Transcriptomic Responses Induced in Muscle and Adipose Tissues of Growing Pigs by Intravenous Infusion of Sodium Butyrate
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Ethics Statement
2.2. Animals, Housing, Experimental Design, and Sampling
2.3. RNA Extraction and Purification
2.4. Library Construction and Sequencing
2.5. Data Analysis
3. Results
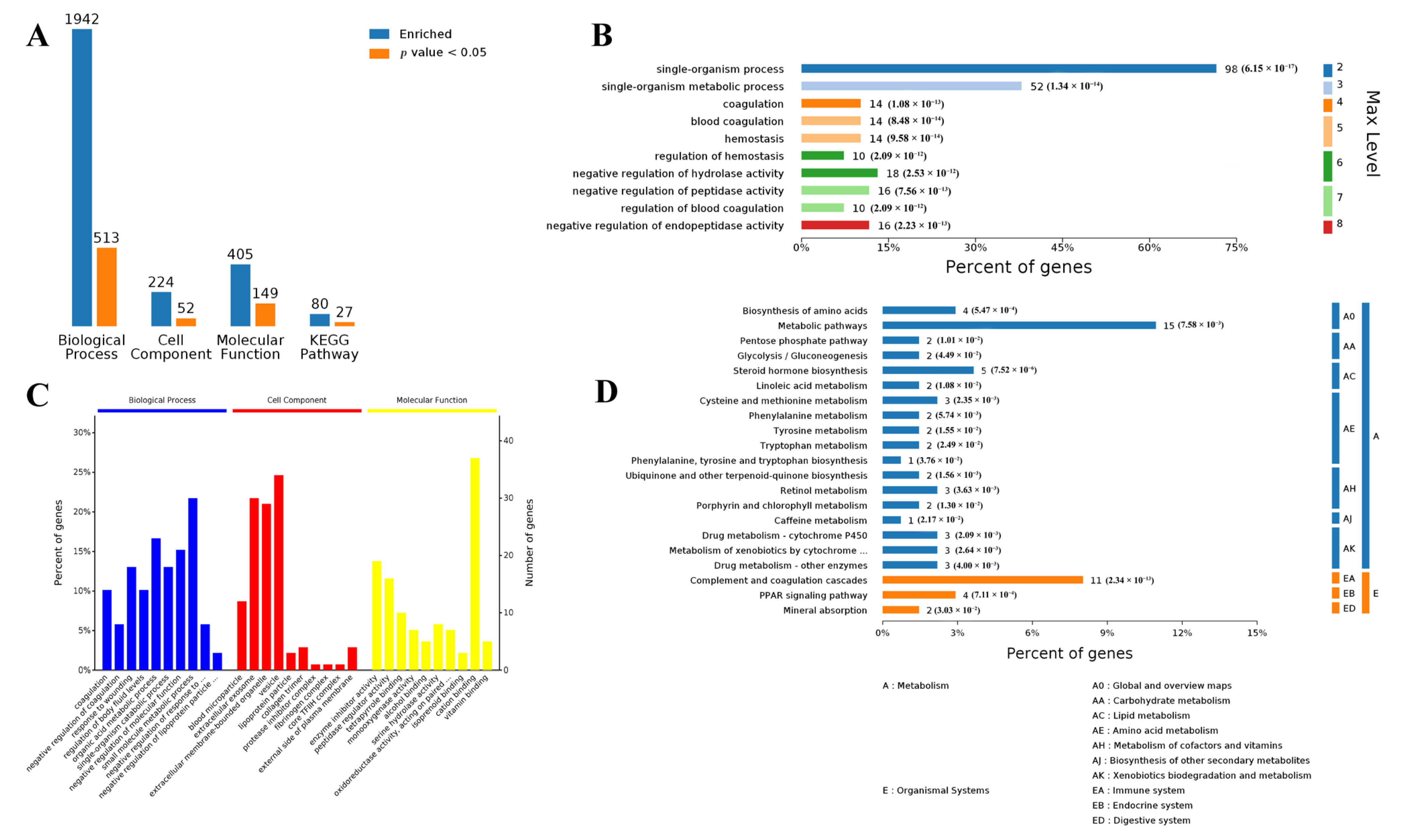
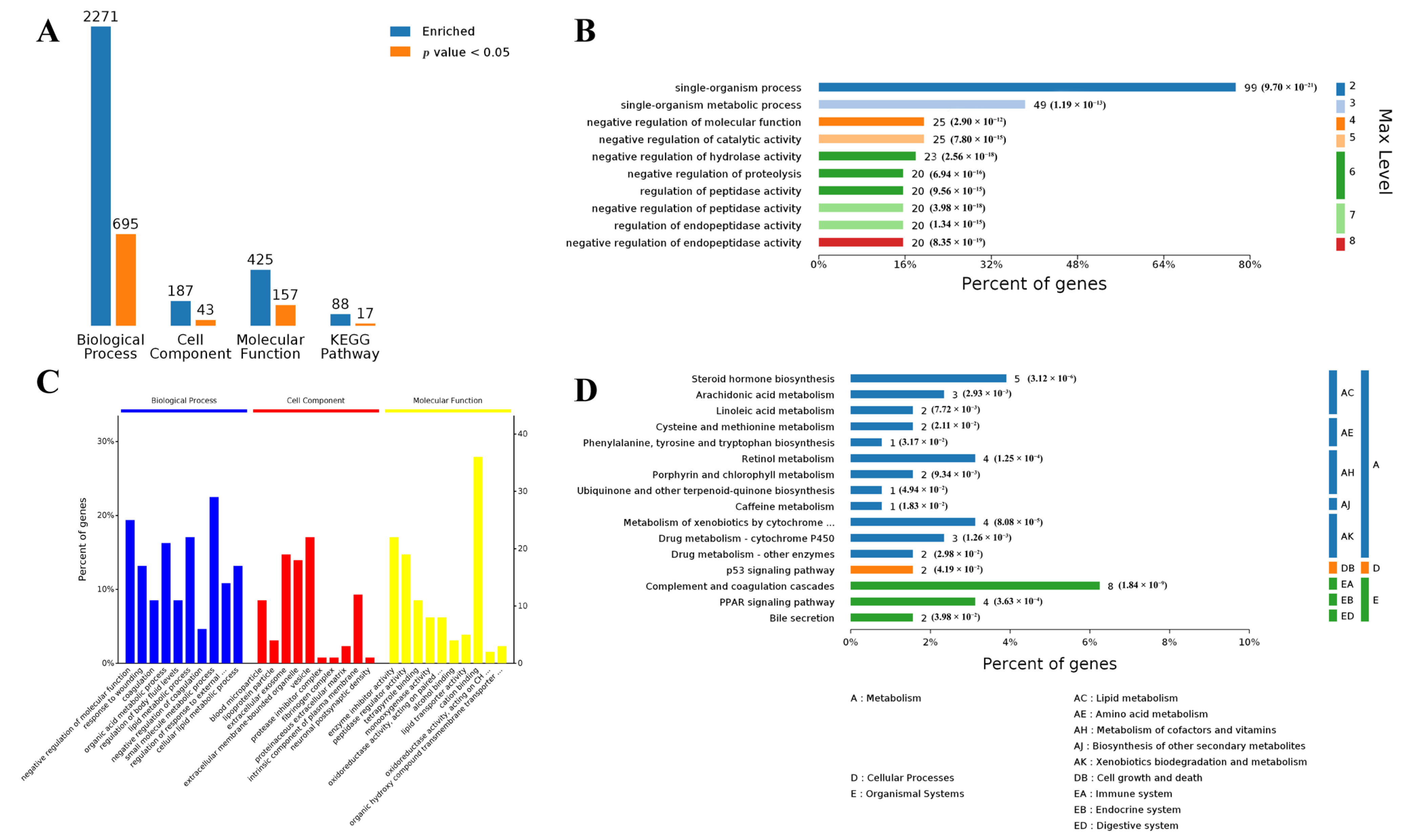
3.1. Differentially Expressed Genes in Muscle and Adipose Tissues
3.2. Functional Analysis of Differentially Expressed Genes in Both Muscle and Adipose Tissues
3.3. Comparison of DEGs Related to Metabolism in Muscle and Adipose Tissues
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Sethi, J.K.; Vidal-Puig, A.J. Thematic review series: Adipocyte biology—Adipose tissue function and plasticity orchestrate nutritional adaptation. J. Lipid Res. 2007, 48, 1253–1262. [Google Scholar] [CrossRef] [Green Version]
- Klaus, S. Adipose tissue as a regulator of energy balance. Curr. Drug Targets 2004, 5, 241–250. [Google Scholar] [CrossRef]
- de Lange, P.; Moreno, M.; Silvestri, E.; Lombardi, A.; Goglia, F.; Lanni, A. Fuel economy in food-deprived skeletal muscle: Signaling pathways and regulatory mechanisms. FASEB J. 2007, 21, 3431–3441. [Google Scholar] [CrossRef] [PubMed]
- Lu, H.; Su, S.; Ajuwon, K.M. Butyrate supplementation to gestatig sows and piglets induces muscle and adipose tissue oxidative genes and improves growth performance. J. Anim. Sci. 2012, 90, 430–432. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gao, Z.G.; Yin, J.; Zhang, J.; Ward, R.E.; Martin, R.J.; Lefevre, M.; Cefalu, W.T.; Ye, J. Butyrate improves insulin sensitivity and increases energy expenditure in mice. Diabetes 2009, 58, 1509–1517. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rumberger, J.M.; Arch, J.R.; Green, A. Butyrate and other short-chain fatty acids increase the rate of lipolysis in 3T3-L1 adipocytes. PeerJ 2014, 2, e611. [Google Scholar] [CrossRef] [Green Version]
- Heimann, E.; Nyman, M.; Degerman, E. Propionic acid and butyric acid inhibit lipolysis and de novo lipogenesis and increase insulin-stimulated glucose uptake in primary rat adipocytes. Adipocyte 2015, 4, 81–88. [Google Scholar] [CrossRef] [Green Version]
- Ohira, H.; Fujioka, Y.; Katagiri, C.; Mamoto, R.; Aoyama-Ishikawa, M.; Amako, K.; Izumi, Y.; Nishiumi, S.; Yoshida, M.; Usami, M.; et al. Butyrate attenuates inflammation and lipolysis generated by the interaction of adipocytes and macrophages. J. Atheroscler. Thromb. 2013, 20, 425–442. [Google Scholar] [CrossRef] [Green Version]
- Canani, R.B.; Costanzo, M.D.; Leone, L.; Pedata, M.; Meli, R.; Calignano, A. Potential beneficial effects of butyrate in intestinal and extraintestinal diseases. World J. Gastroenterol. 2011, 17, 1519–1528. [Google Scholar] [CrossRef]
- Macfarlane, G.; Macfarlane, S. Fermentation in the human large intestine: Its physiologic consequences and the potential contribution of prebiotics. J. Clin. Gastroenterol. 2011, 45, S120–S127. [Google Scholar] [CrossRef] [Green Version]
- Schonfeld, P.; Wojtczak, L. Short- and medium- chain fatty acids in energy metabolism: The cellular perspective. J. Lipid Res. 2016, 57, 943–954. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Choi, W.H.; Um, M.Y.; Ahn, J.; Jung, C.H.; Ha, T.Y. Long-term intake of rice improves insulin sensitivity in mice fed a high-fat diet. Nutrition 2014, 30, 920–927. [Google Scholar] [CrossRef] [PubMed]
- Khan, S.; Jena, G. The role of butyrate, a histone deacetylase inhibitor in diabetes mellitus: Experimental evidence for therapeutic intervention. Epigenomics 2015, 7, 669–680. [Google Scholar] [CrossRef] [PubMed]
- Husted, A.; Trauelsen, M.; Rudenko, O.; Hjorth, S.; Schwartz, T. GPCR-mediated signaling of metabolites. Cell Metab. 2017, 25, 777–796. [Google Scholar] [CrossRef] [Green Version]
- Hong, J.; Jia, Y.; Pan, S.; Jia, L.; Li, H.; Han, Z.; Cai, D.; Zhao, R. Butyrate alleviates high fat diet-induced obesity through activation of adiponectin-mediated pathway and stimulation of mitochondrial function in the skeletal muscle of mice. Oncotarget 2016, 7, 56071–56082. [Google Scholar] [CrossRef] [Green Version]
- Ren, E.; Chen, X.; Yu, S.; Xu, J.; Su, Y.; Zhu, W. Transcriptomic and metabolomic responses induced in the livers of growing pigs by a short-term intravenous infusion of sodium butyrate. Animal 2018, 12, 2318–2326. [Google Scholar] [CrossRef]
- Wang, Z.; Gerstein, M.; Snyder, M. RNA-Seq: A revolutionary tool for transcriptomics. Nat. Rev. Genet. 2009, 10, 57–63. [Google Scholar] [CrossRef]
- Guilloteau, P.; Zabielski, R.; Hammon, H.; Metges, C. Nutritional programming of gastrointestinal tract development. Is the pig a good model for man? Nutr. Res. Rev. 2010, 23, 4–22. [Google Scholar] [CrossRef] [Green Version]
- Chen, X.; Xu, J.; Su, Y.; Zhu, W. Effects of Intravenous Infusion with Sodium Butyrate on Colonic Microbiota, Intestinal Development- and Mucosal Immune-Related Gene Expression in Normal Growing Pigs. Front. Microbiol. 2018, 9, 1652. [Google Scholar] [CrossRef]
- Trapnell, C.; Pachter, L.; Salzberg, S.L. TopHat: Discovering splice junctions with RNA-Seq. Bioinformatics 2009, 25, 1105–1111. [Google Scholar] [CrossRef]
- Trapnell, C.; Williams, B.A.; Pertea, G.; Mortazavi, A.; Kwan, G.; Baren, M.J.; Salzberg, S.L.; Wold, B.J.; Pachter, L. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat. Biotechnol. 2010, 28, 511–515. [Google Scholar] [CrossRef] [Green Version]
- Benjamini, Y.; Hochberg, Y. Controlling the false discovery rate—a practical and powerful approach to multiple testing. J. R. Stat. Soc. Ser. B-Stat. Methodol. 1995, 57, 289–300. [Google Scholar] [CrossRef]
- Arakaki, T.L.; Pezza, J.A.; Cronin, M.A.; Hopkins, C.E.; Zimmer, D.B.; Tolan, D.R.; Allen, K.N. Structure of human brain fructose 1,6-(bis)phosphate aldolase: Linking isozyme structure with function. Protein Sci. 2004, 13, 3077–3084. [Google Scholar] [CrossRef] [PubMed]
- Langellotti, S.; Romano, M.; Guarnaccia, C.; Granata, V.; Orrù, S.; Zagari, A.; Baralle, F.E.; Salvatore, F. A novel anti-aldolase C antibody specifically interacts with residues 85–102 of the protein. mAbs 2014, 6, 708–717. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schönfeld, P.; Wojtczak, A.B.; Geelen, M.J.; Kunz, W.; Wojtczak, L. On the mechanism of the so-called uncoupling effect of medium- and short-chain fatty acids. Biochim. Biophys. Acta 1988, 936, 280–288. [Google Scholar] [CrossRef] [Green Version]
- Nobes, C.D.; Hay, W.W., Jr.; Brand, M.D. The mechanism of stimulation of respiration by fatty acids in isolated hepatocytes. J. Biol. Chem. 1990, 256, 12910–12915. [Google Scholar] [CrossRef]
- González-Manchón, C.; Ayuso, M.S.; Parrilla, R. Control of hepatic gluconeogenesis: Role of fatty acid oxidation. Arch. Biochem. Biophys. 1989, 271, 1–9. [Google Scholar] [CrossRef]
- Scholte, H.R.; Groot, P.H. Organ and intracellular localization of short-chain acyl-CoA synthetases in rat and guinea-pig. Biochim. Biophys. Acta 1975, 409, 283–296. [Google Scholar] [CrossRef]
- Debeer, L.J.; Mannaerts, G.; De Schepper, P.J. Effects of octanoate and oleate on energy metabolism in the perfused rat liver. Eur. J. Biochem. 1974, 47, 591–600. [Google Scholar] [CrossRef]
- Soliman, M.M.; Ahmed, M.M.; Salah-Eldin, A.E.; Abdel-Aal, A.A. Butyrate regulates leptin expression through different signaling pathways in adipocytes. J. Vet. Sci. 2011, 12, 319–323. [Google Scholar] [CrossRef]
- Dobrzyn, P.; Dobrzyn, A.; Miyazaki, M.; Cohen, P.; Asilmaz, E.; Hardie, D.G.; Friedman, J.M.; Ntambi, J.M. Stearoyl-CoA desaturase 1 deficiency increases fatty acid oxidation by activating AMP-activated protein kinase in liver. Proc. Natl. Acad. Sci. USA 2004, 101, 6409–6414. [Google Scholar] [CrossRef] [Green Version]
- De Vadder, F.; Kovatcheva-Datchary, P.; Goncalves, D.; Vinera, J.; Zitoun, C.; Duchampt, A.; Bäckhed, F.; Mithieux, G. Microbiota-generated metabolites promote metabolic benefits via gut-brain neural circuits. Cell 2014, 156, 84–96. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mollica, M.P.; Mattace Raso, G.; Cavaliere, G.; Trinchese, G.; De Filippo, C.; Aceto, S.; Prisco, M.; Pirozzi, C.; Guida, F.D.; Lama, A.; et al. Butyrate Regulates Liver Mitochondrial Function, Efficiency, and Dynamics in Insulin-Resistant Obese Mice. Diabetes 2017, 66, 1405–1418. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, Z.; Yi, C.X.; Katiraei, S.; Kooijman, S.; Zhou, E.; Chung, C.K.; Gao, Y.; Heuvel, J.K.; Meijer, O.C.; Berbée, J.F.P.; et al. Butyrate reduces appetite and activates brown adipose tissue via the gut-brain neural circuit. Gut 2018, 67, 1269–1279. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dunning, K.R.; Anastasi, M.R.; Zhang, V.J.; Russell, D.L.; Robker, R.L. Regulation of Fatty Acid Oxidation in Mouse Cumulus-Oocyte Complexes during Maturation and Modulation by PPAR Agonists. PLoS ONE 2014, 9, e87327. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Brasaemle, D.L. Thematic review series: Adipocyte biology. The perilipin family of structural lipid droplet proteins: Stabilization of lipid droplets and control of lipolysis. J. Lipid. Res. 2007, 48, 2547–2559. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shijun, L.; Khan, R.; Raza, S.H.A.; Jieyun, H.; Chugang, M.; Kaster, N.; Gong, C.; Chunping, Z.; Schreurs, N.M.; Linsen, Z. Function and characterization of the promoter region of perilipin 1 (PLIN1): Roles of E2F1, PLAG1, C/EBPβ, and SMAD3 in bovine adipocytes. Genomics 2020, 112, 2400–2409. [Google Scholar] [CrossRef]
- Grahn, T.H.; Zhang, Y.; Lee, M.J.; Sommer, A.G.; Mostoslavsky, G.; Fried, S.K.; Greenberg, A.S.; Puri, V. FSP27 and PLIN1 interaction promotes the formation of large lipid droplets in human adipocytes. Biochem. Biophys. Res. Commun. 2013, 432, 296–301. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Georgiadi, A.; Wang, Y.; Stienstra, R.; Tjeerdema, N.; Janssen, A.; Stalenhoef, A.; Vliet, J.A.; Roos, A.; Tamsma, J.T.; Smit, J.W.A.; et al. Overexpression of angiopoietin-like protein 4 protects against atherosclerosis development. Arterioscler. Thromb. Vasc. Biol. 2013, 33, 1529–1537. [Google Scholar] [CrossRef] [Green Version]


| Tissues | Genes 1 | Major Metabolic Types |
|---|---|---|
| muscle | PPP1R3B, PCK1, APOA2, ALDOC, UGT2B31, LOC100623255, ANKH, MAT1A, PRPS2, ITIH2, ITIH1 | Carbohydrate metabolism |
| PLIN1, ANKRD23, CES3, LOC10073013, HSD17B13, PCK1, CYP2D25, ETNK2, ACSM4, ADH4, CYP1A2, TTR, CYP2E1, APOC3, CPS1, GC, CYP2C33, CYP2C49, LOC100739741, APOA2, APOC2, MYLIP, ARX, MLC1, ZNF703, LOC100620829, LOC100623140, FASN | Lipid metabolism | |
| PAH, HPD, TAT, TDO2, MAT1A, CPS1, bhmt, SGK1, HRG, APOA2 | Amino acid metabolism | |
| adipose | LEP, C3, IGF1, SLC2A4, APOA2, LPL, UGT2B31, LOC100623255, UPP1, ITIH2, ITIH1, LBP | Carbohydrate metabolism |
| APOC3, APOC2, PNPLA3, APOA2, LPL, LOC100737013, CYP2C49, LEP, LOC100510957, TTR, DHCR24, LOC100739741, CYP1A2, STAR, CYP2D25, CYP2E1, CPS1, CYP2C33, GNLY, ACSM4, C3, GC, LOC100620829, LBP, LOC100623140, SLCO2A1, ANGPTL4 | Lipid metabolism | |
| bhmt, CPS1, TAT, DUOX2, TTR, LEP, SFRP2, PRDM8, HRG, APOA2 | Amino acid metabolism |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, H.; Ren, E.; Xu, R.; Su, Y. Transcriptomic Responses Induced in Muscle and Adipose Tissues of Growing Pigs by Intravenous Infusion of Sodium Butyrate. Biology 2021, 10, 559. https://doi.org/10.3390/biology10060559
Zhang H, Ren E, Xu R, Su Y. Transcriptomic Responses Induced in Muscle and Adipose Tissues of Growing Pigs by Intravenous Infusion of Sodium Butyrate. Biology. 2021; 10(6):559. https://doi.org/10.3390/biology10060559
Chicago/Turabian StyleZhang, He, Erdu Ren, Rongying Xu, and Yong Su. 2021. "Transcriptomic Responses Induced in Muscle and Adipose Tissues of Growing Pigs by Intravenous Infusion of Sodium Butyrate" Biology 10, no. 6: 559. https://doi.org/10.3390/biology10060559
APA StyleZhang, H., Ren, E., Xu, R., & Su, Y. (2021). Transcriptomic Responses Induced in Muscle and Adipose Tissues of Growing Pigs by Intravenous Infusion of Sodium Butyrate. Biology, 10(6), 559. https://doi.org/10.3390/biology10060559







