Diversity of Rainbow Trout Blood B Cells Revealed by Single Cell RNA Sequencing
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Experimental Fish
2.2. Peripheral Blood Leukocyte Isolation and Sorting
2.3. Library Construction and Sequencing
2.4. Alignment and Initial Processing of the Sequencing Data
2.5. Data Integration and Cell Clustering
2.6. Marker Identification and Functional Analysis
3. Results
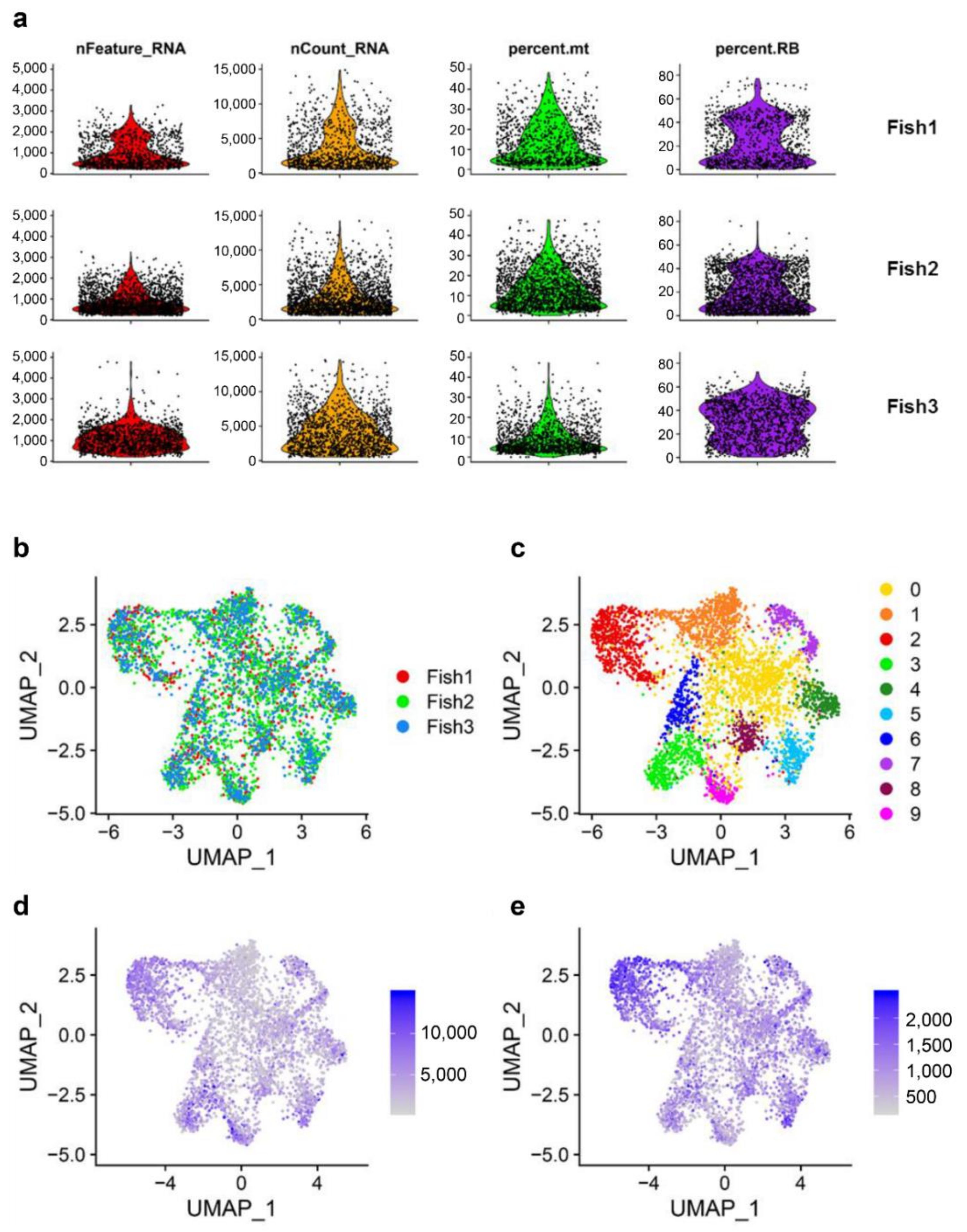
3.1. Single-Cell Transcriptomic Analysis of Rainbow Trout MHC II+ Lymphoid Cells from Peripheral Blood
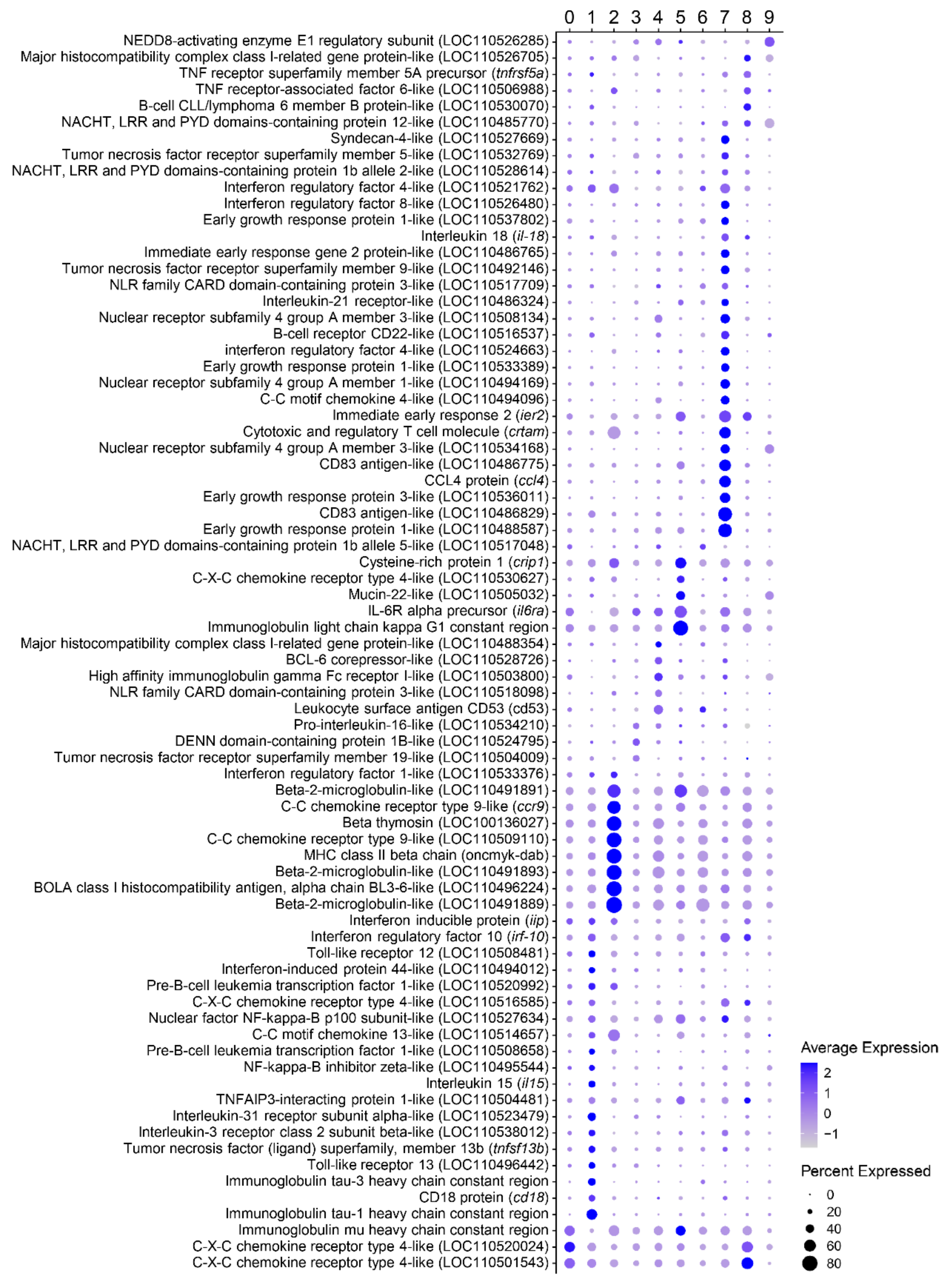
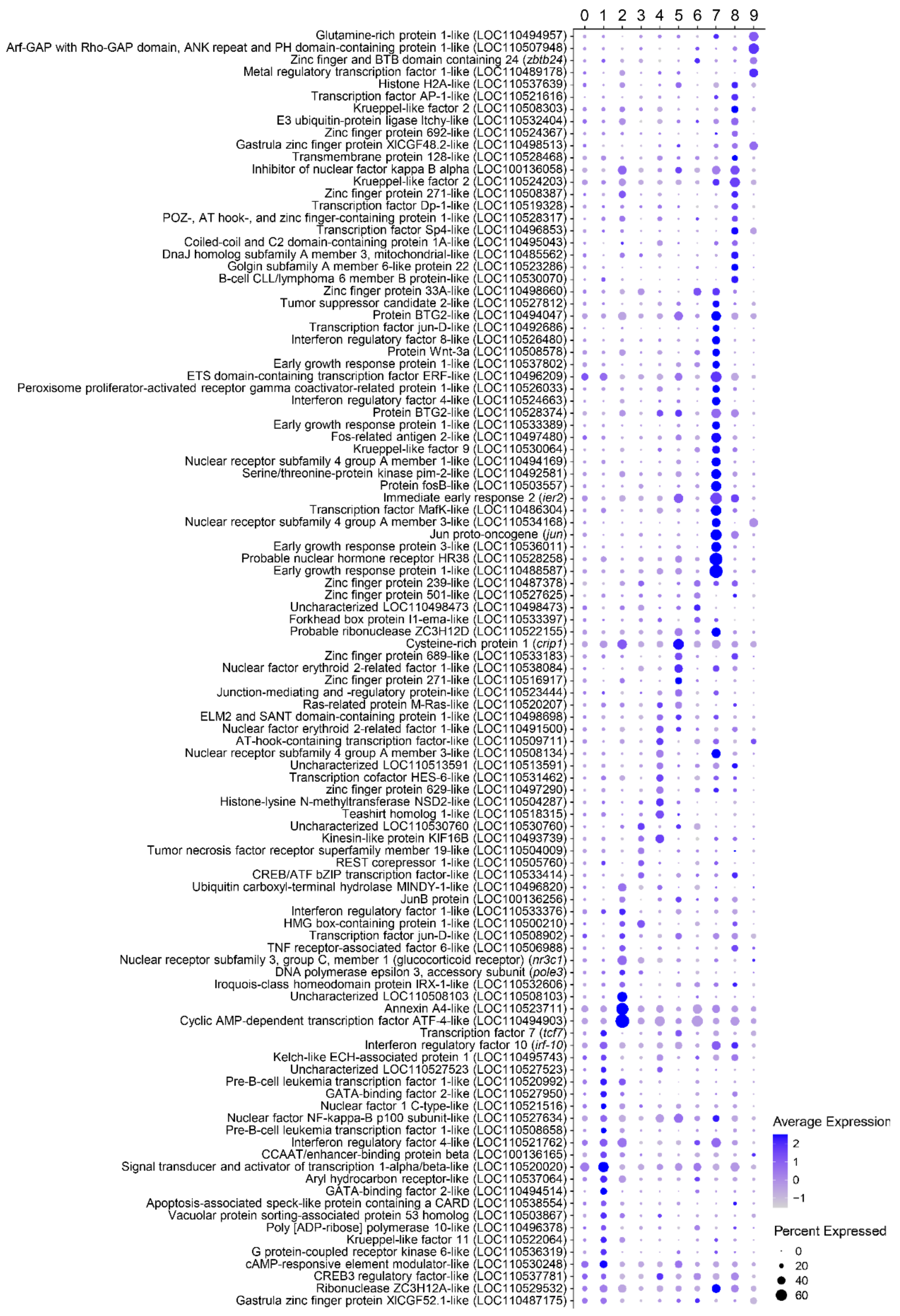
3.2. Characterization of Marker Transcripts Associated with Identified Cell Clusters
3.2.1. Description of Molecular Markers Associated with Different B Cell Subsets
Cluster 0
Cluster 1
Cluster 2
Cluster 3
Cluster 4
Cluster 5
Cluster 6
Cluster 7
Cluster 8
Cluster 9
3.2.2. Long Non-Coding RNAs Associated to B Cell Subsets
4. Discussion
4.1. Cluster 2 May Represent a Subpopulation of Mature B Cells with High Antigen Presenting Capacities
4.2. Cluster 7 May Represent a Subpopulation of Activated B Cells with Innate Properties
4.3. Cells in Cluster 1 Correspond to IgT+ B Cells
4.4. CXCR4-Like 4 Genes as Key Markers for Fish B Cells
4.5. Long Non-Coding RNAs Seem to Regulate Different B Cell Transitional Stages in Fish
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Cooper, M.D.; Alder, M.N. The evolution of adaptive immune systems. Cell 2006, 124, 815–822. [Google Scholar] [CrossRef] [PubMed]
- Press, C.M.; Evensen, Ø. The morphology of the immune system in teleost fishes. Fish Shellfish Immunol. 1999, 9, 309–318. [Google Scholar] [CrossRef]
- Zwollo, P.; Cole, S.; Bromage, E.; Kaattari, S. B cell heterogeneity in the teleost kidney: Evidence for a maturation gradient from anterior to posterior kidney. J. Immunol. 2005, 174, 6608–6616. [Google Scholar] [CrossRef] [PubMed]
- Hansen, J.D.; Landis, E.D.; Phillips, R.B. Discovery of a unique Ig heavy-chain isotype (IgT) in rainbow trout: Implications for a distinctive B cell developmental pathway in teleost fish. Proc. Natl. Acad. Sci. USA 2005, 102, 6919–6924. [Google Scholar] [CrossRef] [PubMed]
- Parra, D.; Takizawa, F.; Sunyer, J.O. Evolution of B cell immunity. Annu. Rev. Anim. Biosci. 2013, 1, 65–97. [Google Scholar] [CrossRef]
- Granja, A.G.; Tafalla, C. Different IgM+ B cell subpopulations residing within the peritoneal cavity of vaccinated rainbow trout are differently regulated by BAFF. Fish Shellfish Immunol. 2019, 85, 9–17. [Google Scholar] [CrossRef]
- Tafalla, C.; González, L.; Castro, R.; Granja, A.G. B cell-activating factor regulates different aspects of B cell functionality and is produced by a subset of splenic B cells in teleost fish. Front. Immunol. 2017, 8, 295. [Google Scholar] [CrossRef]
- Edholm, E.S.; Bengten, E.; Stafford, J.L.; Sahoo, M.; Taylor, E.B.; Miller, N.W.; Wilson, M. Identification of two IgD+ B cell populations in channel catfish, Ictalurus punctatus. J. Immunol. 2010, 185, 4082–4094. [Google Scholar] [CrossRef]
- Castro, R.; Bromage, E.; Abos, B.; Pignatelli, J.; Gonzalez Granja, A.; Luque, A.; Tafalla, C. CCR7 is mainly expressed in teleost gills, where it defines an IgD+IgM- B lymphocyte subset. J. Immunol. 2014, 192, 1257–1266. [Google Scholar] [CrossRef]
- Perdiguero, P.; Martin-Martin, A.; Benedicenti, O.; Diaz-Rosales, P.; Morel, E.; Munoz-Atienza, E.; Garcia-Flores, M.; Simon, R.; Soleto, I.; Cerutti, A.; et al. Teleost IgD+IgM- B cells mount clonally expanded and mildly mutated intestinal IgD responses in the absence of lymphoid follicles. Cell Rep. 2019, 29, 4223–4235. [Google Scholar] [CrossRef]
- Zhang, Y.A.; Salinas, I.; Oriol Sunyer, J. Recent findings on the structure and function of teleost IgT. Fish Shellfish Immunol. 2011, 31, 627–634. [Google Scholar] [CrossRef]
- Zhang, N.; Zhang, X.-J.; Chen, D.-D.; Sunyer, J.O.; Zhang, Y.-A. Molecular characterization and expression analysis of three subclasses of IgT in rainbow trout (Oncorhynchus mykiss). Dev. Comp. Immunol. 2017, 70, 94–105. [Google Scholar] [CrossRef] [PubMed]
- Zwollo, P. Dissecting teleost B cell differentiation using transcription factors. Dev. Comp. Immunol. 2011, 35, 898–905. [Google Scholar] [CrossRef] [PubMed]
- Zwollo, P.; Haines, A.; Rosato, P.; Gumulak-Smith, J. Molecular and cellular analysis of B-cell populations in the rainbow trout using Pax5 and immunoglobulin markers. Dev. Comp. Immunol. 2008, 32, 1482–1496. [Google Scholar] [CrossRef] [PubMed]
- Guslund, N.C.; Solbakken, M.H.; Brieuc, M.S.; Jentoft, S.; Jakobsen, K.S.; Qiao, S.-W. Single-cell transcriptome profiling of immune cell repertoire of the Atlantic cod which naturally lacks the major histocompatibility class II system. Front. Immunol. 2020, 11, 2602. [Google Scholar] [CrossRef] [PubMed]
- Niu, J.; Huang, Y.; Liu, X.; Zhang, Z.; Tang, J.; Wang, B.; Lu, Y.; Cai, J.; Jian, J. Single-cell RNA-seq reveals different subsets of non-specific cytotoxic cells in teleost. Genomics 2020, 112, 5170–5179. [Google Scholar] [CrossRef] [PubMed]
- Abós, B.; Wang, T.; Castro, R.; Granja, A.G.; Leal, E.; Havixbeck, J.; Luque, A.; Barreda, D.R.; Secombes, C.J.; Tafalla, C. Distinct differentiation programs triggered by IL-6 and LPS in teleost IgM+ B cells in the absence of germinal centers. Sci. Rep. 2016, 6, 1–16. [Google Scholar]
- Granja, A.G.; Leal, E.; Pignatelli, J.; Castro, R.; Abos, B.; Kato, G.; Fischer, U.; Tafalla, C. Identification of teleost skin CD8α+ dendritic-like cells, representing a potential common ancestor for mammalian cross-presenting dendritic cells. J. Immunol. 2015, 195, 1825–1837. [Google Scholar] [CrossRef]
- Zheng, G.X.; Terry, J.M.; Belgrader, P.; Ryvkin, P.; Bent, Z.W.; Wilson, R.; Ziraldo, S.B.; Wheeler, T.D.; McDermott, G.P.; Zhu, J.; et al. Massively parallel digital transcriptional profiling of single cells. Nat. Commun. 2017, 8, 14049. [Google Scholar] [CrossRef]
- Satija, R.; Farrell, J.A.; Gennert, D.; Schier, A.F.; Regev, A. Spatial reconstruction of single-cell gene expression data. Nat. Biotechnol. 2015, 33, 495–502. [Google Scholar] [CrossRef]
- Stuart, T.; Butler, A.; Hoffman, P.; Hafemeister, C.; Papalexi, E.; Mauck III, W.M.; Hao, Y.; Stoeckius, M.; Smibert, P.; Satija, R. Comprehensive integration of single-cell data. Cell 2019, 177, 1888–1902.e1821. [Google Scholar] [CrossRef]
- Peñaranda, M.; Michelle, D.; Jensen, I.; Tollersrud, L.G.; Bruun, J.-A.; Jørgensen, J.B. Profiling the atlantic salmon IgM+ B cell surface proteome: Novel information on teleost fish B cell protein repertoire and identification of potential B cell markers. Front. Immunol. 2019, 10, 37. [Google Scholar] [CrossRef]
- Mestas, J.; Hughes, C.C. Of mice and not men: Differences between mouse and human immunology. J. Immunol. 2004, 172, 2731–2738. [Google Scholar] [CrossRef]
- Zhang, X.; Lan, Y.; Xu, J.; Quan, F.; Zhao, E.; Deng, C.; Luo, T.; Xu, L.; Liao, G.; Yan, M. CellMarker: A manually curated resource of cell markers in human and mouse. Nucleic Acids Res. 2019, 47, D721–D728. [Google Scholar] [CrossRef] [PubMed]
- Wurbel, M.-A.; Malissen, M.; Guy-Grand, D.; Meffre, E.; Nussenzweig, M.C.; Richelme, M.; Carrier, A.; Malissen, B. Mice lacking the CCR9 CC-chemokine receptor show a mild impairment of early T-and B-cell development and a reduction in T-cell receptor γδ+ gut intraepithelial lymphocytes. Blood 2001, 98, 2626–2632. [Google Scholar] [CrossRef]
- Pabst, O.; Ohl, L.; Wendland, M.; Wurbel, M.-A.; Kremmer, E.; Malissen, B.; Förster, R. Chemokine receptor CCR9 contributes to the localization of plasma cells to the small intestine. J. Exp. Med. 2004, 199, 411–416. [Google Scholar] [CrossRef]
- Demberg, T.; Mohanram, V.; Venzon, D.; Robert-Guroff, M. Phenotypes and distribution of mucosal memory B-cell populations in the SIV/SHIV rhesus macaque model. Clin. Immunol. 2014, 153, 264–276. [Google Scholar] [CrossRef]
- So, J.-S. Roles of endoplasmic reticulum stress in immune responses. Mol. Cells 2018, 41, 705. [Google Scholar]
- Li, J.; Barreda, D.R.; Zhang, Y.-A.; Boshra, H.; Gelman, A.E.; LaPatra, S.; Tort, L.; Sunyer, J.O. B lymphocytes from early vertebrates have potent phagocytic and microbicidal abilities. Nat. Immunol. 2006, 7, 1116–1124. [Google Scholar] [CrossRef]
- Sunyer, J.O. Evolutionary and functional relationships of B cells from fish and mammals: Insights into their novel roles in phagocytosis and presentation of particulate antigen. Infect. Disord. Drug Targets 2012, 12, 200–212. [Google Scholar] [CrossRef]
- Sunyer, J.O. Fishing for mammalian paradigms in the teleost immune system. Nat. Immunol. 2013, 14, 320. [Google Scholar] [CrossRef]
- Haugland, G.T.; Jordal, A.-E.O.; Wergeland, H.I. Characterization of small, mononuclear blood cells from salmon having high phagocytic capacity and ability to differentiate into dendritic like cells. PLoS ONE 2012, 7, e49260. [Google Scholar] [CrossRef]
- Granja, A.G.; Perdiguero, P.; Martín-Martín, A.; Díaz-Rosales, P.; Soleto, I.; Tafalla, C. Rainbow trout IgM+ B cells preferentially respond to thymus-independent antigens but are activated by CD40L. Front. Immunol. 2019, 10, 2902. [Google Scholar] [CrossRef]
- Mackenzie, S.; Liarte, C.; Iliev, D.; Planas, J.; Tort, L.; Goetz, F. Characterization of a highly inducible novel CC chemokine from differentiated rainbow trout (Oncorhynchus mykiss) macrophages. Immunogenetics 2004, 56, 611–615. [Google Scholar] [CrossRef]
- Laing, K.J.; Secombes, C.J. Chemokines. Dev. Comp. Immunol. 2004, 28, 443–460. [Google Scholar] [CrossRef]
- Peatman, E.; Liu, Z. Evolution of CC chemokines in teleost fish: A case study in gene duplication and implications for immune diversity. Immunogenetics 2007, 59, 613–623. [Google Scholar] [CrossRef]
- Siveke, J.T.; Hamann, A. Cutting edge: T helper 1 and T helper 2 cells respond differentially to chemokines. J. Immunol. 1998, 160, 550–554. [Google Scholar] [PubMed]
- Abós, B.; Castro, R.; Granja, A.G.; Havixbeck, J.J.; Barreda, D.R.; Tafalla, C. Early activation of teleost B cells in response to rhabdovirus infection. J. Virol. 2015, 89, 1768–1780. [Google Scholar] [CrossRef]
- Ma, S.; Turetsky, A.; Trinh, L.; Lu, R. IFN regulatory factor 4 and 8 promote Ig light chain κ locus activation in pre-B cell development. J. Immunol. 2006, 177, 7898–7904. [Google Scholar] [CrossRef]
- Sciammas, R.; Shaffer, A.; Schatz, J.H.; Zhao, H.; Staudt, L.M.; Singh, H. Graded expression of interferon regulatory factor-4 coordinates isotype switching with plasma cell differentiation. Immunity 2006, 25, 225–236. [Google Scholar] [CrossRef]
- Gururajan, M.; Simmons, A.; Dasu, T.; Spear, B.T.; Calulot, C.; Robertson, D.A.; Wiest, D.L.; Monroe, J.G.; Bondada, S. Early growth response genes regulate B cell development, proliferation, and immune response. J. Immunol. 2008, 181, 4590–4602. [Google Scholar] [CrossRef]
- Oh, Y.-K.; Jang, E.; Paik, D.-J.; Youn, J. Early growth response-1 plays a non-redundant role in the differentiation of B cells into plasma cells. Immune Netw. 2015, 15, 161–166. [Google Scholar] [CrossRef] [PubMed]
- Li, S.; Miao, T.; Sebastian, M.; Bhullar, P.; Ghaffari, E.; Liu, M.; Symonds, A.L.; Wang, P. The transcription factors Egr2 and Egr3 are essential for the control of inflammation and antigen-induced proliferation of B and T cells. Immunity 2012, 37, 685–696. [Google Scholar] [CrossRef]
- Ubieta, K.; Garcia, M.; Grötsch, B.; Uebe, S.; Weber, G.F.; Stein, M.; Ekici, A.; Schett, G.; Mielenz, D.; Bozec, A. Fra-2 regulates B cell development by enhancing IRF4 and Foxo1 transcription. J. Exp. Med. 2017, 214, 2059–2071. [Google Scholar] [CrossRef]
- Labbaye, C.; Valtieri, M.; Barberi, T.; Meccia, E.; Masella, B.; Pelosi, E.; Condorelli, G.; Testa, U.; Peschle, C. Differential expression and functional role of GATA-2, NF-E2, and GATA-1 in normal adult hematopoiesis. J. Clin. Investig. 1995, 95, 2346–2358. [Google Scholar] [CrossRef] [PubMed]
- Bresnick, E.H.; Katsumura, K.R.; Lee, H.-Y.; Johnson, K.D.; Perkins, A.S. Master regulatory GATA transcription factors: Mechanistic principles and emerging links to hematologic malignancies. Nucleic Acids Res. 2012, 40, 5819–5831. [Google Scholar] [CrossRef]
- Sanyal, M.; Tung, J.W.; Karsunky, H.; Zeng, H.; Selleri, L.; Weissman, I.L.; Herzenberg, L.A.; Cleary, M.L. B-cell development fails in the absence of the Pbx1 proto-oncogene. Blood 2007, 109, 4191–4199. [Google Scholar] [CrossRef]
- Zhang, Y.-A.; Salinas, I.; Li, J.; Parra, D.; Bjork, S.; Xu, Z.; LaPatra, S.E.; Bartholomew, J.; Sunyer, J.O. IgT, a primitive immunoglobulin class specialized in mucosal immunity. Nat. Immunol. 2010, 11, 827–835. [Google Scholar] [CrossRef]
- Asea, A.; Rehli, M.; Kabingu, E.; Boch, J.A.; Baré, O.; Auron, P.E.; Stevenson, M.A.; Calderwood, S.K. Novel signal transduction pathway utilized by extracellular HSP70 role of Toll-like receptor (TLR) 2 and TLR4. J. Biol. Chem. 2002, 277, 15028–15034. [Google Scholar] [CrossRef]
- Becker, T.; Hartl, F.-U.; Wieland, F. CD40, an extracellular receptor for binding and uptake of Hsp70–peptide complexes. J. Cell Biol. 2002, 158, 1277–1285. [Google Scholar] [CrossRef]
- Oldenburg, M.; Krüger, A.; Ferstl, R.; Kaufmann, A.; Nees, G.; Sigmund, A.; Bathke, B.; Lauterbach, H.; Suter, M.; Dreher, S. TLR13 recognizes bacterial 23S rRNA devoid of erythromycin resistance–forming modification. Science 2012, 337, 1111–1115. [Google Scholar] [CrossRef]
- Zhang, D.; Zhang, G.; Hayden, M.S.; Greenblatt, M.B.; Bussey, C.; Flavell, R.A.; Ghosh, S. A toll-like receptor that prevents infection by uropathogenic bacteria. Science 2004, 303, 1522–1526. [Google Scholar] [CrossRef]
- Armitage, R.J.; Macduff, B.M.; Eisenman, J.; Paxton, R.; Grabstein, K.H. IL-15 has stimulatory activity for the induction of B cell proliferation and differentiation. J. Immunol. 1995, 154, 483–490. [Google Scholar]
- Bernasconi, N.L.; Traggiai, E.; Lanzavecchia, A. Maintenance of serological memory by polyclonal activation of human memory B cells. Science 2002, 298, 2199–2202. [Google Scholar] [CrossRef]
- Mackay, F.; Browning, J.L. BAFF: A fundamental survival factor for B cells. Nat. Rev. Immunol. 2002, 2, 465–475. [Google Scholar] [CrossRef]
- Ma, Q.; Jones, D.; Springer, T.A. The chemokine receptor CXCR4 is required for the retention of B lineage and granulocytic precursors within the bone marrow microenvironment. Immunity 1999, 10, 463–471. [Google Scholar] [CrossRef]
- Muehlinghaus, G.; Cigliano, L.; Huehn, S.; Peddinghaus, A.; Leyendeckers, H.; Hauser, A.E.; Hiepe, F.; Radbruch, A.; Arce, S.; Manz, R.A. Regulation of CXCR3 and CXCR4 expression during terminal differentiation of memory B cells into plasma cells. Blood 2005, 105, 3965–3971. [Google Scholar] [CrossRef] [PubMed]
- Nie, Y.; Waite, J.; Brewer, F.; Sunshine, M.-J.; Littman, D.R.; Zou, Y.-R. The role of CXCR4 in maintaining peripheral B cell compartments and humoral immunity. J. Exp. Med. 2004, 200, 1145–1156. [Google Scholar] [CrossRef] [PubMed]
- Chong, S.W.; Emelyanov, A.; Gong, Z.; Korzh, V. Expression pattern of two zebrafish genes, cxcr4a and cxcr4b. Mech. Dev. 2001, 109, 347–354. [Google Scholar] [CrossRef]
- Lu, W.-J.; Zhou, L.; Gao, F.-X.; Sun, Z.-H.; Li, Z.; Liu, X.-C.; Li, S.-S.; Wang, Y.; Gui, J.-F. Divergent expression patterns and function of two cxcr4 paralogs in hermaphroditic Epinephelus coioides. Int. J. Mol. Sci. 2018, 19, 2943. [Google Scholar] [CrossRef]
- Lu, X.-J.; Zhu, K.; Shen, H.-X.; Nie, L.; Chen, J. CXCR4s in teleosts: Two paralogous chemokine receptors and their roles in hematopoietic stem/progenitor cell homeostasis. J. Immunol. 2020, 204, 1225–1241. [Google Scholar] [CrossRef]
- Lu, W.-J.; Zhou, L.; Gao, F.-X.; Zhou, Y.-L.; Li, Z.; Zhang, X.-J.; Wang, Y.; Gui, J.-F. Dynamic and differential expression of duplicated cxcr4/cxcl12 genes facilitates antiviral response in hexaploid gibel carp. Front. Immunol. 2020, 11, 2176. [Google Scholar] [CrossRef]
- Marques, A.C.; Hughes, J.; Graham, B.; Kowalczyk, M.S.; Higgs, D.R.; Ponting, C.P. Chromatin signatures at transcriptional start sites separate two equally populated yet distinct classes of intergenic long noncoding RNAs. Genome Biol. 2013, 14, R131. [Google Scholar] [CrossRef]
- Winkle, M.; Kluiver, J.; Diepstra, A.; van den Berg, A. Emerging roles for long noncoding RNAs in B-cell development and malignancy. Crit. Rev. Oncol./Hematol. 2017, 120, 77–85. [Google Scholar] [CrossRef] [PubMed]
- Casero, D.; Sandoval, S.; Seet, C.S.; Scholes, J.; Zhu, Y.; Ha, V.L.; Luong, A.; Parekh, C.; Crooks, G.M. Long non-coding RNA profiling of human lymphoid progenitor cells reveals transcriptional divergence of B cell and T cell lineages. Nat. Immunol. 2015, 16, 1282. [Google Scholar] [CrossRef]
- Agirre, X.; Meydan, C.; Jiang, Y.; Garate, L.; Doane, A.S.; Li, Z.; Verma, A.; Paiva, B.; Martín-Subero, J.I.; Elemento, O. Long non-coding RNAs discriminate the stages and gene regulatory states of human humoral immune response. Nat. Commun. 2019, 10, 1–16. [Google Scholar] [CrossRef]
- Brazão, T.F.; Johnson, J.S.; Müller, J.; Heger, A.; Ponting, C.P.; Tybulewicz, V.L. Long noncoding RNAs in B-cell development and activation. Blood 2016, 128, e10–e19. [Google Scholar] [CrossRef] [PubMed]
- Petri, A.; Dybkær, K.; Bøgsted, M.; Thrue, C.A.; Hagedorn, P.H.; Schmitz, A.; Bødker, J.S.; Johnsen, H.E.; Kauppinen, S. Long noncoding RNA expression during human B-cell development. PLoS ONE 2015, 10, e0138236. [Google Scholar] [CrossRef]
- Vives-Bauza, C.; Zhou, C.; Huang, Y.; Cui, M.; De Vries, R.L.; Kim, J.; May, J.; Tocilescu, M.A.; Liu, W.; Ko, H.S. PINK1-dependent recruitment of Parkin to mitochondria in mitophagy. Proc. Natl. Acad. Sci. USA 2010, 107, 378–383. [Google Scholar] [CrossRef]
- Xu, Y.; Shen, J.; Ran, Z. Emerging views of mitophagy in immunity and autoimmune diseases. Autophagy 2020, 16, 3–17. [Google Scholar] [CrossRef] [PubMed]
- Gkikas, I.; Palikaras, K.; Tavernarakis, N. The role of mitophagy in innate immunity. Front. Immunol. 2018, 9, 1283. [Google Scholar] [CrossRef] [PubMed]
- Cho, D.-H.; Kim, J.K.; Jo, E.-K. Mitophagy and Innate Immunity in Infection. Mol. Cells 2020, 43, 10. [Google Scholar]
- Zhang, X.; Yuan, D.; Sun, Q.; Xu, L.; Lee, E.; Lewis, A.J.; Zuckerbraun, B.S.; Rosengart, M.R. Calcium/calmodulin-dependent protein kinase regulates the PINK1/Parkin and DJ-1 pathways of mitophagy during sepsis. FASEB J. 2017, 31, 4382–4395. [Google Scholar] [CrossRef]
- Ellis, G.I.; Zhi, L.; Akundi, R.; Büeler, H.; Marti, F. Mitochondrial and cytosolic roles of PINK 1 shape induced regulatory T-cell development and function. Eur. J. Immunol. 2013, 43, 3355–3360. [Google Scholar] [CrossRef]
- De Haan, N.; Falck, D.; Wuhrer, M. Monitoring of immunoglobulin N-and O-glycosylation in health and disease. Glycobiology 2020, 30, 226–240. [Google Scholar] [CrossRef]
- Collin, M. Antibody glycosylation as an immunological key in health and disease. Glycobiology 2020, 30, 200. [Google Scholar] [CrossRef] [PubMed]
- Arnold, J.N.; Wormald, M.R.; Sim, R.B.; Rudd, P.M.; Dwek, R.A. The impact of glycosylation on the biological function and structure of human immunoglobulins. Annu. Rev. Immunol. 2007, 25, 21–50. [Google Scholar] [CrossRef] [PubMed]
- Van de Bovenkamp, F.S.; Derksen, N.I.; Ooijevaar-de Heer, P.; van Schie, K.A.; Kruithof, S.; Berkowska, M.A.; van der Schoot, C.E.; Ijspeert, H.; van der Burg, M.; Gils, A. Adaptive antibody diversification through N-linked glycosylation of the immunoglobulin variable region. Proc. Natl. Acad. Sci. USA 2019, 115, 1901–1906. [Google Scholar] [CrossRef]
- Koers, J.; Derksen, N.I.; Ooijevaar-de Heer, P.; Nota, B.; van de Bovenkamp, F.S.; Vidarsson, G.; Rispens, T. Biased N-glycosylation site distribution and acquisition across the antibody V region during B cell maturation. J. Immunol. 2019, 202, 2220–2228. [Google Scholar] [CrossRef]
- Wang, J.; Balog, C.I.; Stavenhagen, K.; Koeleman, C.A.; Scherer, H.U.; Selman, M.H.; Deelder, A.M.; Huizinga, T.W.; Toes, R.E.; Wuhrer, M. Fc-glycosylation of IgG1 is modulated by B-cell stimuli. Mol. Cell. Proteomics 2011, 10. [Google Scholar] [CrossRef]
- Weisel, F.J.; Mullett, S.J.; Elsner, R.A.; Menk, A.V.; Trivedi, N.; Luo, W.; Wikenheiser, D.; Hawse, W.F.; Chikina, M.; Smita, S. Germinal center B cells selectively oxidize fatty acids for energy while conducting minimal glycolysis. Nat. Immunol. 2020, 21, 331–342. [Google Scholar] [CrossRef] [PubMed]
- Edgar, R.; Domrachev, M.; Lash, A.E. Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 2002, 30, 207–210. [Google Scholar] [CrossRef] [PubMed]



Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Perdiguero, P.; Morel, E.; Tafalla, C. Diversity of Rainbow Trout Blood B Cells Revealed by Single Cell RNA Sequencing. Biology 2021, 10, 511. https://doi.org/10.3390/biology10060511
Perdiguero P, Morel E, Tafalla C. Diversity of Rainbow Trout Blood B Cells Revealed by Single Cell RNA Sequencing. Biology. 2021; 10(6):511. https://doi.org/10.3390/biology10060511
Chicago/Turabian StylePerdiguero, Pedro, Esther Morel, and Carolina Tafalla. 2021. "Diversity of Rainbow Trout Blood B Cells Revealed by Single Cell RNA Sequencing" Biology 10, no. 6: 511. https://doi.org/10.3390/biology10060511
APA StylePerdiguero, P., Morel, E., & Tafalla, C. (2021). Diversity of Rainbow Trout Blood B Cells Revealed by Single Cell RNA Sequencing. Biology, 10(6), 511. https://doi.org/10.3390/biology10060511





