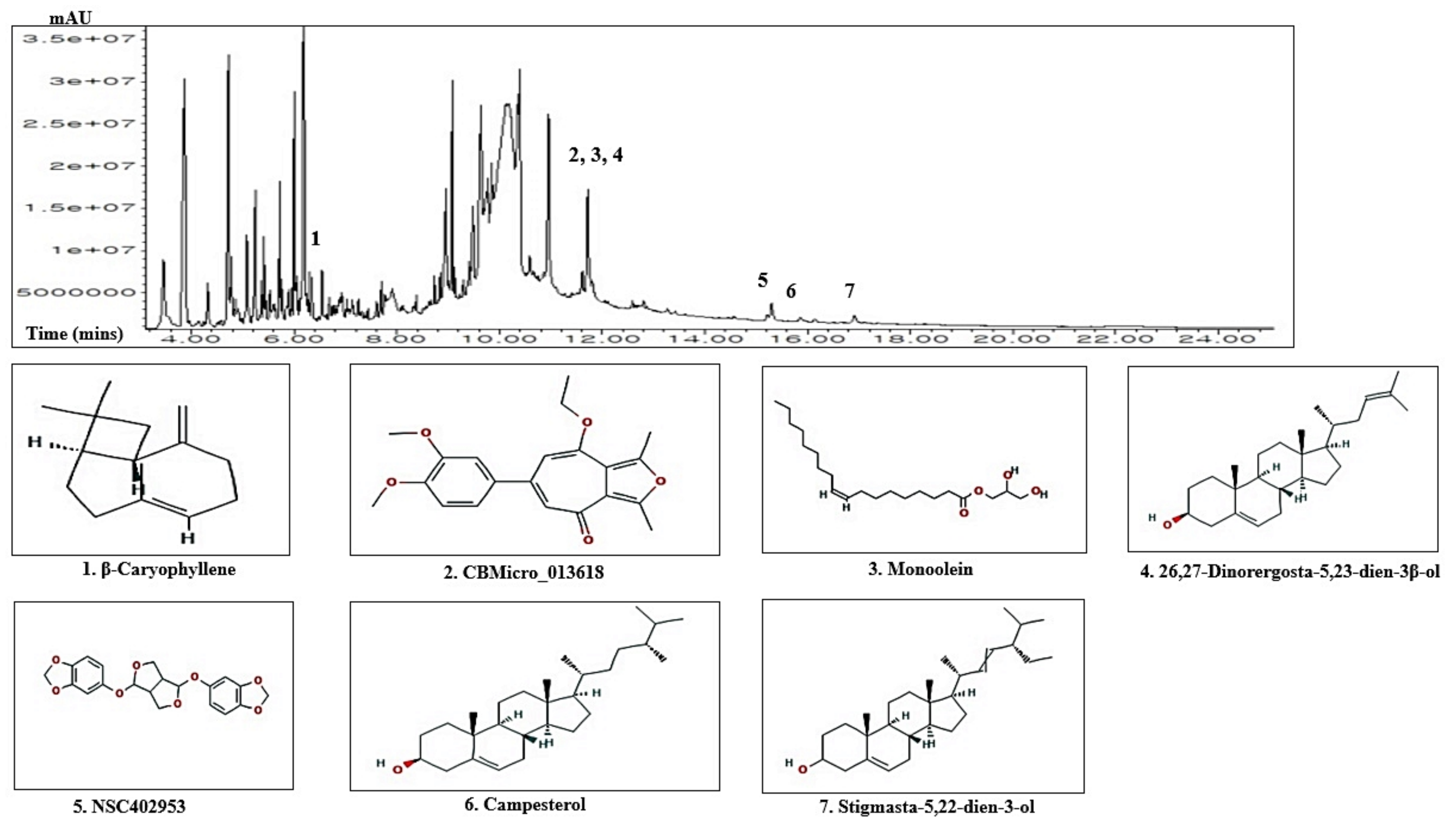


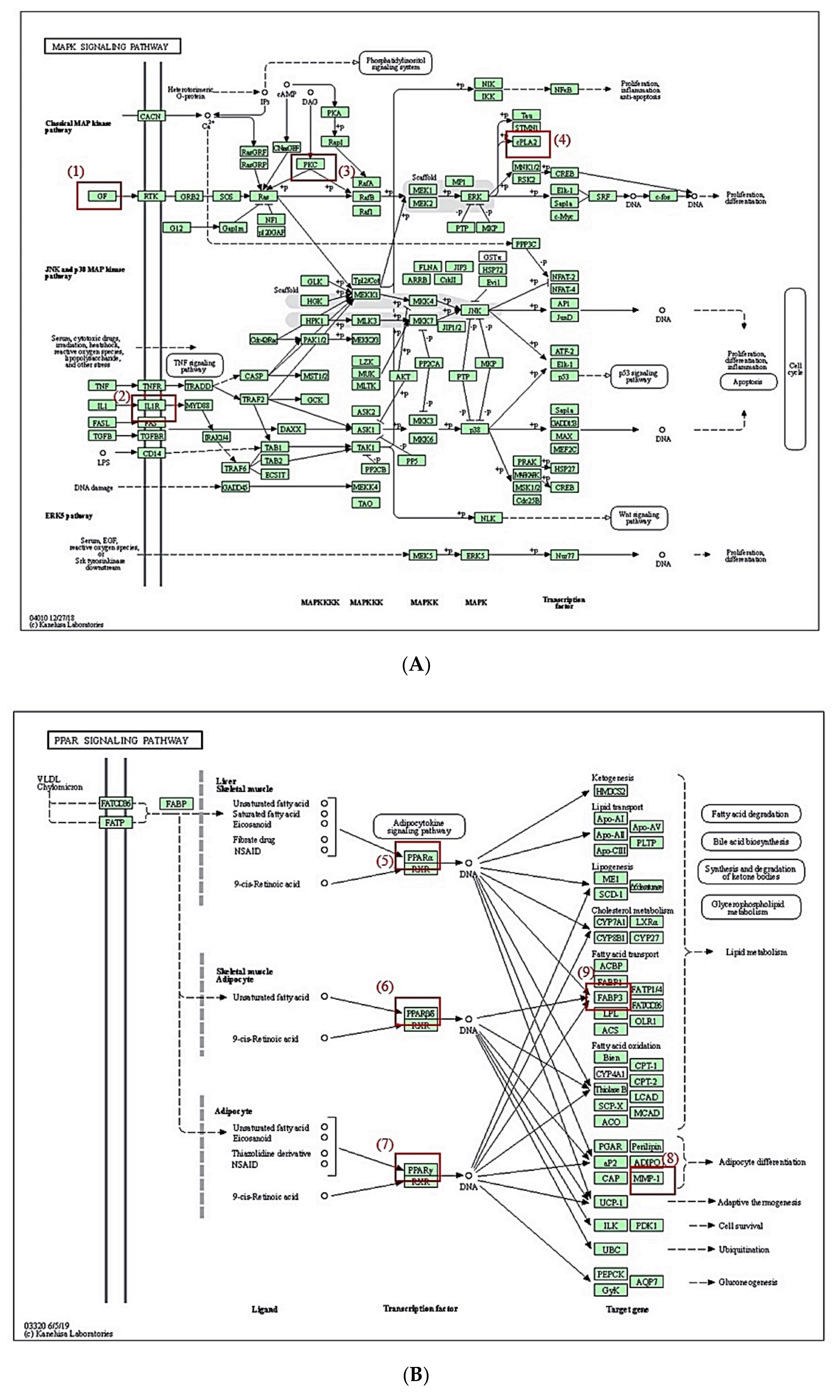


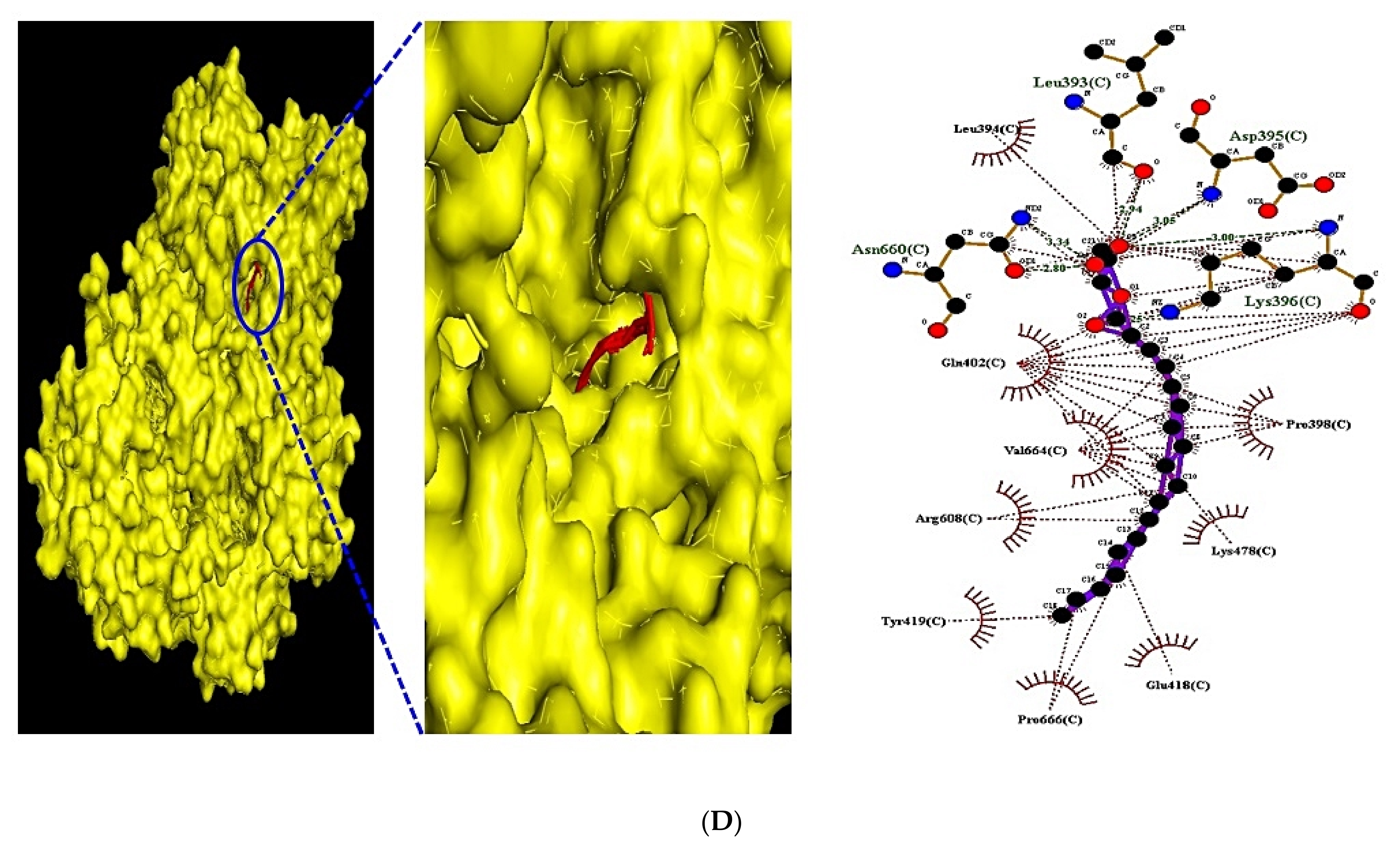
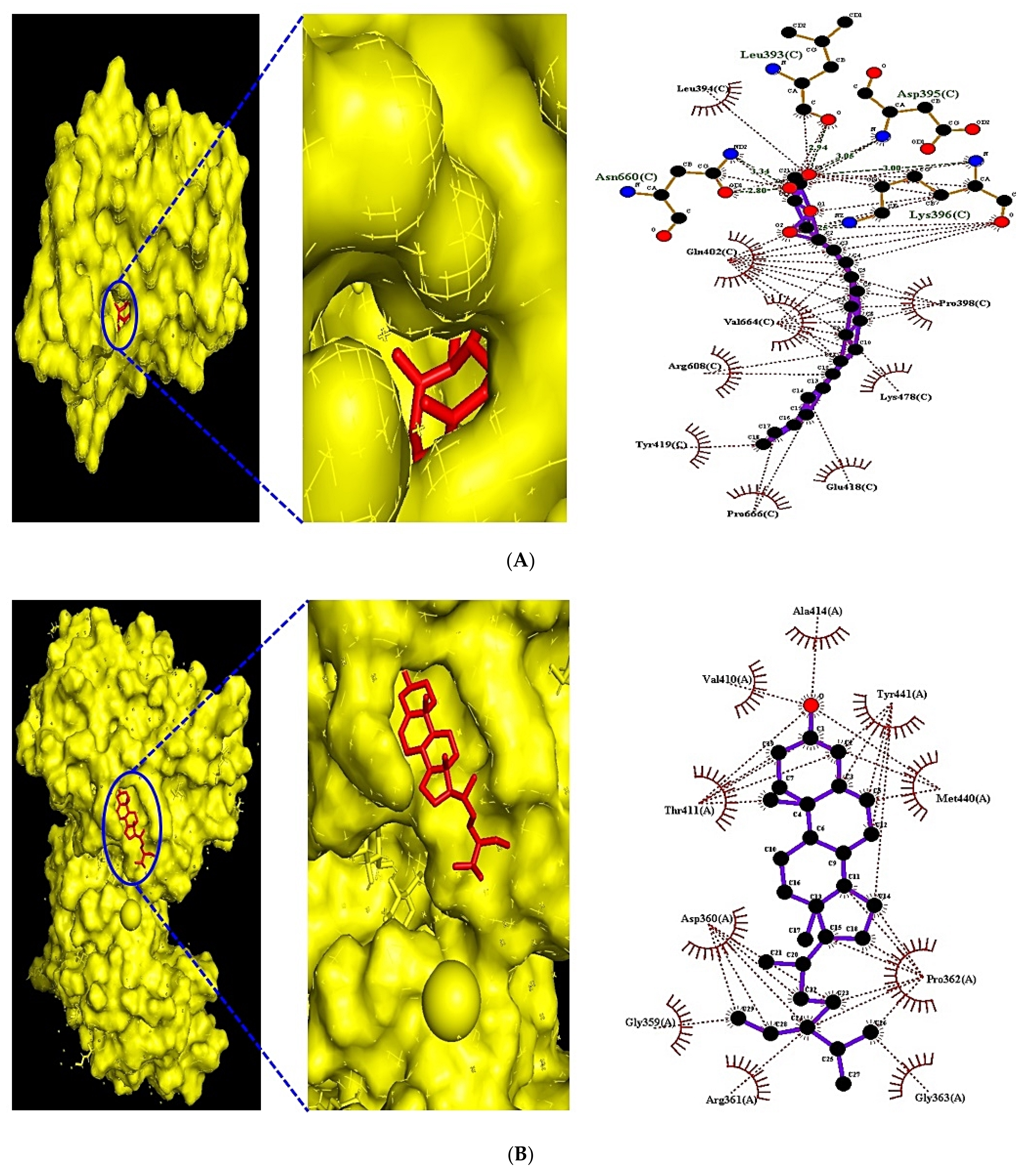
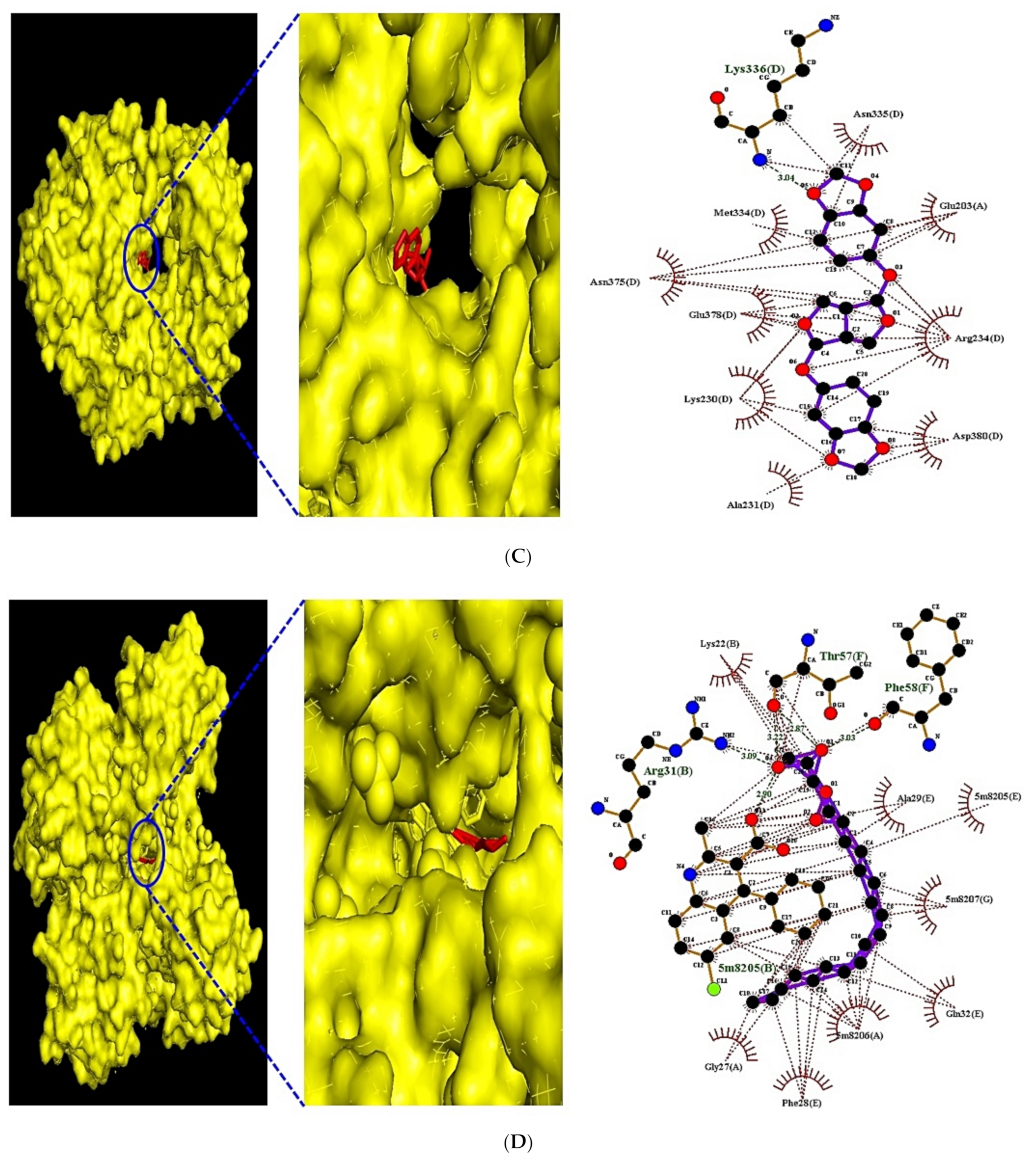

| No. | Compounds | Pubchem ID | RT (mins) | Area (%) | Pharmacological Activities (Reference) |
|---|---|---|---|---|---|
| 1 | Myrcene | 31253 | 3.462 | 1.49 | Antibacterial, Antioxidant, Fungicide |
| 2 | 3(5)-[[1,2-Dihydroxy-3-propoxy]methyl]-4-hydroxy-1H-pyrazole-5(3)-carboxamide | 135747301 | 3.683 | 0.09 | No reported |
| 3 | β-Phellandrene | 11142 | 3.866 | 4.28 | Fungicide |
| 4 | Hex-3-yne | 13568 | 3.971 | 0.10 | No reported |
| 5 | 3-Hydroxycyclohexanone | 439950 | 4.087 | 0.06 | No reported |
| 6 | Isopropyl hexanoate | 16832 | 4.145 | 0.12 | No reported |
| 7 | Terpinolene | 11463 | 4.250, 4.318 | 0.59 | Fungicide, Antioxidant |
| 8 | Vinylcyclooctane | 93331 | 4.520 | 0.07 | No reported |
| 9 | 2-Tetradecynoic acid | 324386 | 4.587 | 0.10 | No reported |
| 10 | Citronellal | 7794 | 4.721 | 2.75 | Antibacterial, Fungicide |
| 11 | 3-Hydroxy-2,3-dihydromaltol | 119838 | 4.779 | 0.67 | No reported |
| 12 | Pulegol | 92793 | 4.856 | 0.29 | No reported |
| 13 | Octanoic acid | 379 | 4.923 | 0.24 | Candidicide, Fungicide |
| 14 | (E)-4-Undecenal | 5283357 | 4.981 | 0.10 | No reported |
| 15 | 4-Isopropyl-2-cyclohexenone | 92780 | 5.087 | 1.04 | No reported |
| 16 | Citronellol | 8842 | 5.241 | 1.20 | Antibacterial, Candidicide, Sedative |
| 17 | (E)-beta-Ocimene | 5281553 | 5.366 | 0.39 | Insecticide |
| 18 | 3,7-Dimethylocta-2,6-dien-1-ol | 4458 | 5.414 | 0.65 | No reported |
| 19 | Spiro[4 .4]nona-1,3-diene, 1,2-dimethyl- | 570800 | 5.452 | 0.21 | No reported |
| 20 | Piperitone | 6987 | 5.529 | 0.41 | Antiasthmatic |
| 21 | Nonanoic acid | 8158 | 5.606 | 0.42 | Perfumery |
| 22 | 8,8-Dimethoxy-2,6-dimethyloct-2-ene | 102507 | 5.721 | 0.98 | No reported |
| 23 | p-Isopropylbenzyl formate | 105515 | 5.760 | 0.40 | No reported |
| 24 | Citronellic acid | 10402 | 5.895 | 0.55 | No reported |
| 25 | α-Terpinene | 7462 | 5.952 | 0.24 | Antispasmodic |
| 26 | 2,6-Octadiene, 2,6-dimethyl- | 5365898 | 6.000 | 1.60 | No reported |
| 27 | Terpinyl propionate | 62328 | 6.048 | 0.43 | No reported |
| 28 | Geranyl acetate | 1549026 | 6.193 | 4.60 | Sedative |
| 29 | 3-Methylcyclohexene | 11573 | 6.250 | 0.21 | No reported |
| 30 | 1,4-Dimethyl-4beta-methoxy-2,5-cyclohexadien-1α-ol | 12561656 | 6.298 | 0.33 | No reported |
| 31 | 2-Propenoic acid, 3-phenyl-, methyl ester | 7644 | 6.346 | 0.49 | No reported |
| 32 | 6-Methylenespiro[4.5]decane | 564762 | 6.471 | 0.07 | No reported |
| 33 | β-Caryophyllene | 5281515 | 6.539 | 0.67 | Antibacterial, Antiinflammation |
| 34 | Bergamotane | 86000267 | 6.625 | 0.09 | No reported |
| 35 | 3-Methyl-4,7-dioxo-oct-2-enal | 5363705 | 6.693 | 0.30 | No reported |
| 36 | 2,6-Dimethyl-3,5,7-octatriene-2-ol, Z,Z- | 5363692 | 6.779 | 0.24 | No reported |
| 37 | 2-Dodecenoic acid | 5282729 | 6.818 | 0.12 | No reported |
| 38 | 1,6,10-Dodecatrien-3-ol, 3,7,11-trimethyl- | 8888 | 6.885 | 0.35 | No reported |
| 39 | 1-Methyldecahydronaphthalene | 34193 | 6.943 | 0.46 | No reported |
| 40 | Cadina-1(10),4-diene | 10223 | 7.029 | 0.34 | No reported |
| 41 | 2-(4-Methylcyclohexyl)prop-2-en-1-ol | 543946 | 7.135 | 0.42 | No reported |
| 42 | Tetradec-13-enal | 522841 | 7.250 | 0.24 | No reported |
| 43 | 9-Octadecenoic acid | 965 | 7.308 | 0.15 | Antiinflammation, Antileukotriene |
| 44 | 1,2-Di-but-2-enyl-cyclohexane | 5367574 | 7.375 | 0.10 | No reported |
| 45 | 4,12,12-trimethyl-9-methylene-5-oxatricyclo[8.2.0.04,6]dodecane | 73555586 | 7.433 | 0.11 | No reported |
| 46 | 3,4-O-Isopropylidene-d-galactose | 54504880 | 7.568 | 0.08 | No reported |
| 47 | 2-Hexenoic acid, 6-cyclohexyl- | 5367614 | 7.616 | 0.22 | No reported |
| 48 | Heptadec-8-ene | 520230 | 7.693 | 0.35 | No reported |
| 49 | Octane | 356 | 7.779 | 0.35 | No reported |
| 50 | Myristic acid | 11005 | 7.827, 8.096 | 0.44 | Anticancer, Antioxidant |
| 51 | D-(-)-Kinic Acid | 1064 | 7.914 | 1.35 | No reported |
| 52 | Nonadecanoic acid | 12591 | 8.135 | 0.35 | No reported |
| 53 | 10-Bromoundecanoic acid | 543401 | 8.337, 8.385 | 0.77 | No reported |
| 54 | Stearic acid | 5281 | 8.520 | 0.23 | Hypocholesterolemic |
| 55 | Cysteamine S-sulfate | 76242 | 8.587, 9.231, 9.298 | 1.27 | No reported |
| 56 | Limonene dioxide | 232703 | 8.635 | 0.23 | No reported |
| 57 | 2,6-Dimethyl-4-nitro-3-phenyl-cyclohexanone | 562366 | 8.664 | 0.26 | No reported |
| 58 | Methyl palmitate | 8181 | 8.731 | 0.46 | No reported |
| 59 | 2,6-Dimethyl-1,3,6-heptatriene | 5368331 | 8.846 | 0.68 | No reported |
| 60 | Palmitic acid | 985 | 8.962, 9.020 | 2.65 | Antioxidant, Pesticide |
| 61 | Neral | 643779 | 9.077 | 1.58 | Antibacterial, Antispasmodic |
| 62 | 2-Methyl-6-methylene-1,7-octadien-3-one | 93231 | 9.125 | 0.75 | No reported |
| 63 | Bis(3-benzyl-2,4-pentanedionato)palladium(II) | 5363840 | 9.423 | 1.03 | No reported |
| 64 | Pentamethylbenzenesulfonyl chloride | 590180 | 9.491, 9.635 | 6.52 | No reported |
| 65 | Myrtenal | 61130 | 9.769 | 3.77 | Antimalarial, Antiplasmodial |
| 66 | N,N-Dimethyl-2-phenylethen-1-amine | 23277871 | 10.154, 10.183 | 20.61 | No reported |
| 67 | Allyl(chloromethyl)dimethylsilane | 556526 | 10.394 | 7.64 | No reported |
| 68 | Cyclohexene, 4-(4-ethylcyclohexyl)-1-pentyl- | 543386 | 10.596 | 1.64 | No reported |
| 69 | 3-Epicycloeucalenol | 543796 | 10.654 | 1.09 | No reported |
| 70 | 2,5-Furandione, 3-dodecenyl- | 5362708 | 10.750 | 0.61 | No reported |
| 71 | 1-cinnamyl-3-methylindole-2-carbaldehyde | N/A | 10.875 | 1.35 | No reported |
| 72 | Glyceryl palmitate | 14900 | 10.962 | 4.82 | No reported |
| 73 | 2-Methyl-Z,Z-3,13-octadecadienol | 5364412 | 11.414 | 0.37 | No reported |
| 74 | Pentadeca-2,3,6,9,12,13-hexaen-8-one, 2,5,5,11,11,14-hexamethyl- | 5370200 | 11.519 | 0.51 | No reported |
| 75 | CBMicro_013618 | 1109374 | 11.616 | 1.04 | No reported |
| 76 | Monoolein | 5283468 | 11.721 | 2.02 | Antioxidant |
| 77 | Cyclohexene, 4-(4-ethylcyclohexyl)-1-pentyl- | 543386 | 11.808, 12.596 | 1.56 | No reported |
| 78 | Cedrane-8,13-diol | 188457 | 12.654 | 0.12 | No reported |
| 79 | 26,27-Dinorergosta-5,23-dien-3β-ol | 22213488 | 12.721 | 0.18 | No reported |
| 80 | Cholest-4-en-3-one, 14-methyl- | 277841 | 13.279 | 0.07 | No reported |
| 81 | (+)-Sesamolin | 585998 | 15.221 | 0.08 | No reported |
| 82 | NSC402953 | 345349 | 15.308 | 0.32 | No reported |
| 83 | Campesterol | 173183 | 15.866 | 0.12 | Antioxidant, Hypocholesterolemic |
| 84 | Stigmasta-5,22-dien-3-ol | 53870683 | 16.144 | 0.06 | Antimicrobial, Antioxidant, Antidiabetic |
| 85 | Clionasterol | 457801 | 16.923 | 0.15 | Anticancer, Antidaibetic, Antioxidant |
| No. | Compounds | Lipinski Rules | Lipinski’s Violations | Bioavailability Score | TPSA(Ų) | |||
|---|---|---|---|---|---|---|---|---|
| MW | HBA | HBD | MLog P | |||||
| <500 | <10 | ≤5 | ≤4.15 | ≤1 | > 0.1 | <140 | ||
| 1 | Myrcene | 136.23 | 0 | 0 | 3.56 | 0 | 0.55 | 0.00 |
| 2 | 3(5)-[[1,2-Dihydroxy-3-propoxy]methyl]-4-hydroxy-1H-pyrazole-5(3)-carboxamide | 231.21 | 6 | 5 | −2.70 | 0 | 0.55 | 141.69 |
| 3 | β-Phellandrene | 136.23 | 0 | 0 | 3.27 | 0 | 0.55 | 0.00 |
| 4 | Hex-3-yne | 82.14 | 0 | 0 | 3.37 | 0 | 0.55 | 0.00 |
| 5 | 3-Hydroxycyclohexanone | 114.14 | 2 | 1 | 0.07 | 0 | 0.55 | 37.30 |
| 6 | Isopropyl hexanoate | 158.24 | 2 | 0 | 2.28 | 0 | 0.55 | 26.30 |
| 7 | Terpinolene | 136.23 | 0 | 0 | 3.27 | 0 | 0.55 | 0.00 |
| 8 | Vinylcyclooctane | 138.25 | 0 | 0 | 4.29 | 1 | 0.55 | 0.00 |
| 9 | 2-Tetradecynoic acid | 224.34 | 2 | 1 | 3.58 | 0 | 0.85 | 37.30 |
| 10 | Citronellal | 154.25 | 1 | 0 | 2.59 | 0 | 0.55 | 17.07 |
| 11 | 3-Hydroxy-2,3-dihydromaltol | 144.13 | 4 | 2 | −1.77 | 0 | 0.85 | 66.76 |
| 12 | Pulegol | 154.25 | 1 | 1 | 2.30 | 0 | 0.55 | 20.23 |
| 13 | Octanoic acid | 144.21 | 2 | 1 | 1.96 | 0 | 0.85 | 37.30 |
| 14 | (E)-4-Undecenal | 168.28 | 1 | 0 | 2.88 | 0 | 0.55 | 17.07 |
| 15 | 4-Isopropyl-2-cyclohexenone | 138.21 | 1 | 0 | 1.89 | 0 | 0.55 | 17.07 |
| 16 | Citronellol | 156.27 | 1 | 1 | 2.70 | 0 | 0.55 | 20.23 |
| 17 | (E)--Ocimene | 136.23 | 0 | 0 | 3.56 | 0 | 0.55 | 0.00 |
| 18 | 3,7-Dimethylocta-2,6-dien-1-ol | 154.25 | 1 | 1 | 2.59 | 0 | 0.55 | 20.23 |
| 19 | Spiro[4.4]nona-1,3-diene, 1,2-dimethyl- | 148.24 | 0 | 0 | 3.56 | 0 | 0.55 | 0.00 |
| 20 | Piperitone | 152.23 | 1 | 0 | 2.20 | 0 | 0.55 | 17.07 |
| 21 | Nonanoic acid | 158.24 | 2 | 1 | 2.28 | 0 | 0.85 | 37.30 |
| 22 | 8,8-Dimethoxy-2,6-dimethyloct-2-ene | 200.32 | 2 | 0 | 2.75 | 0 | 0.55 | 18.46 |
| 23 | p-Isopropylbenzyl formate | 178.23 | 2 | 0 | 2.58 | 0 | 0.55 | 26.30 |
| 24 | Citronellic acid | 170.25 | 2 | 1 | 2.47 | 0 | 0.85 | 37.30 |
| 25 | alpha-Terpinene | 136.23 | 0 | 0 | 3.27 | 0 | 0.55 | 0.00 |
| 26 | 2,6-Octadiene, 2,6-dimethyl- | 138.25 | 0 | 0 | 3.66 | 0 | 0.55 | 0.00 |
| 27 | Terpinyl propionate | 210.31 | 2 | 0 | 2.92 | 0 | 0.55 | 26.30 |
| 28 | Geranyl acetate | 196.29 | 2 | 0 | 2.95 | 0 | 0.55 | 26.30 |
| 29 | 3-Methylcyclohexene | 96.17 | 0 | 0 | 3.33 | 0 | 0.55 | 0.00 |
| 30 | 1,4-Dimethyl-4β-methoxy-2,5-cyclohexadien-1α-ol | 154.21 | 2 | 1 | 0.97 | 0 | 0.55 | 29.46 |
| 31 | 2-Propenoic acid, 3-phenyl-, methyl ester | 162.19 | 2 | 0 | 2.20 | 0 | 0.55 | 26.30 |
| 32 | 6-Methylenespiro[4.5]decane | 150.26 | 0 | 0 | 4.58 | 1 | 0.55 | 0.00 |
| 33 | beta-Caryophyllene | 204.35 | 0 | 0 | 4.63 | 1 | 0.55 | 0.00 |
| 34 | Bergamotane | 208.38 | 0 | 0 | 5.80 | 1 | 0.55 | 0.00 |
| 35 | 3-Methyl-4,7-dioxo-oct-2-enal | 168.19 | 3 | 0 | 0.29 | 0 | 0.55 | 51.21 |
| 36 | 2,6-Dimethyl-3,5,7-octatriene-2-ol, Z,Z- | 152.23 | 1 | 1 | 2.49 | 0 | 0.55 | 20.23 |
| 37 | 2-Dodecenoic acid | 198.30 | 2 | 1 | 3.04 | 0 | 0.85 | 37.30 |
| 38 | 1,6,10-Dodecatrien-3-ol, 3,7,11-trimethyl- | 222.37 | 1 | 1 | 3.86 | 0 | 0.55 | 20.23 |
| 39 | 1-Methyldecahydronaphthalene | 152.28 | 0 | 0 | 4.72 | 1 | 0.55 | 0.00 |
| 40 | Cadina-1(10),4-diene | 204.35 | 1 | 0 | 4.63 | 1 | 0.55 | 0.00 |
| 41 | 2-(4-Methylcyclohexyl)prop-2-en-1-ol | 154.25 | 1 | 1 | 2.30 | 0 | 0.55 | 20.23 |
| 42 | Tetradec-13-enal | 210.36 | 1 | 0 | 3.70 | 0 | 0.55 | 17.07 |
| 43 | 9-Octadecenoic acid | 282.46 | 2 | 1 | 4.57 | 1 | 0.85 | 37.30 |
| 44 | 1,2-Di-but-2-enyl-cyclohexane | 192.34 | 0 | 0 | 4.37 | 1 | 0.55 | 0.00 |
| 45 | 4,12,12-trimethyl-9-methylene-5-oxatricyclo[8.2.0.04,6]dodecane | 220.35 | 1 | 0 | 3.67 | 0 | 0.55 | 12.53 |
| 46 | 3,4-O-Isopropylidene-d-galactose | 220.22 | 6 | 3 | −1.34 | 0 | 0.55 | 88.38 |
| 47 | 2-Hexenoic acid, 6-cyclohexyl- | 196.29 | 2 | 1 | 2.65 | 0 | 0.85 | 37.30 |
| 48 | Heptadec-8-ene | 238.45 | 0 | 0 | 6.54 | 1 | 0.55 | 0.00 |
| 49 | Octane | 114.23 | 0 | 0 | 4.20 | 1 | 0.55 | 0.00 |
| 50 | Myristic acid | 228.37 | 2 | 1 | 3.69 | 0 | 0.85 | 37.30 |
| 51 | D-(-)-Kinic Acid | 192.17 | 6 | 5 | −2.14 | 0 | 0.55 | 118.22 |
| 52 | Nonadecanoic acid | 298.50 | 2 | 1 | 4.91 | 1 | 0.85 | 37.30 |
| 53 | 10-Bromoundecanoic acid | 265.19 | 2 | 1 | 3.29 | 0 | 0.85 | 37.30 |
| 54 | Stearic acid | 284.48 | 2 | 1 | 4.67 | 1 | 0.85 | 37.30 |
| 55 | Cysteamine S-sulfate | 157.21 | 4 | 2 | −1.51 | 0 | 0.55 | 114.07 |
| 56 | Limonene dioxide | 168.23 | 2 | 0 | 1.52 | 0 | 0.55 | 25.06 |
| 57 | 2,6-Dimethyl-4-nitro-3-phenyl-cyclohexanone | 247.29 | 3 | 0 | 1.66 | 0 | 0.55 | 62.89 |
| 58 | Methyl palmitate | 270.45 | 2 | 0 | 4.44 | 1 | 0.55 | 26.30 |
| 59 | 2,6-Dimethyl-1,3,6-heptatriene | 122.21 | 0 | 0 | 3.26 | 0 | 0.55 | 0.00 |
| 60 | Palmitic acid | 256.42 | 2 | 1 | 4.19 | 1 | 0.85 | 37.30 |
| 61 | Neral | 152.23 | 1 | 0 | 2.49 | 0 | 0.55 | 17.07 |
| 62 | 2-Methyl-6-methylene-1,7-octadien-3-one | 150.22 | 1 | 0 | 2.40 | 0 | 0.55 | 17.07 |
| 63 | Bis(3-benzyl-2,4-pentanedionato)palladium(II) | 486.90 | 4 | 2 | 2.69 | 0 | 0.85 | 74.60 |
| 64 | Pentamethylbenzenesulfonyl chloride | 246.75 | 2 | 0 | 3.04 | 0 | 0.55 | 42.52 |
| 65 | Myrtenal | 150.22 | 1 | 0 | 2.20 | 0 | 0.55 | 17.07 |
| 66 | N,N-Dimethyl-2-phenylethen-1-amine | 147.22 | 0 | 0 | 2.40 | 0 | 0.55 | 3.24 |
| 67 | Allyl(chloromethyl)dimethylsilane | 148.71 | 0 | 0 | 2.81 | 0 | 0.55 | 0.00 |
| 68 | Cyclohexene, 4-(4-ethylcyclohexyl)-1-pentyl- | 262.47 | 0 | 0 | 6.61 | 1 | 0.55 | 0.00 |
| 69 | 3-Epicycloeucalenol | 426.72 | 1 | 1 | 6.92 | 1 | 0.55 | 20.23 |
| 70 | 2,5-Furandione, 3-dodecenyl- | 266.38 | 3 | 0 | 3.53 | 0 | 0.55 | 43.37 |
| 71 | 1-cinnamyl-3-methylindole-2-carbaldehyde | 275.34 | 1 | 0 | 3.20 | 0 | 0.55 | 22.00 |
| 72 | Glyceryl palmitate | 330.50 | 4 | 2 | 3.18 | 0 | 0.55 | 66.76 |
| 73 | 2-Methyl-Z,Z-3,13-octadecadienol | 280.49 | 1 | 1 | 4.91 | 1 | 0.55 | 20.23 |
| 74 | Pentadeca-2,3,6,9,12,13-hexaen-8-one, 2,5,5,11,11,14-hexamethyl- | 298.46 | 1 | 0 | 4.93 | 1 | 0.55 | 17.07 |
| 75 | CBMicro_013618 | 354.40 | 5 | 0 | 1.66 | 0 | 0.55 | 57.90 |
| 76 | Monoolein | 356.54 | 4 | 2 | 3.52 | 0 | 0.55 | 66.76 |
| 77 | Cyclohexene, 4-(4-ethylcyclohexyl)-1-pentyl- | 262.47 | 0 | 0 | 6.61 | 1 | 0.55 | 0.00 |
| 78 | Cedrane-8,13-diol | 238.37 | 2 | 2 | 2.88 | 0 | 0.55 | 40.46 |
| 79 | 26,27-Dinorergosta-5,23-dien-3β-ol | 370.61 | 1 | 1 | 6.03 | 1 | 0.55 | 20.23 |
| 80 | Cholest-4-en-3-one, 14-methyl- | 398.66 | 1 | 0 | 6.43 | 1 | 0.55 | 17.07 |
| 81 | (+)-Sesamolin | 370.35 | 7 | 0 | 1.85 | 0 | 0.55 | 64.61 |
| 82 | NSC402953 | 386.35 | 8 | 0 | 1.74 | 0 | 0.55 | 73.84 |
| 83 | Campesterol | 400.68 | 1 | 1 | 6.54 | 1 | 0.55 | 20.23 |
| 84 | Stigmasta-5,22-dien-3-ol | 412.69 | 1 | 1 | 6.62 | 1 | 0.55 | 20.23 |
| 85 | Clionasterol | 414.71 | 1 | 1 | 6.73 | 1 | 0.55 | 20.23 |
| No. | Target | Degree of Value | No. | Target | Degree of Value |
|---|---|---|---|---|---|
| 1 | VEGFA | 42 | 51 | ENPP2 | 9 |
| 2 | MMP9 | 28 | 52 | PPARA | 8 |
| 3 | TLR4 | 23 | 53 | PLA2G2A | 8 |
| 4 | IL2 | 23 | 54 | NLRP3 | 8 |
| 5 | FGF2 | 23 | 55 | MAOA | 8 |
| 6 | PPARG | 22 | 56 | F3 | 8 |
| 7 | ESR1 | 21 | 57 | EDNRA | 8 |
| 8 | PTPRC | 19 | 58 | AKR1B1 | 8 |
| 9 | AR | 19 | 59 | SHBG | 7 |
| 10 | CXCR3 | 18 | 60 | PTPN6 | 7 |
| 11 | MMP2 | 17 | 61 | PRKCA | 7 |
| 12 | CNR1 | 17 | 62 | DHFR | 7 |
| 13 | LPAR3 | 16 | 63 | TYR | 6 |
| 14 | ABCG2 | 16 | 64 | NR1I3 | 6 |
| 15 | NR3C1 | 15 | 65 | MAOB | 6 |
| 16 | LPAR2 | 15 | 66 | HMGCR | 6 |
| 17 | LPAR1 | 15 | 67 | HDAC6 | 6 |
| 18 | HDAC1 | 15 | 68 | ESR2 | 6 |
| 19 | ABCB1 | 14 | 69 | RORC | 5 |
| 20 | S1PR1 | 14 | 70 | HSD11B1 | 5 |
| 21 | CYP2C19 | 14 | 71 | GSR | 5 |
| 22 | CYP1A2 | 14 | 72 | FGF1 | 5 |
| 23 | CYP19A1 | 14 | 73 | BCHE | 5 |
| 24 | ALOX5 | 14 | 74 | RARB | 4 |
| 25 | ABCB1 | 14 | 75 | HSD11B2 | 4 |
| 26 | PLA2G1B | 13 | 76 | GRK6 | 4 |
| 27 | TNFRSF1A | 12 | 77 | GPR35 | 4 |
| 28 | PLA2G4A | 12 | 78 | GPBAR1 | 4 |
| 29 | GABBR2 | 12 | 79 | DHCR7 | 4 |
| 30 | GABBR1 | 12 | 80 | PTGER2 | 3 |
| 31 | CNR2 | 12 | 81 | PPARD | 3 |
| 32 | PTGES | 11 | 82 | PDE4B | 3 |
| 33 | PTGER4 | 11 | 83 | IL6ST | 3 |
| 34 | NOS2 | 11 | 84 | HEXB | 3 |
| 35 | MTNR1B | 11 | 85 | CES2 | 3 |
| 36 | LTB4R | 11 | 86 | AKR1B10 | 3 |
| 37 | GLI1 | 11 | 87 | TTR | 2 |
| 38 | EDNRB | 11 | 88 | RORA | 2 |
| 39 | TLR9 | 10 | 89 | KCNA3 | 2 |
| 40 | S1PR3 | 10 | 90 | FABP3 | 2 |
| 41 | MMP1 | 10 | 91 | CTRB1 | 2 |
| 42 | HDAC3 | 10 | 92 | CPA1 | 2 |
| 43 | G6PD | 10 | 93 | CA2 | 2 |
| 44 | CYP17A1 | 10 | 94 | ASAH1 | 2 |
| 45 | ALOX12 | 10 | 95 | ACP1 | 2 |
| 46 | ALDH1A1 | 10 | 96 | PDE4D | 1 |
| 47 | VDR | 9 | 97 | PAM | 1 |
| 48 | TRPV1 | 9 | 98 | HEXA | 1 |
| 49 | SLC6A4 | 9 | 99 | GSTK1 | 1 |
| 50 | SHH | 9 |
| KEGG ID & Description | Target Genes | False Discovery Rate |
|---|---|---|
| hsa04933:AGE-RAGE signaling pathway in diabetic complications | PRKCA,MMP2,VEGFA,F3 | 0.01010 |
| hsa04926:Relaxin signaling pathway | PRKCA,VEGFA,EDNRB,MMP1,MMP2,MMP9, NOS2 | 0.00018 |
| hsa04919:Thyroid hormone signaling pathway | PRKCA,ESR1,HDAC1,HDAC3 | 0.01460 |
| hsa04917:Prolactin signaling pathway | ESR1,ESR2,CYP17A1 | 0.02200 |
| hsa04915:Estrogen signaling pathway | GABBR1,GABBR2,MMP2,MMP9,ESR1,ESR2 | 0.00110 |
| hsa04912:GnRH signaling pathway | PRKCA,MMP2,PLA2G4A | 0.03510 |
| hsa04660:T cell receptor signaling pathway | IL2,PTPRC,PTPN6 | 0.03950 |
| hsa04644:Fc epsilon RI signaling pathway | PRKCA,ALOX5,PLA2G4A | 0.02070 |
| hsa04370:VEGF signaling pathway | VEGFA,PRKCA,PLA2G4A | 0.01690 |
| hsa04151:PI3K-Akt signaling pathway | PRKCA,VEGFA,IL2,TLR4,FGF1,FGF2,LPAR1,LPAR2,LPAR3 | 0.00110 |
| hsa04072:Phospholipase D signaling pathway | PRKCA,MMP2,LPAR1,LPAR2,LPAR3 | 0.00660 |
| hsa04071:Sphingolipid signaling pathway | PRKCA,S1PR1,S1PR3,ASAH1,TNFRSF1A | 0.00340 |
| hsa04066:HIF-1 signaling pathway | PRKCA,VEGFA,TLR4,NOS2 | 0.01010 |
| hsa04024:cAMP signaling pathway | PPARA,EDNRA,PDE4B,PDE4D,GABBR1,GABBR2,PTGER2,GLI1 | 0.00023 |
| hsa04020:Calcium signaling pathway | PRKCA,NOS2,EDNRB,EDNRA | 0.03850 |
| hsa04015:Rap1 signaling pathway | PRKCA,VEGFA,FGF1,FGF2,CNR1,LPAR1,LPAR2,LPAR3 | 0.00025 |
| hsa04014:Ras signaling pathway | PRKCA,VEGFA,FGF1,FGF2,PLA2G2A,PLA2G1B | 0.00200 |
| hsa04010:MAPK signaling pathway | PRKCA,VEGFA,FGF1,FGF2,TNFRSF1A,PLA2G4A | 0.01810 |
| hsa03320:PPAR signaling pathway | PPARA,PPARD,PPARG,FABP3,MMP1 | 0.00070 |
| No. | Target | Degree of Value | No. | Target | Degree of Value |
|---|---|---|---|---|---|
| 1 | PRKCA | 14 | 21 | TLR4 | 2 |
| 2 | VEGFA | 8 | 22 | IL2 | 2 |
| 3 | MMP2 | 5 | 23 | PPARD | 1 |
| 4 | PLA2G4A | 4 | 24 | PPARG | 1 |
| 5 | FGF2 | 4 | 25 | FABP3 | 1 |
| 6 | FGF1 | 4 | 26 | ALOX5 | 1 |
| 7 | NOS2 | 3 | 27 | CYP17A1 | 1 |
| 8 | ESR1 | 3 | 28 | S1PR1 | 1 |
| 9 | LPAR1 | 3 | 29 | S1PR3 | 1 |
| 10 | LPAR2 | 3 | 30 | ASAH1 | 1 |
| 11 | LPAR3 | 3 | 31 | GLI1 | 1 |
| 12 | PPARA | 2 | 32 | PTGER2 | 1 |
| 13 | MMP1 | 2 | 33 | F3 | 1 |
| 14 | EDNRB | 2 | 34 | CNR1 | 1 |
| 15 | MMP9 | 2 | 35 | HDAC1 | 1 |
| 16 | GABBR2 | 2 | 36 | HDAC3 | 1 |
| 17 | GABBR1 | 2 | 37 | PLA2G2A | 1 |
| 18 | ESR2 | 2 | 38 | PLA2G1B | 1 |
| 19 | TNFRSF1A | 2 | 39 | PTPRC | 1 |
| 20 | EDNRA | 2 | 40 | PTPN6 | 1 |
| Grid Box | Hydrogen Bond Interactions | Hydrophobic Interactions | |||||
|---|---|---|---|---|---|---|---|
| Protein | Ligand | PubChem ID | Binding Energy (kcal/mol) | Center | Dimension | Amino Acid Residue | Amino Acid Residue |
| FGF1 (PDB ID:3OJ2) | (★) Campesterol | 173183 | −8.4 | x 9.051 | x 40 | Asp283, Ser252 | Arg251,Phe172, Ile257 |
| y 22.527 | y 40 | Ser220, Leu258, Ile204 | |||||
| z −0.061 | z 40 | Ala260, Gln259, Tyr281 | |||||
| 3,4-O-Isopropylidene-d-galactose | 54504880 | −6.1 | x 9.051 | x 40 | Arg255, Thr174, Phe172 | Asn350, Asn173, Ala349 | |
| y 22.527 | y 40 | Asn107, Gln348 | |||||
| z −0.061 | z 40 | ||||||
| Positive control | (a) Suramin sodium | 8514 | −19.1 | x 9.051 | x 40 | Ser282, Lys27 | Arg203, Ile204, Ala260 |
| y 22.527 | y 40 | Gln259, Leu258, Tyr281 | |||||
| z −0.061 | z 40 | Asn22, Tyr23, Asp283 | |||||
| Val249, Pro149, Glu250 | |||||||
| His254, Ser252, Ile257 | |||||||
| Phe172, Val222, Ser220 | |||||||
| FGF2 (PDB ID:1IIL) | (★) 26,27-Dinorergosta-5,23-dien-3β-ol | 22213488 | −8.0 | x 26.785 | x 40 | Thr139, Ser137 | Glu323, Ser122, Trp123 |
| y 14.360 | y 40 | Lys313, Leu312, Ile329 | |||||
| z −1.182 | z 40 | Leu327, Tyr328 | |||||
| Campesterol | 173183 | −7.9 | x 26.785 | x 40 | Ser137 | Thr139, Glu323, Lys313 | |
| y 14.360 | y 40 | Asp336, Tyr328, Ile329 | |||||
| z −1.182 | z 40 | Leu312, Leu327, Ser122 | |||||
| Trp123, Thr319 | |||||||
| Stigmasta-5,22-dien-3-ol | 53870683 | −7.8 | x 26.785 | x 40 | Tyr340, Asp336 | Ile329, Leu312, Ser122 | |
| y 14.360 | y 40 | Trp123, Ser137, Thr319 | |||||
| z −1.182 | z 40 | Glu323, Asn318, Lys313 | |||||
| Leu327 | |||||||
| 3,4-O-Isopropylidene-d-galactose | 54504880 | −5.6 | x 26.785 | x 40 | Glu323, Trp123, Ser137 | Ser122 | |
| y 14.360 | y 40 | Thr139, Lys313 | |||||
| z −1.182 | z 40 | ||||||
| Positive control | (b) NSC172285 | 299405 | −14.7 | x 26.785 | x 40 | Tyr207 | Val209, Asp99, Lys119 |
| y 14.360 | y 40 | Lys199, Gln200, Glu201 | |||||
| z −1.182 | z 40 | ||||||
| (b) NSC37204 | 235612 | −9.5 | x 26.785 | x 40 | Thr358, Arg210, Thr121 | Val209, Asn265, Lys119 | |
| y 14.360 | y 40 | Arg118, Glu201 | Asp99, Gln200, Trp356 | ||||
| z −1.182 | z 40 | ||||||
| VEGFA (PDB ID: 3V2A) | (★) 3,4-O-Isopropylidene-d-galactose | 54504880 | −5.3 | x 38.009 | x 40 | Gly312, Ser310 | Gly255, Glu44, Ser311 |
| y −10.962 | y 40 | Ile256, Asp257, Lys84 | |||||
| z 12.171 | z 40 | Pro85 | |||||
| Glyceryl palmitate | 14900 | −5.2 | x 38.009 | x 40 | Pro40, Asp276 | Arg275, Phe36, Lys286 | |
| y −10.962 | y 40 | Lys48, Asn253, Ile46 | |||||
| z 12.171 | z 40 | ||||||
| Monoolein | 5283468 | −5.1 | x 38.009 | x 40 | Asp276, Pro40 | Arg275, Asp34, Asn253 | |
| y −10.962 | y 40 | Lys48, Phe47, Ile46 | |||||
| z 12.171 | z 40 | Phe36, Lys286 | |||||
| Methyl palmitate | 8181 | −4.0 | x 38.009 | x 40 | n/a | Pro40, Arg275, Phe36 | |
| y −10.962 | y 40 | Ile46, Asn253, Lys286 | |||||
| z 12.171 | z 40 | Asp276 | |||||
| Isopropyl hexanoate | 16832 | −3.9 | x 38.009 | x 40 | n/a | Pro85, Ser310, Gly312 | |
| y −10.962 | y 40 | Glu44, Ser311, Gly255 | |||||
| z 12.171 | z 40 | Gln87, Lys84, Asp257 | |||||
| Positive control | (c) BAW2881 | 16004702 | −7.6 | x 38.009 | x 40 | n/a | Lys286, Asp34, Ser50 |
| y −10.962 | y 40 | Asp276, Pro40, Phe36 | |||||
| z 12.171 | z 40 | Ile46 | |||||
| TNFRSF1A (PDB ID: 1NCF) | (★) CBMicro_013618 | 1109374 | −6.8 | x 21.259 | x 40 | Lys132, Gln133 | Glu109, Tyr106, Gln130 |
| y 14.648 | y 40 | Gln133 | |||||
| z 34.77 | z 40 | ||||||
| 2-Propenoic acid, 3-phenyl-, methyl ester | 7644 | −5.0 | x 21.259 | x 40 | Lys35 | Ala62, Glu64, His34 | |
| y 14.648 | y 40 | Lys35, Glu64 | |||||
| z 34.77 | z 40 | ||||||
| Positive control | (d) Enamine_004209 | 2340496 | −5.3 | x 21.259 | x 40 | Glu109, Cys96, Tyr106 | Asn110, Ph112, Val95 |
| y 14.648 | y 40 | Gln82, Ser74, Thr94 | |||||
| z 34.77 | z 40 | Arg77, Arg132 | |||||
| PLA2G4A (PDB ID: 1BCI) | (★) Stearic acid | 5281 | −4.5 | x −0.058 | x 40 | Gly33, Lys32 | Pro42, Val30, Ile67 |
| y 0.077 | y 40 | Val127, Thr31, Gln126 | |||||
| z 0.285 | z 40 | ||||||
| Methyl palmitate | 8181 | −3.9 | x −0.058 | x 40 | Lys58 | Phe77, Pro54, Thr53 | |
| y 0.077 | y 40 | Leu79, Tyr16, Glu76 | |||||
| z 0.285 | z 40 | Ile78 | |||||
| Palmitic acid | 985 | −3.8 | x −0.058 | x 40 | Thr53 | Leu79, Phe77, Ile78 | |
| y 0.077 | y 40 | Tyr16, Glu76, Pro54 | |||||
| z 0.285 | z 40 | ||||||
| Myristic acid | 11005 | −3.3 | x −0.058 | x 40 | n/a | Asp55, Pro54, Tyr16 | |
| y 0.077 | y 40 | Phe77, Ile78, Thr53 | |||||
| z 0.285 | z 40 | ||||||
| Positive control | (e) Berberine | 2353 | −6.6 | x −0.058 | x 40 | n/a | Arg59, Asp99, Asn95 |
| y 0.077 | y 40 | His62, Phe63, Asn64 | |||||
| z 0.285 | z 40 | Arg61, Ala94, Tyr45 | |||||
| PRKCA (PDB ID: 3IW4) | (★) Monoolein | 5283468 | −6.7 | x −14.059 | x 40 | Asn660, Leu393, Asp395 | Pro398, Lys478, Glu418 |
| y 38.224 | y 40 | Lys396 | Pro666, Tyr419, Arg608 | ||||
| z 32.319 | z 40 | Val664, Gln402 | |||||
| Glyceryl palmitate | 14900 | −6.6 | x −14.059 | x 40 | Asp395, Lys396, Leu393 | Leu394, Gln402, Lys478 | |
| y 38.224 | y 40 | Asn660 | Arg608, Pro666, Ile667 | ||||
| z 32.319 | z 40 | Val664, Pro398, Pro397 | |||||
| Stearic acid | 5281 | −6.3 | x −14.059 | x 40 | Lys396, Leu393 | Pro397, Pro398, Lys478 | |
| y 38.224 | y 40 | Arg608, Ile667, Pro666 | |||||
| z 32.319 | z 40 | His665, Val664, Gln402 | |||||
| Asn660, Leu394 | |||||||
| Nonadecanoic acid | 12591 | −6.2 | x −14.059 | x 40 | Leu393, Lys396 | Asn660, Pro397, Pro398 | |
| y 38.224 | y 40 | Lys478, Pro666, Arg608 | |||||
| z 32.319 | z 40 | Glu418, Val664, Gln402 | |||||
| Leu394 | |||||||
| 1,6,10-Dodecatrien-3-ol, 3,7,11-trimethyl- | 8888 | −6.2 | x −14.059 | x 40 | Lys372, Gln408, Gln650 | Val410, Thr409, Gly540 | |
| y 38.224 | y 40 | Ile645, Asp539, Asp503 | |||||
| z 32.319 | z 40 | Phe538, Glu543 | |||||
| 2,5-Furandione, 3-dodecenyl- | 5362708 | −6.1 | x −14.059 | x 40 | Lys396, Asp395 | Asn660, Gln402, Pro397 | |
| y 38.224 | y 40 | Pro398, Glu552, Gln662 | |||||
| z 32.319 | z 40 | Val664, Leu394 | |||||
| 2,6-Dimethyl-3,5,7-octatriene-2-ol, Z,Z- | 5363692 | −5.3 | x −14.059 | x 40 | Gly540 | Val410, Ile645, Asp503 | |
| y 38.224 | y 40 | Pro502, Glu543, Gln650 | |||||
| z 32.319 | z 40 | Leu546, Asp542 | |||||
| 2-Dodecenoic acid | 5282729 | −5.1 | x −14.059 | x 40 | Lys396, Asn660, Leu393 | Leu394, Gln662, Glu552 | |
| y 38.224 | y 40 | Val664, Gln402 | |||||
| z 32.319 | z 40 | ||||||
| 9-Octadecenoic acid | 965 | −5.0 | x −14.059 | x 40 | Leu393, Lys396 | Asn660, Glu552, Gln548 | |
| y 38.224 | y 40 | His553, Ser549, Gln662 | |||||
| z 32.319 | z 40 | Val664, Gln402, Leu394 | |||||
| Octanoic acid | 379 | −5.0 | x −14.059 | x 40 | Asn660, Lys396, Gln402 | Pro397, Lys478, Pro398 | |
| y 38.224 | y 40 | Val664, Glu552, Arg608 | |||||
| z 32.319 | z 40 | ||||||
| Methyl palmitate | 8181 | −5.0 | x −14.059 | x 40 | n/a | Gln377, Asn647, Asp373 | |
| y 38.224 | y 40 | Ile648, Asp649, Asn468 | |||||
| z 32.319 | z 40 | Lys465, Phe350, Asp467 | |||||
| Ile376 | |||||||
| Palmitic acid | 985 | −5.0 | x −14.059 | x 40 | Leu393, Asp395, Lys396 | Pro397, Pro398, Val664 | |
| y 38.224 | y 40 | Glu552, His553, Ser549 | |||||
| z 32.319 | z 40 | Gln548,Gln662, Gln402 | |||||
| Leu394, Asn660 | |||||||
| (E)-4-Undecenal | 5283357 | −4.8 | x −14.059 | x 40 | n/a | Gln642, Pro536, Ile645 | |
| y 38.224 | y 40 | Gly540, Val410, Gln650 | |||||
| z 32.319 | z 40 | Glu543, Asp542, Asp503 | |||||
| Leu546 | |||||||
| Myristic acid | 11005 | −4.8 | x −14.059 | x 40 | Lys396, Gln402 | Asp395, Leu393, Leu394 | |
| y 38.224 | y 40 | Pro398, Val664, Gln662 | |||||
| z 32.319 | z 40 | Asn660 | |||||
| Nonanoic acid | 8158 | −4.7 | x −14.059 | x 40 | Leu393, Lys396 | Leu394, Asn660, Pro397 | |
| y 38.224 | y 40 | Gln402, Gln662, Val664 | |||||
| z 32.319 | z 40 | ||||||
| Heptadec-8-ene | 520230 | −4.6 | x −14.059 | x 40 | n/a | Asn647, Asp424, Met426 | |
| y 38.224 | y 40 | Gln377,Ile376, Phe350 | |||||
| z 32.319 | z 40 | Asp467, Asp373 | |||||
| Positive control | (f) Sphingosine | 5280335 | −5.5 | x −14.059 | x 40 | Asn660, Gln662, Lys396 | Pro397, Gln402, Val664 |
| y 38.224 | y 40 | Gln548, Glu552, His553 | |||||
| z 32.319 | z 40 | Leu394, Ser549, Asp395 | |||||
| Grid Box | Hydrogen Bond Interactions | Hydrophobic Interactions | |||||
|---|---|---|---|---|---|---|---|
| Protein | Ligand | PubChem ID | Binding Energy(kcal/mol) | Center | Dimension | Amino Acid Residue | Amino Acid Residue |
| PPARA (PDB ID: 3SP6) | (★) β-Caryophyllene | 5281515 | −8.6 | x 8.006 | x 40 | n/a | Leu321, Leu331, Gly335 |
| y −0.459 | y 40 | Val324, Met220, Tyr334 | |||||
| z 23.392 | z 40 | Ala333, Thr279, Asn219 | |||||
| Thr283 | |||||||
| Cadina-1(10),4-diene | 10223 | −7.4 | x 8.006 | x 40 | n/a | Met220, Leu331, Val324 | |
| y −0.459 | y 40 | Thr279, Thr283, Leu321 | |||||
| z 23.392 | z 40 | Ile317, Met320 | |||||
| 26,27-Dinorergosta-5,23-dien-3beta-ol | 22213488 | −7.0 | x 8.006 | x 40 | Lys345 | Glu356, Asp353, Pro357 | |
| y −0.459 | y 40 | Leu443, His440, Glu439 | |||||
| z 23.392 | z 40 | Leu436, Lys358, Asp360 | |||||
| Clionasterol | 457801 | −6.7 | x 8.006 | x 40 | Lys345 | Asp360, Pro357, Glu439 | |
| y −0.459 | y 40 | His440, Leu443, Asp353 | |||||
| z 23.392 | z 40 | Glu356 | |||||
| Cyclohexene, 4-(4-ethylcyclohexyl)-1-pentyl- | 543386 | −6.6 | x 8.006 | x 40 | n/a | Met320, Phe218, Met220 | |
| y −0.459 | y 40 | Thr279, Val332, Ala333 | |||||
| z 23.392 | z 40 | Tyr334, Thr283, Asn219 | |||||
| Spiro[4.4]nona-1,3-diene, 1,2-dimethyl- | 570800 | −6.5 | x 8.006 | x 40 | n/a | Leu321, Leu331, Val324 | |
| y −0.459 | y 40 | Met320, Asn219, Thr283 | |||||
| z 23.392 | z 40 | Thr279, Met220 | |||||
| 1-Methyldecahydronaphthalene | 34193 | −6.4 | x 8.006 | x 40 | n/a | Met320, Val324, Met220 | |
| y −0.459 | y 40 | Asn219, Thr279, Thr283 | |||||
| z 23.392 | z 40 | Leu321 | |||||
| 1,3,4,5-Tetrahydroxycyclohexanecarboxylic acid | 1064 | −6.3 | x 8.006 | x 40 | Ile317, Glu286, Asn219 | Met320, Met220, Leu321 | |
| y −0.459 | y 40 | Thr283 | |||||
| z 23.392 | z 40 | ||||||
| Stigmasta-5,22-dien-3-ol | 53870683 | −6.3 | x 8.006 | x 40 | n/a | Arg465, Glu462, Ser688 | |
| y −0.459 | y 40 | Val306, Asn303, Thr307 | |||||
| z 23.392 | z 40 | Tyr311, Gly390, Pro389 | |||||
| Lys310, Asp466 | |||||||
| Terpinolene | 11463 | −6.2 | x 8.006 | x 40 | n/a | Thr279, Tyr334, Val324 | |
| y −0.459 | y 40 | Met220, Met320, Leu321 | |||||
| z 23.392 | z 40 | Thr283, Asn219 | |||||
| β-Phellandrene | 11142 | −6.0 | x 8.006 | x 40 | n/a | Leu331, Val324, Leu321 | |
| y −0.459 | y 40 | Ile317, Thr283, Met320 | |||||
| z 23.392 | z 40 | Thr279 | |||||
| Citronellic acid | 10402 | −5.9 | x 8.006 | x 40 | Thr283, Glu286, Met220 | Met320, Asn219, Tyr334 | |
| y −0.459 | y 40 | Gly335, Leu321, Val324 | |||||
| z 23.392 | z 40 | Ile317 | |||||
| Stearic acid | 5281 | −5.7 | x 8.006 | x 40 | n/a | Phe361, Asp432, Leu436 | |
| y −0.459 | y 40 | Glu439, His440, Leu443 | |||||
| z 23.392 | z 40 | Asp353, Gln442, Ile446 | |||||
| Pro357, Lys358 | |||||||
| Monoolein | 5283468 | −5.7 | x 8.006 | x 40 | Asn261, Lys257 | Leu258, His274, Cys275 | |
| y −0.459 | y 40 | Ala333, Val255, Cys278 | |||||
| z 23.392 | z 40 | ||||||
| 2,6-Octadiene, 2,6-dimethyl- | 5365898 | −5.7 | x 8.006 | x 40 | n/a | Met320, Phe218, Leu331 | |
| y −0.459 | y 40 | Val324, Met220, Leu321 | |||||
| z 23.392 | z 40 | ||||||
| Citronellol | 8842 | −5.5 | x 8.006 | x 40 | Thr283 | Leu331, Val324, Ile317 | |
| y −0.459 | y 40 | Leu321, Met320, Thr279 | |||||
| z 23.392 | z 40 | ||||||
| Myrcene | 31253 | −5.5 | x 8.006 | x 40 | n/a | Val332, Val324, Ile317 | |
| y −0.459 | y 40 | Leu321, Met220, Thr283 | |||||
| z 23.392 | z 40 | Met320, Leu331 | |||||
| (E)-β-Ocimene | 5281553 | −5.5 | x 8.006 | x 40 | n/a | Thr283, Ile317, Met320 | |
| y −0.459 | y 40 | Tyr334, Val332, Val324 | |||||
| z 23.392 | z 40 | Gly335, Leu331, Thr279 | |||||
| Leu321 | |||||||
| Citronellal | 7794 | −5.2 | x 8.006 | x 40 | Thr283 | Leu321, Met220, Met320 | |
| y −0.459 | y 40 | Val324, Asn219, Thr279 | |||||
| z 23.392 | z 40 | Ile317 | |||||
| Heptadec-8-ene | 520230 | −5.2 | x 8.006 | x 40 | n/a | Leu321, Ile317, Thr283 | |
| y −0.459 | y 40 | Thr279, Val255, Ala333 | |||||
| z 23.392 | z 40 | Tyr334, Leu331, Val332 | |||||
| Val324 | |||||||
| Myristic acid | 11005 | −5.2 | x 8.006 | x 40 | Tyr334, Ala333,Thr279 | Val332, Met220, Met320 | |
| y −0.459 | y 40 | Ile317, Leu321, Thr283 | |||||
| z 23.392 | z 40 | Asn219 | |||||
| Nonanoic acid | 8158 | −5.1 | x 8.006 | x 40 | Ala333 | Leu331, Leu321, Val332 | |
| y −0.459 | y 40 | Ile317, Thr283, Met320 | |||||
| z 23.392 | z 40 | Val324, Thr279 | |||||
| 10-Bromoundecanoic acid | 543401 | −5.1 | x 8.006 | x 40 | n/a | Met320, Val324, Leu321 | |
| y −0.459 | y 40 | Thr279, Leu331, Val332 | |||||
| z 23.392 | z 40 | Asn219, Tyr334, Met220 | |||||
| 2-Methyl-Z,Z-3,13-octadecadienol | 5364412 | −5.0 | x 8.006 | x 40 | n/a | Leu254, Ala333, Cys275 | |
| y −0.459 | y 40 | Tyr334, Ile317, Met320 | |||||
| z 23.392 | z 40 | Thr283, Leu321, Leu331 | |||||
| Val324, Ala250, Thr279 | |||||||
| Ile241, Val255 | |||||||
| Octanoic acid | 379 | −5.0 | x 8.006 | x 40 | Asn219, Thr283, Met220 | Phe218, Leu321, Val324 | |
| y −0.459 | y 40 | Glu286 | Leu331, Met320 | ||||
| z 23.392 | z 40 | ||||||
| Nonadecanoic acid | 12591 | −4.9 | x 8.006 | x 40 | n/a | Asn336, Leu254, Ala333 | |
| y −0.459 | y 40 | Ala250, Cys275, Val255 | |||||
| z 23.392 | z 40 | Tyr334 | |||||
| 3-Hydroxycyclohexanone | 439950 | −4.9 | x 8.006 | x 40 | Met220 | Met320, Phe218, Asn219 | |
| y −0.459 | y 40 | Glu286 | |||||
| z 23.392 | z 40 | ||||||
| Palmitic acid | 985 | −4.9 | x 8.006 | x 40 | n/a | Glu251, Val332, Ile241 | |
| y −0.459 | y 40 | Ala333, Thr279, Val255 | |||||
| z 23.392 | z 40 | Tyr334, Leu258, Cys275 | |||||
| Ala250, Leu254 | |||||||
| 3-Methylcyclohexene | 11573 | −4.6 | x 8.006 | x 40 | n/a | Met320, Ile317, Leu321 | |
| y −0.459 | y 40 | Thr279, Thr283 | |||||
| z 23.392 | z 40 | ||||||
| 9-Octadecenoic acid | 965 | −4.4 | x 8.006 | x 40 | n/a | Glu251, Ala250, Leu254 | |
| y −0.459 | y 40 | Val255, Ile241, Ala333 | |||||
| z 23.392 | z 40 | Asn336, Tyr334, Cys275 | |||||
| 2-Tetradecynoic acid | 324386 | −4.1 | x 8.006 | x 40 | Thr307 | Glu462, Ser688, Gln691 | |
| y −0.459 | y 40 | Tyr311, Lys310, Asn303 | |||||
| z 23.392 | z 40 | Val306 | |||||
| Methyl palmitate | 8181 | −3.7 | x 8.006 | x 40 | n/a | Tyr311, Gln691, Pro389 | |
| y −0.459 | y 40 | Lys310, Thr307, Asn303 | |||||
| z 23.392 | z 40 | Val306, Ser688, Glu462 | |||||
| Positive control | (a) Clofibrate | 2796 | −6.4 | x 8.006 | x 40 | Thr283 | Ala333, Tyr334, Asn219 |
| y −0.459 | y 40 | Met320, Leu321, Met220 | |||||
| z 23.392 | z 40 | Phe218, Val332, Val324 | |||||
| Thr279 | |||||||
| (a) Gemfibrozil | 3463 | −6.3 | x 8.006 | x 40 | Tyr468 | Tyr464, Lys448, Leu456 | |
| y −0.459 | y 40 | Arg465, Gln442, Ala441 | |||||
| z 23.392 | z 40 | ||||||
| (a) Ciprofibrate | 2763 | −5.4 | x 8.006 | x 40 | Ala333, Thr279 | Lys257, Cys278, Tyr334 | |
| y −0.459 | y 40 | Cys275, Val255, Leu258 | |||||
| z 23.392 | z 40 | ||||||
| (a) Bezafibrate | 39042 | −5.8 | x 8.006 | x 40 | Thr307, Ser688 | Asn303, Glu462, Val306 | |
| y −0.459 | y 40 | Leu690, Lys310, Gly390 | |||||
| z 23.392 | z 40 | ||||||
| (a) Fenofibrate | 3339 | −5.4 | x 8.006 | x 40 | n/a | Gln435, Ala431, Asp360 | |
| y −0.459 | y 40 | Pro357, Leu436, Glu439 | |||||
| z 23.392 | z 40 | Lys364, Phe361, Asp432 | |||||
| PPARD (PDB ID:5U3Q) | (★) Stigmasta-5,22-dien-3-ol | 53870683 | −8.6 | x 39.265 | x 40 | n/a | Ala414, Tyr441, Met440 |
| y −18.736 | y 40 | Pro362, Gly363, Arg361 | |||||
| z 119.392 | z 40 | Gly359, Asp360, Thr411 | |||||
| Val410 | |||||||
| Clionasterol | 457801 | −7.3 | x 39.265 | x 40 | Met440 | Ala414, Tyr441, Tyr284 | |
| y −18.736 | y 40 | Pro362, Arg361, Asp360 | |||||
| z 119.392 | z 40 | Val410, Thr411 | |||||
| 26,27-Dinorergosta-5,23-dien-3beta-ol | 22213488 | −7.3 | x 39.265 | x 40 | n/a | Lys188, Glu262, Lys265 | |
| y −18.736 | y 40 | Ser266, Ser271 | |||||
| z 119.392 | z 40 | ||||||
| Stearic acid | 5281 | −6.8 | x 39.265 | x 40 | n/a | Glu288, Tyr284, Asp439 | |
| y −18.736 | y 40 | Asp360, Val367, Gly359 | |||||
| z 119.392 | z 40 | Leu364, Gly363, Arg361 | |||||
| Pro362, Met440, Thr411 | |||||||
| Tyr441 | |||||||
| 1,3,4,5-Tetrahydroxycyclohexanecarboxylic acid | 1064 | −6.6 | x 39.265 | x 40 | Tyr441, Glu288 | Ala414, Thr411, Val410 | |
| y −18.736 | y 40 | Tyr284, Arg361, Arg407 | |||||
| z 119.392 | z 40 | Met440 | |||||
| Nonadecanoic acid | 12591 | −5.8 | x 39.265 | x 40 | Asp360 | Pro362, Tyr441, Met440 | |
| y −18.736 | y 40 | Val410, Glu288, Arg407 | |||||
| z 119.392 | z 40 | Thr411, Arg361, Tyr284 | |||||
| Citronellal | 7794 | −5.7 | x 39.265 | x 40 | n/a | Asn307, Ala306, Thr252 | |
| y −18.736 | y 40 | Trp228, Arg248, Val305 | |||||
| z 119.392 | z 40 | Gln230, Lys229 | |||||
| Nonanoic acid | 8158 | −5.7 | x 39.265 | x 40 | Thr256 | Glu255, Asn307, Lys229 | |
| y −18.736 | y 40 | Trp228, Ala306, Thr252 | |||||
| z 119.392 | z 40 | Glu259, Asn191 | |||||
| Citronellic acid | 10402 | −5.3 | x 39.265 | x 40 | n/a | Arg361, Tyr441, Thr411 | |
| y −18.736 | y 40 | Met440, Tyr284 | |||||
| z 119.392 | z 40 | ||||||
| Myristic acid | 11005 | −5.2 | x 39.265 | x 40 | Tyr441 | Ala414, Glu288, Pro362 | |
| y −18.736 | y 40 | Arg361, Asp439, Tyr284 | |||||
| z 119.392 | z 40 | Arg407, Thr411, Met440 | |||||
| Val410 | |||||||
| Octanoic acid | 379 | −5.1 | x 39.265 | x 40 | Lys229, Gln230, Arg248 | Cys251, Trp228, Val305 | |
| y −18.736 | y 40 | Ala306, Thr252 | |||||
| z 119.392 | z 40 | ||||||
| 9-Octadecenoic acid | 965 | −5.0 | x 39.265 | x 40 | n/a | Met440, Thr411, Tyr441 | |
| y −18.736 | y 40 | Pro362, Tyr284, Arg361 | |||||
| z 119.392 | z 40 | Val410 | |||||
| 3-Hydroxycyclohexanone | 439950 | −5.0 | x 39.265 | x 40 | Arg407, Thr411 | Val410, Met440, Tyr441 | |
| y −18.736 | y 40 | ||||||
| z 119.392 | z 40 | ||||||
| Citronellol | 8842 | −4.8 | x 39.265 | x 40 | Arg361 | Asp439, Met440, Tyr441 | |
| y −18.736 | y 40 | Tyr284, Val410 | |||||
| z 119.392 | z 40 | ||||||
| Palmitic acid | 985 | −4.6 | x 39.265 | x 40 | n/a | Tyr441, Pro362, Arg361 | |
| y −18.736 | y 40 | Val410, Tyr284, Glu288 | |||||
| z 119.392 | z 40 | Met440, Thr411, Ala414 | |||||
| Arg407 | |||||||
| 10-Bromoundecanoic acid | 543401 | −4.6 | x 39.265 | x 40 | Arg361, Tyr284 | Tyr441, Met440, Pro362 | |
| y −18.736 | y 40 | Thr411, Glu288 | |||||
| z 119.392 | z 40 | ||||||
| Methyl palmitate | 8181 | −4.5 | x 39.265 | x 40 | n/a | Thr411, Tyr441, Pro362 | |
| y −18.736 | y 40 | Met440, Arg361, Tyr284 | |||||
| z 119.392 | z 40 | Glu288 | |||||
| Positive control | |||||||
| (b) Cardarine | 9803963 | −8.5 | x 39.265 | x 40 | Ser271, Ser272 | Lys265, Glu262, Ser266 | |
| y −18.736 | y 40 | Lys265, Ser271, Pro268 | |||||
| z 119.392 | z 40 | Ser269 | |||||
| PPARG (PDB ID: 3E00) | (★) NSC402953 | 345349 | −8.2 | x 2.075 | x 40 | Lys336 | Asn335, Glu203, Arg234 |
| y 31.910 | y 40 | Asp380, Ala231, Lys230 | |||||
| z 18.503 | z 40 | Glu378, Asn375, Met334 | |||||
| 26,27-Dinorergosta-5,23-dien-3beta-ol | 22213488 | −8.0 | x 2.075 | x 40 | Asn375 | Val372, Asn335, Val205 | |
| y 31.910 | y 40 | Val163, Glu207, Glu208 | |||||
| z 18.503 | z 40 | ||||||
| Stigmasta-5,22-dien-3-ol | 53870683 | −7.8 | x 2.075 | x 40 | Asn375 | Arg202, Glu203, Lys336 | |
| y 31.910 | y 40 | Val163, Arg164, Gln206 | |||||
| z 18.503 | z 40 | Lys165, Glu208, Glu207 | |||||
| Val372, Asn335 | |||||||
| Clionasterol | 457801 | −7.7 | x 2.075 | x 40 | Asn375 | Asn335, Val372, Lys336 | |
| y 31.910 | y 40 | Val163, Arg164, Glu208 | |||||
| z 18.503 | z 40 | Asp166, Glu207, Arg202 | |||||
| Glu203 | |||||||
| Terpinyl propionate | 62328 | −6.6 | x 2.075 | x 40 | Glu343 | Leu340, Ile341, Leu333 | |
| y 31.910 | y 40 | Leu228, Met329, Arg288 | |||||
| z 18.503 | z 40 | ||||||
| Nonadecanoic acid | 12591 | −5.9 | x 2.075 | x 40 | Glu351 | Lys354, Thr168, Tyr189 | |
| y 31.910 | y 40 | Tyr169, Thr162, Leu167 | |||||
| z 18.503 | z 40 | Tyr192, Arg202, Arg350 | |||||
| Asp337, Lys336, Gln193 | |||||||
| Stearic acid | 5281 | −5.9 | x 2.075 | x 40 | Ser342, Cys285 | Ile341, Phe226, Ile296 | |
| y 31.910 | y 40 | Glu295, Ala292, Met329 | |||||
| z 18.503 | z 40 | Leu333, Leu228, Arg288 | |||||
| Citronellic acid | 10402 | −5.2 | x 2.075 | x 40 | n/a | Glu295, Arg288, Ala292 | |
| y 31.910 | y 40 | Leu228, Leu333, Met329 | |||||
| z 18.503 | z 40 | ||||||
| Palmitic acid | 985 | −5.1 | x 2.075 | x 40 | Glu291, Arg288 | Glu343, Leu333, Leu228 | |
| y 31.910 | y 40 | Met229, Ala292, Ile326 | |||||
| z 18.503 | z 40 | Glu295 | |||||
| Myristic acid | 11005 | −5.1 | x 2.075 | x 40 | Glu343 | Arg288, Ser342, Leu333 | |
| y 31.910 | y 40 | Met329, Leu228, Glu295 | |||||
| z 18.503 | z 40 | Pro227, Ala292 | |||||
| Nonanoic acid | 8158 | −5.0 | x 2.075 | x 40 | Lys354 | Arg350, Glu351, Tyr169 | |
| y 31.910 | y 40 | Tyr192, Tyr189, Leu167 | |||||
| z 18.503 | z 40 | Gln193, Asp337, Lys336 | |||||
| Citronellal | 7794 | −4.9 | x 2.075 | x 40 | Gln193 | Tyr189, Arg202, Leu167 | |
| y 31.910 | y 40 | Thr162, Tyr192, Asp337 | |||||
| z 18.503 | z 40 | Lys336, Arg350, Lys354 | |||||
| Glu351 | |||||||
| (E)-4-Undecenal | 5283357 | −4.9 | x 2.075 | x 40 | Tyr169 | Leu167, Tyr192, Lys336 | |
| y 31.910 | y 40 | Arg350, Glu351, Asp337 | |||||
| z 18.503 | z 40 | Gln193, Lys354, Tyr189 | |||||
| Octanoic acid | 379 | −4.7 | x 2.075 | x 40 | Arg202, Leu167 | Asp337, Thr168, Tyr192 | |
| y 31.910 | y 40 | Tyr169, Gln193, Tyr189 | |||||
| z 18.503 | z 40 | Glu351, Lys336 | |||||
| 9-Octadecenoic acid | 965 | −4.1 | x 2.075 | x 40 | Arg164, Glu208 | Glu207, Glu203, Asp166 | |
| y 31.910 | y 40 | Lys336, Arg202, Val372 | |||||
| z 18.503 | z 40 | Asn375, Val163 | |||||
| Methyl palmitate | 8181 | −3.8 | x 2.075 | x 40 | Glu291, Arg288 | Glu343, Leu333, Leu330 | |
| y 31.910 | y 40 | Leu228, Met329, Ala292 | |||||
| z 18.503 | z 40 | Ile326, Glu295 | |||||
| Positive control | (c) Pioglitazone | 4829 | −7.7 | x 2.075 | x 40 | Arg288 | Ile326, Leu333, Met329 |
| y 31.910 | y 40 | Ala292, Ile341, Cys285 | |||||
| z 18.503 | z 40 | Ser342, Glu343, Glu295 | |||||
| Leu228 | |||||||
| (c) Rosiglitazone | 77999 | −7.4 | x 2.075 | x 40 | Tyr169 | Glu351, Tyr189, Gln193 | |
| y 31.910 | y 40 | Thr168, Leu167, Glu369 | |||||
| z 18.503 | z 40 | Lys373, Val372, Arg350 | |||||
| Lys336, Tyr192, Asp337 | |||||||
| (c) Lobeglitazone | 9826451 | −7.3 | x 2.075 | x 40 | Arg234 | Val372, Asn375, Met334 | |
| y 31.910 | y 40 | Val163, Lys230, Glu203 | |||||
| z 18.503 | z 40 | Lys157, Val205, Arg164 | |||||
| Arg202, Asp166, Lys336 | |||||||
| FABP3 (PDB ID: 5HZ9) | (★) Monoolein | 5283468 | −8.9 | x −1.215 | x 40 | Arg31, Thr57, Phe58 | Ala29, Gln32, Phe28 |
| y 46.730 | y 40 | Gly27 | |||||
| z −15.099 | z 40 | ||||||
| Glyceryl palmitate | 14900 | −8.7 | x −1.215 | x 40 | Arg31 | Thr57, Phe58, Ala29 | |
| y 46.730 | y 40 | Gln32, Phe28, Lys22 | |||||
| z −15.099 | z 40 | ||||||
| Nonadecanoic acid | 12591 | −8.3 | x −1.215 | x 40 | Lys22 | Ala29, Gln32, Phe28 | |
| y 46.730 | y 40 | Gly27, Gly25 | |||||
| z −15.099 | z 40 | ||||||
| Stearic acid | 5281 | −8.3 | x −1.215 | x 40 | n/a | Gly27, Asp78, Thr122 | |
| y 46.730 | y 40 | Phe58, Ala29, Phe28 | |||||
| z −15.099 | z 40 | Lys22, Lys59, Asp77 | |||||
| Thr30 | |||||||
| 2-Dodecen-1-ylsuccinic anhydride | 5362708 | −7.8 | x −1.215 | x 40 | Thr30, Gly27 | Gly25, Gln32, Ala29 | |
| y 46.730 | y 40 | Phe28 | |||||
| z −15.099 | z 40 | ||||||
| Heptadec-8-ene | 520230 | −7.6 | x −1.215 | x 40 | n/a | Phe28, Met36, Thr57 | |
| y 46.730 | y 40 | Val33, Ala29, Gln32 | |||||
| z −15.099 | z 40 | ||||||
| 2-Methyl-Z,Z-3,13-octadecadienol | 5364412 | −7.4 | x −1.215 | x 40 | n/a | Lys22, Gly25, Gly27 | |
| y 46.730 | y 40 | Gln32, Phe28, Ala29 | |||||
| z −15.099 | z 40 | ||||||
| 2-Tetradecynoic acid | 324386 | −7.4 | x −1.215 | x 40 | Arg31, Lys22 | Phe58, Ala29, Gln32 | |
| y 46.730 | y 40 | Phe28, Thr57, Lys59 | |||||
| z −15.099 | z 40 | ||||||
| Methyl palmitate | 8181 | −7.1 | x −1.215 | x 40 | n/a | Phe28, Gly27, Gly25 | |
| y 46.730 | y 40 | Gln32, Ala29 | |||||
| z −15.099 | z 40 | ||||||
| 9-Octadecenoic acid | 637517 | −7.1 | x −1.215 | x 40 | n/a | Ala29, Gly25, Phe28 | |
| y 46.730 | y 40 | Gly27, Gly25, Gln32 | |||||
| z −15.099 | z 40 | ||||||
| Myristic acid | 11005 | −6.9 | x −1.215 | x 40 | Phe58 | Ala29, Phe28, Gly27 | |
| y 46.730 | y 40 | Lys22 | |||||
| z −15.099 | z 40 | ||||||
| 2-Dodecenoic acid | 5282729 | −6.8 | x −1.215 | x 40 | Arg31 | Lys59, Phe58, Thr57 | |
| y 46.730 | y 40 | Ala29, Gln32, Phe28 | |||||
| z −15.099 | z 40 | Lys22 | |||||
| Citronellic acid | 10402 | −6.6 | x −1.215 | x 40 | Phe58 | Lys22, Met36, Ala29 | |
| y 46.730 | y 40 | Thr57 | |||||
| z −15.099 | z 40 | ||||||
| Palmitic acid | 985 | −6.5 | x −1.215 | x 40 | n/a | Val33, Thr57, Lys22 | |
| y 46.730 | y 40 | Phe58, Ala29, Gln32 | |||||
| z −15.099 | z 40 | ||||||
| (E)-4-Undecenal | 5283357 | −6.5 | x −1.215 | x 40 | n/a | Gln32, Ala29, Gly25 | |
| y 46.730 | y 40 | Gly27, Phe28 | |||||
| z −15.099 | z 40 | ||||||
| Isopropyl hexanoate | 16832 | −6.4 | x −1.215 | x 40 | n/a | Ala29, Phe28, Val33 | |
| y 46.730 | y 40 | ||||||
| z −15.099 | z 40 | ||||||
| Octane | 356 | −6.1 | x −1.215 | x 40 | n/a | Phe28 | |
| y 46.730 | y 40 | ||||||
| z −15.099 | z 40 | ||||||
| 10-Bromoundecanoic acid | 543401 | −6.1 | x −1.215 | x 40 | n/a | Thr57, Ala29, Gln32 | |
| y 46.730 | y 40 | Phe28, Lys22, Thr57 | |||||
| z −15.099 | z 40 | ||||||
| Nonanoic acid | 8158 | −6.0 | x −1.215 | x 40 | n/a | Gln32, Phe28 | |
| y 46.730 | y 40 | ||||||
| z −15.099 | z 40 | ||||||
| Octanoic acid | 379 | −5.8 | x −1.215 | x 40 | n/a | Gln32, Val33, Gly25 | |
| y 46.730 | y 40 | Gly27, Phe28, Ala29 | |||||
| z −15.099 | z 40 | ||||||
| 1,3,4,5-Tetrahydroxycyclohexanecarboxylic acid | 1064 | −5.6 | x −1.215 | x 40 | Asn307, Ala306, Lys229 | Val305, Thr252, Trp228 | |
| y 46.730 | y 40 | ||||||
| z −15.099 | z 40 | ||||||
| Citronellol | 8842 | −5.4 | x −1.215 | x 40 | n/a | Ala306, Glu255, Asn307 | |
| y 46.730 | y 40 | Thr252, Trp228, Arg248 | |||||
| z −15.099 | z 40 | Gln230, Lys229 | |||||
| 3-Hydroxycyclohexanone | 439950 | −5.3 | x −1.215 | x 40 | Thr54, Arg107, His94 | Thr61, Leu52, Ile63 | |
| y 46.730 | y 40 | Glu73, Leu105 | |||||
| z −15.099 | z 40 | ||||||
| Neral | 643779 | −5.0 | x −1.215 | x 40 | n/a | Arg361, Thr411, Tyr441 | |
| y 46.730 | y 40 | Met440 | |||||
| z −15.099 | z 40 | ||||||
| Citronellal | 7794 | −4.9 | x −1.215 | x 40 | n/a | Thr411, Met440, Pro362 | |
| y 46.730 | y 40 | Asp360, Val410, Tyr284 | |||||
| z −15.099 | z 40 | ||||||
| MMP1 (PDB ID: 1SU3) | (★) 2-Propenoic acid, 3-phenyl-, methyl ester | 7644 | −5.0 | x 34.394 | x 40 | Arg399, Tyr411, Tyr397 | Asp418, Phe436, Tyr390 |
| y −44.313 | y 40 | Phe419, Phe447, Pro449 | |||||
| z 37.396 | z 40 | Lys413 | |||||
| Geranyl acetate | 1549026 | −4.5 | x 34.394 | x 40 | Tyr411 | Arg399, Asp418, Pro449 | |
| y −44.313 | y 40 | Phe447, Lys452, Phe419 | |||||
| z 37.396 | z 40 | Tyr390, Tyr397 | |||||
| 2-Dodecenoic acid | 5282729 | −4.3 | x 34.394 | x 40 | Tyr397, Tyr411 | Lys413, Tyr390, Pro449 | |
| y −44.313 | y 40 | Phe419, Phe447, Asp418 | |||||
| z 37.396 | z 40 | ||||||
| Citronellal | 7794 | −4.0 | x 34.394 | x 40 | Arg399 | Phe419, Asp418, Tyr397 | |
| y −44.313 | y 40 | Phe436, Tyr390, Glu383 | |||||
| z 37.396 | z 40 | Pro449 | |||||
| (E)-4-Undecenal | 5283357 | −3.6 | x 34.394 | x 40 | n/a | Lys452, Asp418, Pro449 | |
| y −44.313 | y 40 | Phe436, Phe419, Phe447 | |||||
| z 37.396 | z 40 | Tyr390, Tyr397 | |||||
| Positive control | (d) Batimastat | 5362422 | −6.7 | x 34.394 | x 40 | Thr112, His113, Lys396 | Pro146, Glu110, Arg108 |
| y −44.313 | y 40 | Thr373 | Pro371, Trp398, Pro412 | ||||
| z 37.396 | z 40 | Val393 | |||||
| (d) Ilomastat | 132519 | −6.5 | x 34.394 | x 40 | His113 | Thr145, Ser142, Thr148 | |
| y −44.313 | y 40 | Leu147, Lys413, His417 | |||||
| z 37.396 | z 40 | Met414, Pro412, Gln264 | |||||
| Pro146 | |||||||
| Parameters | Compound Name | ||||||
|---|---|---|---|---|---|---|---|
| Campesterol | 26,27-Dinorergosta-5,23-dien-3β-ol | CBMicro_013618 | Monoolein | β-Caryophyllene | Stigmasta-5,22-dien-3-ol | NSC0402953 | |
| Ames toxicity | NAT | NAT | NAT | NAT | NAT | NAT | NAT |
| Carcinogens | NC | NC | NC | NC | NC | NC | NC |
| Acute oral toxicity | I | I | III | IV | III | I | III |
| Rat acute toxicity | 2.8078 | 2.8078 | 2.5735 | 1.0526 | 1.4345 | 2.6561 | 1.9796 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Oh, K.; Adnan, M.; Cho, D. Uncovering Mechanisms of Zanthoxylum piperitum Fruits for the Alleviation of Rheumatoid Arthritis Based on Network Pharmacology. Biology 2021, 10, 703. https://doi.org/10.3390/biology10080703
Oh K, Adnan M, Cho D. Uncovering Mechanisms of Zanthoxylum piperitum Fruits for the Alleviation of Rheumatoid Arthritis Based on Network Pharmacology. Biology. 2021; 10(8):703. https://doi.org/10.3390/biology10080703
Chicago/Turabian StyleOh, Kikwang, Md. Adnan, and Dongha Cho. 2021. "Uncovering Mechanisms of Zanthoxylum piperitum Fruits for the Alleviation of Rheumatoid Arthritis Based on Network Pharmacology" Biology 10, no. 8: 703. https://doi.org/10.3390/biology10080703
APA StyleOh, K., Adnan, M., & Cho, D. (2021). Uncovering Mechanisms of Zanthoxylum piperitum Fruits for the Alleviation of Rheumatoid Arthritis Based on Network Pharmacology. Biology, 10(8), 703. https://doi.org/10.3390/biology10080703








