Targeting the Mild-Hypoxia Driving Force for Metabolic and Muscle Transcriptional Reprogramming of Gilthead Sea Bream (Sparus aurata) Juveniles
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Ethics Statement
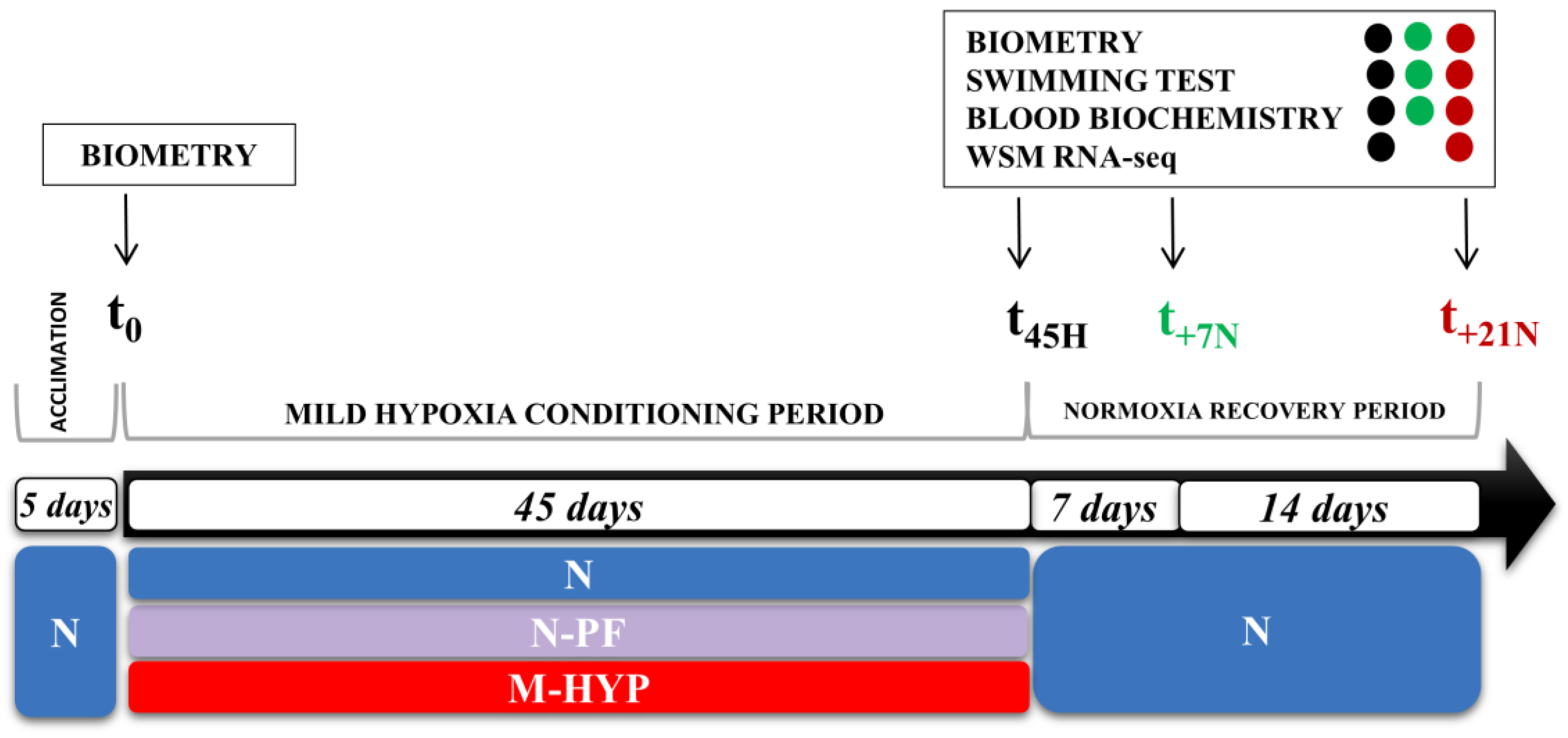
2.2. Experimental Setup of Hypoxia Conditioning
2.3. Swim Tunnel Respirometer
2.4. Blood Biochemistry
2.5. Illumina Sequencing and Sample Quality Assessment
2.6. Statistics
3. Results
3.1. Growth Performance during Mild-Hypoxia and Normoxia Restoration
3.2. Blood Patterns at the End of the Mild-Hypoxia Conditioning Period
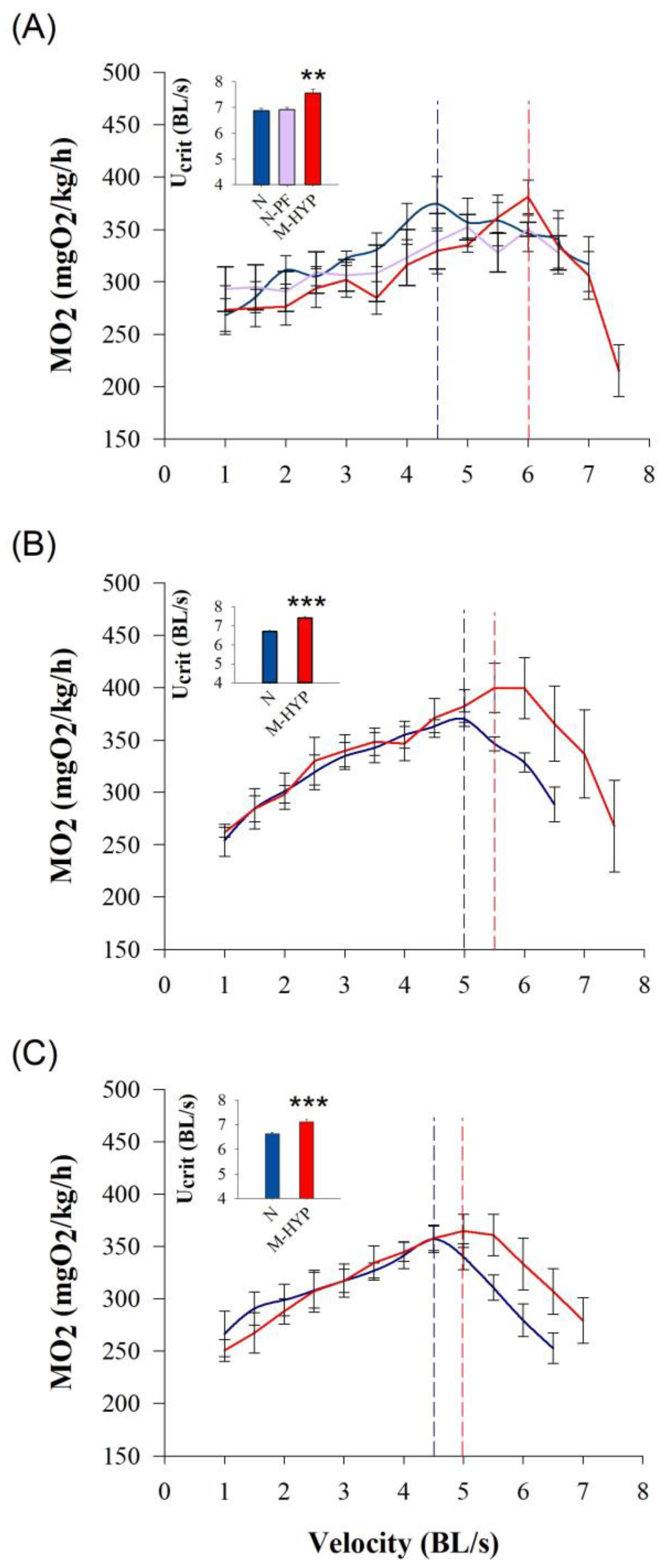
3.3. Swim Tests: Critical Swimming and Blood Patterns after Exhaustive Exercise
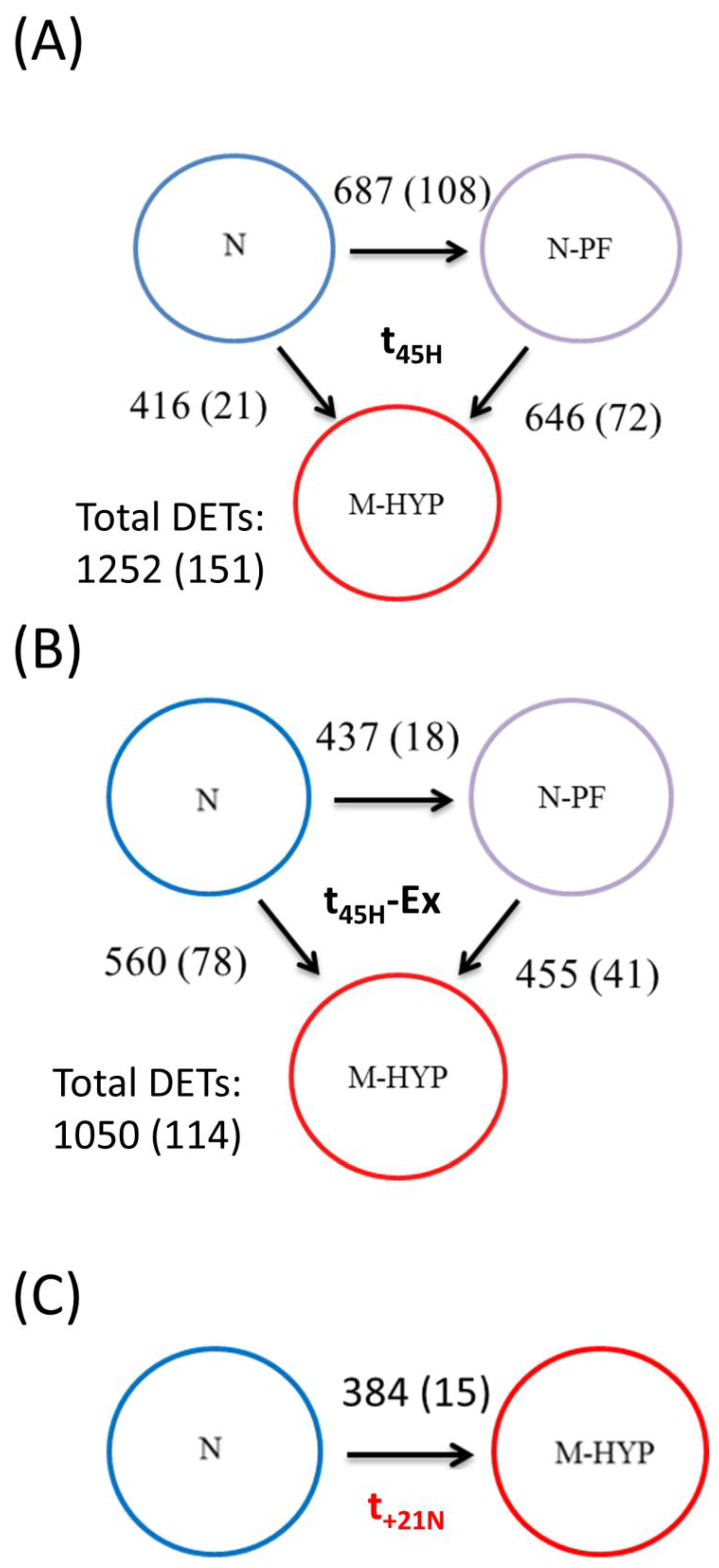
3.4. Analysis of RNA-seq Libraries and DE Genes by Stringent FDR
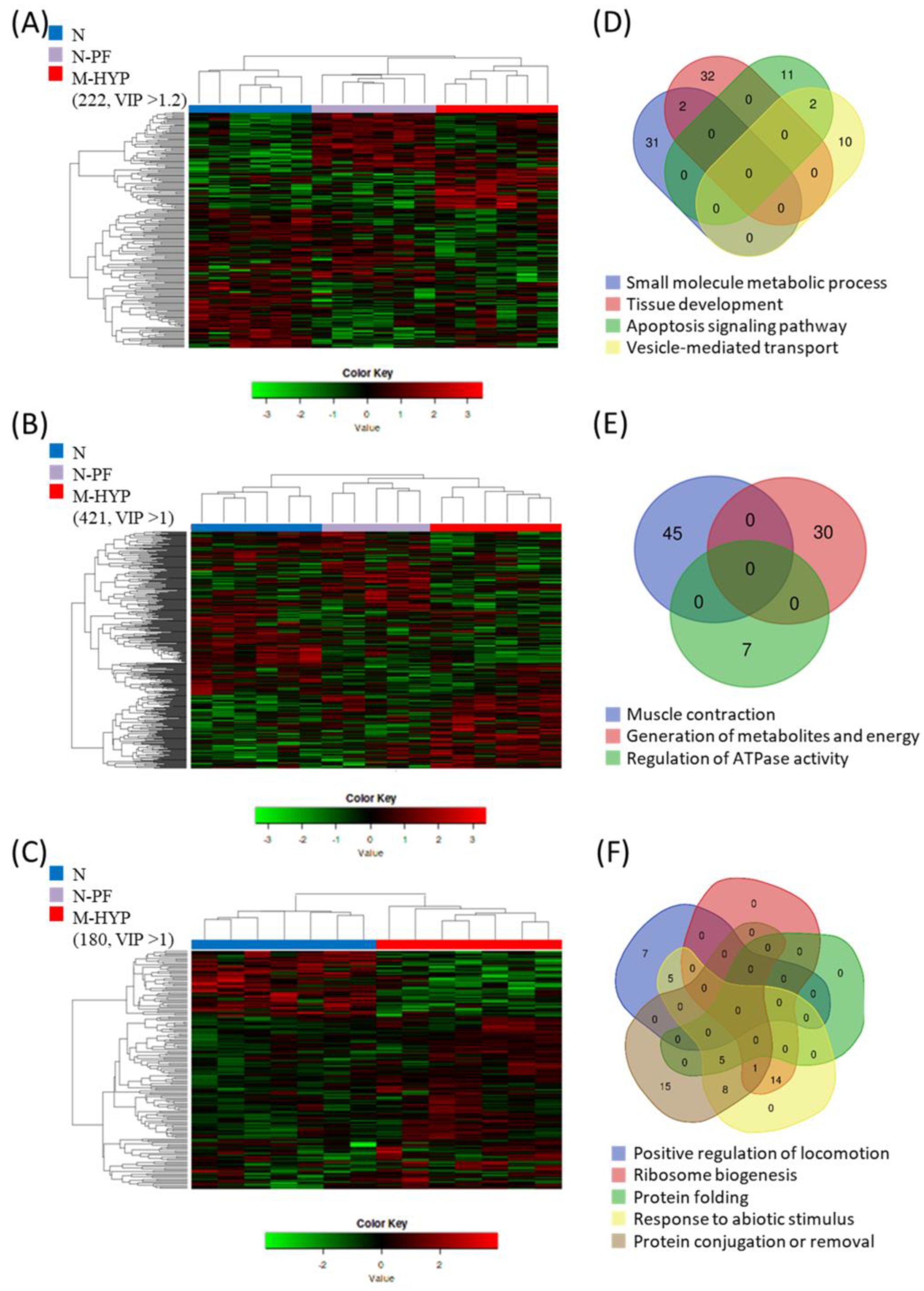
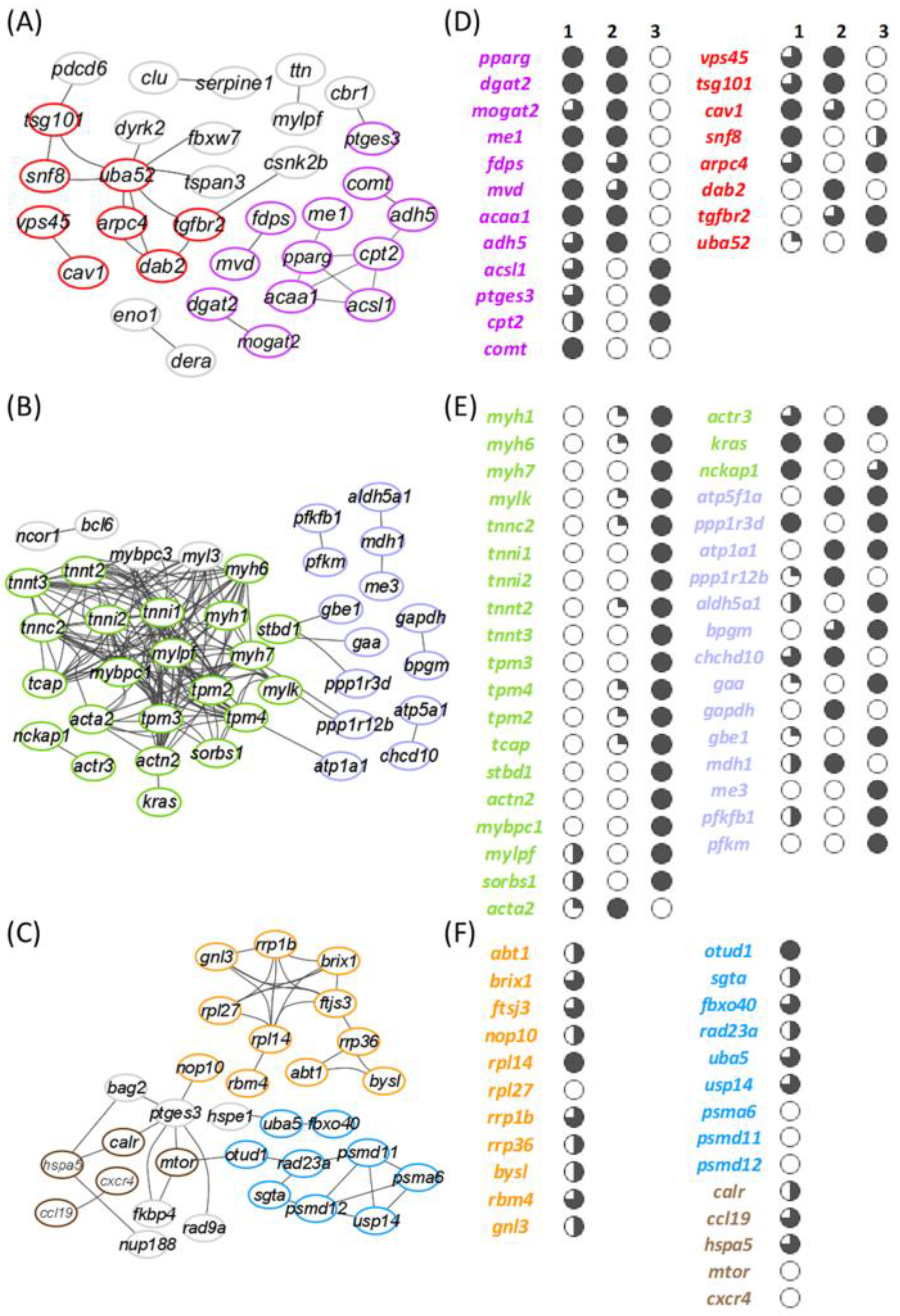
3.5. Discriminant Classifiers and Enriched GO Terms
3.6. Linking Enriched Processes with Gene Expression Patterns
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Hoppeler, H.; Vogt, M. Muscle tissue adaptations to hypoxia. J. Exp. Biol. 2001, 204, 3133–3139. [Google Scholar] [CrossRef]
- Murray, A.J. Metabolic adaptation of skeletal muscle to high altitude hypoxia: How new technologies could resolve the controversies. Genome Med. 2009, 1, 117. [Google Scholar] [CrossRef] [PubMed]
- Gamboa, J.L.; Andrade, F.H. Muscle endurance and mitochondrial function after chronic normobaric hypoxia: Contrast of respiratory and limb muscles. Eur. J. Physiol. 2012, 463, 327–338. [Google Scholar] [CrossRef]
- Larson, J.; Drew, K.L.; Folkow, L.P.; Milton, S.L.; Park, T.J. No oxygen? No problem! Intrinsic brain tolerance to hypoxia in vertebrates. J. Exp. Biol. 2014, 217, 1024–1039. [Google Scholar] [CrossRef] [PubMed]
- Storey, K.B. Regulation of hypometabolism: Insights into epigenetic controls. J. Exp. Biol. 2015, 218, 150–159. [Google Scholar] [CrossRef]
- Wu, H.; Chen, W.; Zhao, F.; Zhou, Q.; Reinach, P.S.; Deng, L.; Ma, L.; Luo, S.; Srinivasalu, N.; Pan, M.; et al. Scleral hypoxia is a target for myopia control. Proc. Natl. Acad. Sci. USA 2018, 115, E7091–E7100. [Google Scholar] [CrossRef] [PubMed]
- Brown, C.J.; Rupert, J.L. Hypoxia and environmental epigenetics. High Alt. Med. Biol. 2014, 15, 323–330. [Google Scholar] [CrossRef]
- Julian, C.G. Epigenomis and human adaptation to high altitude. J. Appl. Physiol. 2017, 123, 1362–1370. [Google Scholar] [CrossRef] [PubMed]
- Richards, J.G. Physiological, behavioral and biochemical adaptations of intertidal fishes to hypoxia. J. Exp. Biol. 2011, 214, 191–199. [Google Scholar] [CrossRef] [PubMed]
- Zhu, C.D.; Wang, Z.H.; Yan, B. Strategies for hypoxia adaptation in fish species: A review. J. Comp. Physiol. B Biochem. Syst. Environ. Physiol. 2013, 183, 1005–1013. [Google Scholar] [CrossRef]
- Deutsch, C.; Ferrel, A.; Seibel, B.; Pörtner, H.O.; Huey, R.B. Ecophysiology. Climate change tightens a metabolic constraint on marine habitats. Science 2015, 348, 1132–1135. [Google Scholar] [CrossRef]
- Schmidtko, S.; Stramma, L.; Visbeck, M. Decline in global oceanic oxygen content during the past five decades. Nature 2017, 542, 335–339. [Google Scholar] [CrossRef] [PubMed]
- Oschlies, A.; Brandt, P.; Stramma, L.; Schmidtko, S. Drivers and mechanisms of ocean deoxygenation. Nat. Geosci. 2018, 11, 467–473. [Google Scholar] [CrossRef]
- FAO. The State of World Fisheries and Aquaculture 2018. Contributing to Food Security and Nutrition for All; Food and Agriculture Organization of the United Nations: Rome, Italy, 2018. [Google Scholar]
- Sae-Lim, P.; Kause, A.; Mulder, H.A.; Olesen, I. Breeding and genetics symposium: Climate change and selective breeding in aquaculture. J. Anim. Sci. 2017, 95, 1801–1812. [Google Scholar] [CrossRef]
- Abdel-Tawwab, M.; Monier, M.N.; Hoseinifar, S.H.; Faggio, C. Fish response to hypoxia stress: Growth, physiological, and immunological biomarkers. Fish Physiol. Biochem. 2019, 45, 997–1013. [Google Scholar] [CrossRef]
- Remen, M.; Nederlof, M.A.J.; Folkedal, O.; Thorsheim, G.; Sitjà-Bobadilla, A.; Pérez-Sánchez, J.; Oppedal, F.; Olsen, R.E. Effect of temperature on the metabolism, behaviour and oxygen requirements of Sparus aurata. Aquacult. Environ. Interact. 2015, 7, 115–123. [Google Scholar] [CrossRef]
- Remen, M.; Sievers, M.; Torgersen, T.; Oppedal, F. The oxygen threshold for maximal feed intake of Atlantic salmon post-smolts is highly temperature-dependent. Aquaculture 2016, 464, 582–592. [Google Scholar] [CrossRef]
- Araújo-Luna, R.; Ribeiro, L.; Bergheim, A.; Pousão-Ferreira, P. The impact of different rearing condition on gilthead seabream welfare: Dissolved oxygen levels and stocking densities. Aquacult. Res. 2018, 49, 3845–3855. [Google Scholar] [CrossRef]
- Martos-Sitcha, J.A.; Simó-Mirabet, P.; de Las Heras, V.; Calduch-Giner, J.À.; Pérez-Sánchez, J. Tissue-specific orchestration of gilthead sea bream resilience to hypoxia and high stocking density. Front. Physiol. 2019, 10, 840. [Google Scholar] [CrossRef]
- Parma, L.; Pelusio, N.F.; Gisbert, E.; Esteban, M.A.; D’amico, F.; Soverini, M.; Candela, M.; Dondi, F.; Gatta, P.P.; Bonaldo, A. Effects of rearing density on growth, digestive conditions, welfare indicators and gut bacterial community of gilthead sea bream (Sparus aurata, L. 1758) fed different fishmeal and fish oil dietary levels. Aquaculture 2020, 518, 734854. [Google Scholar] [CrossRef]
- Calduch-Giner, J.À.; Davey, G.; Saera-Vila, A.; Houeix, B.; Talbot, A.; Prunet, P.; Cairns, M.T.; Pérez-Sánchez, J. Use of microarray technology to assess the time course of liver stress response after confinement exposure in gilthead sea bream (Sparus aurata L.). BMC Genom. 2010, 11, 193. [Google Scholar] [CrossRef]
- McBryan, T.L.; Healy, T.M.; Haakons, K.L.; Schulte, P.M. Warm acclimation improves hypoxia tolerance in Fundulus heteroclitus. J. Exp. Biol. 2016, 219, 474–484. [Google Scholar] [CrossRef] [PubMed]
- Healy, T.M.; Chung, D.J.; Crowther, K.G.; Schulte, P.M. Metabolic and regulatory responses involved in cold acclimation in Atlantic killifish, Fundulus heteroclitus. J. Comp. Physiol. B. 2017, 187, 463–475. [Google Scholar] [CrossRef]
- Salin, K.; Auer, S.K.; Villasevil, E.M.; Anderson, G.J.; Cairns, A.G.; Mullen, W.; Hartley, R.C.; Metcalfe, N.B. Using the MitoB method to assess levels of reactive oxygen species in ecological studies of oxidative stress. Sci. Rep. 2017, 7, 41228. [Google Scholar] [CrossRef]
- Vanderplancke, G.; Claireaux, G.; Quazuguel, P.; Huelvan, C.; Corporeau, C.; Mazurais, D.; Zambonino-Infante, J.L. Exposure to chronic moderate hypoxia impacts physiological and developmental traits of European sea bass (Dicentrarchus labrax) larvae. Fish Physiol. Biochem. 2015, 41, 233–242. [Google Scholar] [CrossRef]
- Wang, S.Y.; Lau, K.; Lai, K.P.; Zhang, J.W.; Tse, A.C.; Li, J.W.; Tong, Y.; Chan, T.F.; Wong, C.K.; Chiu, J.M.; et al. Hypoxia causes transgenerational impairments in reproduction of fish. Nat. Commun. 2016, 7, 12114. [Google Scholar] [CrossRef] [PubMed]
- Manchenkov, T.; Pasillas, M.P.; Haddad, G.G.; Imam, F.B. Novel genes critical for hypoxic preconditioning in zebrafish are regulators of insulin and glucose metabolism. G3-Genes Genom. Genet. 2015, 5, 1107–1116. [Google Scholar] [CrossRef] [PubMed]
- Sinex, J.A.; Chapman, R.F. Hypoxic training methods for improving endurance exercise performance. J. Sport Health Sci. 2015, 4, 4. [Google Scholar] [CrossRef]
- Hawley, J.A.; Lundby, C.; Cotter, J.D.; Burke, L.M. Maximizing cellular adaptation to endurance exercise in skeletal muscle. Cell Metab. 2018, 27, 962–976. [Google Scholar] [CrossRef]
- Fu, S.-J.; Brauner, C.J.; Cao, Z.-D.; Richards, J.G.; Peng, J.-L.; Dhillon, R.; Wang, Y.-X. The effect of acclimation to hypoxia and sustained exercise on subsequent hypoxia tolerance and swimming performance in goldfish (Carassius auratus). J. Exp. Biol. 2011, 214, 2080–2088. [Google Scholar] [CrossRef]
- Magnoni, L.J.; Martos-Sitcha, J.A.; Queiroz, A.; Calduch-Giner, J.A.; Magalhàes Gonçalves, J.F.; Rocha, C.M.R.; Abreu, H.T.; Schrama, J.W.; Ozorio, R.O.A.; Pérez-Sánchez, J. Dietary supplementation of heat-treated Gracilaria and Ulva seaweeds enhanced acute hypoxia tolerance in gilthead seabream (Sparus aurata). Biol. Open 2017, 6, 897–908. [Google Scholar] [CrossRef] [PubMed]
- Martos-Sitcha, J.A.; Bermejo-Nogales, A.; Calduch-Giner, J.A.; Pérez-Sánchez, J. Gene expression profiling of whole blood cells supports a more efficient mitochondrial respiration in hypoxia-challenged gilthead sea bream (Sparus aurata). Front. Zool. 2017, 14, 34. [Google Scholar] [CrossRef] [PubMed]
- Perera, E.; Rosell-Moll, E.; Naya-Català, F.; Simó-Mirabet, P.; Calduch-Giner, J.; Pérez-Sánchez, J. Effects of genetics and early-life mild hypoxia on size variation in farmed gilthead sea bream (Sparus aurata). Fish Physiol. Biochem. 2020. [Google Scholar] [CrossRef] [PubMed]
- Martos-Sitcha, J.A.; Simó-Mirabet, P.; Piazzon, M.C.; de las Heras, V.; Calduch-Giner, J.A.; Puyalto, M.; Tinsley, J.; Makol, A.; Sitjà-Bobadilla, A.; Pérez-Sánchez, J. Dietary sodium heptanoate helps to improve feed efficiency, growth hormone status and swimming performance in gilthead sea bream (Sparus aurata). Aquac. Nutr. 2018, 24, 1638–1651. [Google Scholar] [CrossRef]
- Martínez-Barberá, J.P.; Pendón, C.; Martí-Palanca, H.; Calduch-Giner, J.À.; Rodríguez, R.B.; Valdivia, M.M.; Pérez-Sánchez, J. The use of recombinant gilthead sea bream (Sparus aurata) growth hormone for radioiodination and standard preparation in radioimmunoassay. Comp. Biochem. Physiol. A Physiol. 1995, 110, 335–340. [Google Scholar] [CrossRef]
- Vega-Rubín de Celis, S.; Gómez-Requeni, P.; Pérez-Sánchez, J. Production and characterization of recombinantly derived peptides and antibodies for accurate determinations of somatolactin, growth hormone and insulin-like growth factor-I in European sea bass (Dicentrarchus labrax). Gen. Comp. Endocrinol. 2004, 139, 266–277. [Google Scholar] [CrossRef] [PubMed]
- Schmieder, R.; Edwards, R. Quality control and preprocessing of metagenomic datasets. Bioinformatics 2011, 27, 863–864. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.; Pertea, G.; Trapnell, C.; Pimentel, H.; Kelley, R.; Salzberg, S.L. TopHat2: Accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 2013, 14, R36. [Google Scholar] [CrossRef] [PubMed]
- Pérez-Sánchez, J.; Naya-Català, F.; Soriano, B.; Piazzon, M.C.; Hafez, A.; Gabaldón, T.; Llorens, C.; Sitjà-Bobadilla, A.; Calduch-Giner, J.A. Genome sequencing and transcriptome analysis reveal recent species-specific gene duplications in the plastic gilthead sea bream (Sparus aurata). Front. Mar. Sci. 2019, 6, 760. [Google Scholar] [CrossRef]
- Trapnell, C.; Roberts, A.; Goff, L.; Pertea, G.; Kim, D.; Kelley, D.R.; Pimentel, H.; Salzberg, S.L.; Rinn, J.L.; Pachter, L. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat. Protoc. 2017, 7, 562–578. [Google Scholar] [CrossRef]
- Li, H.; Ma, M.L.; Luo, S.; Zhang, R.M.; Han, P.; Hu, W. Metabolic responses to ethanol in Saccharomyces cerevisiae using a gas chromatography tandem mass spectrometry-based metabolomics approach. Int. J. Biochem. Cell Biol. 2012, 44, 1087–1096. [Google Scholar] [CrossRef]
- Kieffer, D.A.; Piccolo, B.D.; Vaziri, N.D.; Liu, S.; Lau, W.L.; Khazaeli, M.; Nazertehrani, S.; Moore, M.E.; Marco, M.L.; Martin, R.J.; et al. Resistant starch alters gut microbiome and metabolomic profiles concurrent with amelioration of chronic kidney disease in rats. Am. J. Physiol. Renal Physiol. 2016, 310, F857–F871. [Google Scholar] [CrossRef]
- Ge, S.X.; Jung, D.; Yao, R. ShinyGO: A graphical gene-set enrichment tool for animals and plants. Bioinformatics 2020, 36, 2628–2629. [Google Scholar] [CrossRef] [PubMed]
- Szklarczyk, D.; Gable, A.L.; Lyon, D.; Junge, A.; Wyder, S.; Huerta-Cepas, J.; Simonovic, M.; Doncheva, N.T.; Morris, J.H.; Bork, P.; et al. STRING v11: Protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 2019, 47, D607–D613. [Google Scholar] [CrossRef] [PubMed]
- Futami, R.; Muñoz-Pomer, A.; Viu, J.M.; Domínguez-Escribà, L.C.L.; Bernet, G.P.; Sempere, J.M.; Moya, A.; Llorens, C. GPRO: The professional tool for management, functional analysis and annotation of omic sequences and databases. Biotech. Bioinf. 2011, 1, 1–5. [Google Scholar]
- Garcia, H.E.; Gordon, L.I. Oxygen solubility in seawater: Better fitting equations. Limnol. Oceanogr. 1992, 37, 1307–1312. [Google Scholar] [CrossRef]
- Intergovernmental Panel on Climate Change. Climate Change 2014-Impacts, Adaptation and Vulnerability: Part A: Global and Sectoral Aspects: Working Group II Contribution to the IPCC Fifth Assessment Report; Cambridge University Press: Cambridge, UK, 2014. [Google Scholar]
- Pichavant, K.; Person-Le-Ruyet, J.; Bayon, N.L.; Severe, A.; Roux, A.L.; Boeuf, G. Comparative effects of long-term hypoxia on growth, feeding and oxygen consumption in juvenile turbot and European sea bass. J. Fish Biol. 2001, 59, 875–883. [Google Scholar] [CrossRef]
- Cadiz, L.; Zambonino-Infante, J.L.; Quazuguel, P.; Madec, L.; Le Delliou, H.; Mazurais, D. Metabolic response to hypoxia in European sea bass (Dicentrarchus labrax) displays developmental plasticity. Comp. Biochem. Physiol. 2018, 215, 1–9. [Google Scholar] [CrossRef]
- Vikeså, V.; Nankervis, L.; Hevrøy, E.M. High dietary energy level stimulates growth hormone receptor and feed utilization in large Atlantic salmon (Salmo salar L.) under hypoxic conditions. Aquac. Nutr. 2017, 23, 1193–1203. [Google Scholar] [CrossRef]
- Dam, A.V.; Pauly, D. Simulation of the effects of oxygen on food consumption and growth of Nile tilapia, Oreochromis niloticus (L.). Aquacult. Res. 1995, 26, 427–440. [Google Scholar] [CrossRef]
- Saravanan, S.; Geurden, I.; Figueiredo-Silva, A.C.; Kaushik, S.J.; Haidar, M.N.; Verreth, J.A.; Schrama, J.W. Control of voluntary feed intake in fish: A role for dietary oxygen demand in Nile tilapia (Oreochromis niloticus) fed diets with different macronutrient profiles. Br. J. Nutr. 2012, 108, 1519–1529. [Google Scholar] [CrossRef]
- Brett, J.R. Environmental factors and growth. In Fish Physiology, 1st ed.; Hoar, W.S., Randall, D.J., Brett, J.R., Eds.; Academic Press: London, UK, 1977; Volume VIII, pp. 599–675. [Google Scholar]
- Pérez-Sánchez, J.; Martí-Palanca, H.; Kaushik, S. Ration size and protein intake affect growth hormone (GH) levels, hepatic GH-binding and plasma insulin-like growth factor-I immunoreactivity in a marine teleost, gilthead sea bream (Sparus aurata). J. Nutr. 1995, 125, 546–552. [Google Scholar] [PubMed]
- Monternier, P.A.; Marmillot, V.; Rouanet, J.L.; Roussel, D. Mitochondrial phenotypic flexibility enhances energy savings during winter fast in king penguin chicks. J. Exp. Biol. 2014, 217, 2691–2697. [Google Scholar] [CrossRef] [PubMed]
- Salin, K.; Villasevil, E.M.; Auer, S.K.; Anderson, G.J.; Selman, C.; Metcalfe, N.B.; Chinopoulos, C. Simultaneous measurement of mitochondrial respiration and ATP production in tissue homogenates and calculation of effective P/O ratios. Phys. Rep. 2016, 4, e13007. [Google Scholar] [CrossRef] [PubMed]
- Pérez-Sánchez, J.; Simó-Mirabet, P.; Naya-Català, F.; Martos-Sitcha, J.A.; Perera, E.; Bermejo-Nogales, A.; Benedito-Palos, L.; Calduch-Giner, J.À. Somatotropic axis regulation unravels the differential effect of nutritional and environmental factors in growth performance of marine farmed fish. Front. Endocrinol. 2018, 9, 687. [Google Scholar] [CrossRef] [PubMed]
- Metcalfe, N.B.; Van Leeuwen, T.E.; Killen, S.S. Does individual variation in metabolic phenotype predict fish behavior and performance? J. Fish Biol. 2016, 88, 298–321. [Google Scholar] [CrossRef] [PubMed]
- Knap, P.W.; Kause, A. Phenotyping for genetic improvement of feed efficiency in fish: Lessons from pig breeding. Front. Genet. 2018, 9, 184. [Google Scholar] [CrossRef] [PubMed]
- Perera, E.; Simó-Mirabet, P.; Shin, H.S.; Rosell-Moll, E.; Naya-Català, F.; De las Heras, V.; Martos-Sitcha, J.A.; Karalazos, V.; Armero, E.; Arizcun, M.; et al. Selection for growth is associated in gilthead sea bream (Sparus aurata) with diet flexibility, changes in growth patterns and higher intestine plasticity. Aquaculture 2019, 507, 349–360. [Google Scholar] [CrossRef]
- Piazzon, M.C.; Naya-Català, F.; Perera, E.; Palenzuela, O.; Sitjà-Bobadilla, A.; Pérez-Sànchez, J. Genetic selection for growth drives differences in intestinal microbiota composition and parasite disease resistance in gilthead sea bream. Microbiome 2020, 8, 168. [Google Scholar] [CrossRef]
- Anttila, K.; Mänttäri, S. Ultrastructural differences and histochemical characteristics in swimming muscles between wild and reared Atlantic salmon. Acta Physiol. 2009, 196, 249–257. [Google Scholar] [CrossRef]
- Zhang, Y. Estimating Aerobic and Anaerobic Capacities Using the Respiratory Assessment Paradigm: A Validation Using Atlantic Salmon (Salmo salar) and European Sea Bass (Dicentrarchus labrax); The University of British Columbia: Vancouver, BC, Canada, 2016. [Google Scholar]
- Bellringer, K.L.; Thorgaard, G.H.; Carter, P.A. Domestication is associated with reduced burst swimming performance and increased body size in clonal rainbow trout lines. Aquaculture 2014, 420–421, 154–159. [Google Scholar] [CrossRef]
- Claireaux, G.; McKenzie, D.J.; Genge, A.G.; Chatelier, A.; Aubin, J.; Farrell, A.P. Linking swimming performance, cardiac pumping ability and cardiac anatomy in rainbow trout. J. Exp. Biol. 2005, 208, 1775–1784. [Google Scholar] [CrossRef]
- Blasco, J.; Moya, A.; Millán-Cubillo, A.; Vélez, E.J.; Capilla, E.; Pérez-Sánchez, J.; Gutiérrez, J.; Fernández-Borrás, J. Growth-promoting effects of sustained swimming in fingerlings of gilthead sea bream (Sparus aurata L.). J. Comp. Physiol. B 2015, 185, 859–868. [Google Scholar] [CrossRef] [PubMed]
- Vélez, E.J.; Lutfi, E.; Azizi, S.; Perelló, M.; Salmerón, C.; Riera-Codina, M.; Ibarz, A.; Fernández-Borràs, J.; Blasco, J.; Capilla, E.; et al. Understanding fish muscle growth regulation to optimize aquaculture production. Aquaculture 2017, 467, 28–40. [Google Scholar] [CrossRef]
- Palstra, A.P.; Kals, J.; Böhm, T.; Bastiaansen, J.; Komen, H. Swimming performance and oxygen consumption as non-lethal indicators of production traits in atlantic salmon and gilthead seabream. Front. Physiol. 2020, 11, 759. [Google Scholar] [CrossRef]
- Palstra, A.P.; Planas, J.V. Swimming Physiology of Fish: Towards Using Exercise to Farm a Fit Fish in Sustainable Aquaculture, 1st ed.; Springer: Berlin, Germany, 2013; pp. 1–429. [Google Scholar]
- Palstra, A.P.; Beltran, S.; Burgerhout, E.; Brittijn, S.A.; Magnoni, L.J.; Henkel, C.V.; Jansen, H.J.; van den Thillart, G.E.; Spaink, H.P.; Planas, J.V. Deep RNA sequencing of the skeletal muscle transcriptome in swimming fish. PLoS ONE 2013, 8, e53171. [Google Scholar] [CrossRef] [PubMed]
- Mylonis, I.; Simos, G.; Paraskeva, E. Hypoxia-inducible factors and the regulation of lipid metabolism. Cells 2019, 8, 214. [Google Scholar] [CrossRef]
- Krishnan, J.; Suter, M.; Windak, R.; Krebs, T.; Felley, A.; Montessuit, C.; Tokarska-Schlattner, M.; Aasum, E.; Bogdanova, A.; Perriard, E.; et al. Activation of a HIF1α -PPARγ axis underlies the integration of glycolytic and lipid anabolic pathways in pathologic cardiac hypertrophy. Cell Metab. 2009, 9, 512–524. [Google Scholar] [CrossRef]
- Shi, H.; Zhao, W.; Zhang, C.; Shahzad, K.; Luo, J.; Loor, J.J. Transcriptome-wide analysis reveals the role of PPARγ controlling the lipid metabolism in goat mammary epithelial cells. PPAR Res. 2016, 2016, 9195680. [Google Scholar] [CrossRef]
- Liu, Y.; Ma, Z.; Zhao, C.; Wang, Y.; Wu, G.; Xiao, J.; McClain, C.J.; Li, X.; Feng, W. HIF-1α and HIF-2α are critically involved in hypoxia-induced lipid accumulation in hepatocytes through reducing PGC-1α-mediated fatty acid β-oxidation. Toxicol. Lett. 2014, 226, 117–123. [Google Scholar] [CrossRef] [PubMed]
- Ban, J.J.; Ruthenborg, R.J.; Cho, K.W.; Kim, J.W. Regulation of obesity and insulin resistance by hypoxia-inducible factors. Hypoxia (Auckl) 2014, 2, 171–183. [Google Scholar]
- Gaspar, J.M.; Mendes, N.F.; Corrêa-da-Silva, F.; Lima-Junior, J.C.; Gaspar, R.C.; Ropelle, E.R.; Araujo, E.P.; Carvalho, H.M.; Velloso, L.A. Downregulation of HIF complex in the hypothalamus exacerbates diet-induced obesity. Brain. Behav. Immun. 2018, 73, 550–561. [Google Scholar] [CrossRef] [PubMed]
- Virtue, S.; Vidal-Puig, A. Nothing iffy about HIF in the hypothalamus. PLoS Biol. 2011, 9, e1001116. [Google Scholar] [CrossRef]
- Gaspar, J.M.; Velloso, L.A. Hypoxia inducible factor as a central regulator of metabolism—Implications for the development of obesity. Front. Neurosci. 2018, 12, 813. [Google Scholar] [CrossRef] [PubMed]
- Zhao, L.L.; Sun, J.L.; Liang, J.; Liu, Q.; Luo, J.; Li, Z.Q.; Yan, T.M.; Zhou, J.; Yang, S. Enhancing lipid metabolism and inducing antioxidant and immune responses to adapt to acute hypoxic stress in Schizothorax prenanti. Aquaculture 2020, 519, 734933. [Google Scholar] [CrossRef]
- Listenberger, L.L.; Han, X.; Lewis, S.E.; Cases, S.; Farese, R.V., Jr.; Ory, D.S.; Schaffer, J.E. Triglyceride accumulation protects against fatty acid-induced lipotoxicity. Proc. Natl. Acad. Sci. USA 2003, 100, 3077–3082. [Google Scholar] [CrossRef] [PubMed]
- Mylonis, I.; Sembongi, H.; Befani, C.; Liakos, P.; Siniossoglou, S.; Simos, G. Hypoxia causes triglyceride accumulation by HIF-1-mediated stimulation of lipin 1 expression. J. Cell Sci. 2012, 125, 3485–3493. [Google Scholar] [CrossRef] [PubMed]
- Gracey, A.Y.; Troll, J.V.; Somero, G.N. Hypoxia-induced gene expression profiling in the euryoxic fish Gillichthys mirabilis. Proc. Natl. Acad. Sci. USA 2001, 98, 1993–1998. [Google Scholar] [CrossRef] [PubMed]
- Lewis, C.A.; Brault, C.; Peck, B.; Bensaad, K.; Griffiths, B.; Mitter, R.; Chakravarty, P.; East, P.; Dankworth, B.; Alibhai, D.; et al. SREBP maintains lipid biosynthesis and viability of cancer cells under lipid- and oxygen-deprived conditions and defines a gene signature associated with poor survival in glioblastoma multiforme. Oncogene 2015, 34, 5128–5140. [Google Scholar] [CrossRef]
- Wang, M.S.; Li, Y.; Peng, M.S.; Zhong, L.; Wang, Z.J.; Li, Q.Y.; Tu, X.L.; Dong, Y.; Zhu, C.L.; Wang, L. Genomic analyses reveal potential independent adaptation to high altitude in Tibetan chickens. Mol. Biol. Evol. 2015, 32, 1880–1889. [Google Scholar] [CrossRef] [PubMed]
- Ge, R.L.; Simonson, T.S.; Gordeuk, V.; Prchal, J.T.; McClain, D.A. Metabolic aspects of high-altitude adaptation in Tibetans. Exp. Biol. 2015, 100, 1247–1255. [Google Scholar] [CrossRef]
- Horscroft, J.A.; Kotwica, A.O.; Laner, V.; West, J.A.; Hennis, P.J.; Levett, D.; Howard, D.J.; Fernandez, B.O.; Burgess, S.L.; Ament, Z.; et al. Metabolic basis to Sherpa altitude adaptation. Proc. Natl. Acad. Sci. USA 2017, 114, 6382–6387. [Google Scholar] [CrossRef]
- Ge, R.L.; Cai, Q.; Shen, Y.Y.; San, A.; Ma, L.; Zhang, Y.; Yi, X.; Chen, Y.; Yang, L.; Huang, Y. Draft genome sequence of the Tibetan antelope. Nat. Commun. 2013, 1858, 4. [Google Scholar] [CrossRef]
- Qiu, Q.; Zhang, G.J.; Ma, T.; Qian, W.B.; Wang, J.Y.; Ye, Z.Q.; Cao, C.C.; Hu, Q.J.; Kim, J.; Larkin, D.M.; et al. The yak genome and adaptation to life at high altitude. Nat. Genet. 2012, 44, 946–949. [Google Scholar] [CrossRef] [PubMed]
- Zhao, D.; Zhang, Z.; Cease, A.; Harrison, J.; Kang, L. Efficient utilization of aerobic metabolism helps Tibetan locusts conquer hypoxia. BMC Genom. 2013, 14, 631. [Google Scholar] [CrossRef] [PubMed]
- Kang, J.; Ma, X.; He, S. Evidence of high-altitude adaptation in the glyptosternoid fish, Creteuchiloglanis macropterus from the Nujiang River obtained through transcriptome analysis. BMC Evol. Biol. 2017, 17, 229. [Google Scholar] [CrossRef]
- Wright, J.R.; McCloskey, D.I.; Fitzpatrick, R.C. Effects of muscle perfusion pressure on fatigue and systemic arterial pressure in human subjects. J. Appl. Physiol. 1999, 86, 845–851. [Google Scholar] [CrossRef]
- Wan, J.J.; Qin, Z.; Wang, P.Y.; Sun, Y.; Liu, X. Muscle fatigue: General understanding and treatment. Exp. Mol. Med. 2017, 49, e384. [Google Scholar] [CrossRef]
- Zierath, J.R.; Hawley, J.A. Skeletal muscle fiber type: Influence on contractile and metabolic properties. PLoS Biol. 2004, 2, e348. [Google Scholar] [CrossRef] [PubMed]
- Trappe, S.; Harber, M.; Creer, A.; Gallagher, P.; Slivka, D.; Minchev, K.; Whitsett, D. Single muscle fiber adaptations with marathon training. J. Appl. Phisiol. 2006, 101, 721–727. [Google Scholar] [CrossRef] [PubMed]
- Nikinmaa, M. Oxygen-dependent cellular functions—Why fishes and their aquatic environment are a prime choice of study. Comp. Biochem. Physiol. Part A Mol. Integr. Physiol. 2002, 133, 1–16. [Google Scholar] [CrossRef]
- Ton, C.; Stamatiou, D.; Liew, C.C. Gene expression profile of zebrafish exposed to hypoxia during development. Physiol. Genom. 2003, 13, 97–106. [Google Scholar] [CrossRef]
- Zhong, X.P.; Wang, D.; Zhang, Y.B.; Gui, J.F. Identification and characterization of hypoxia-induced genes in Carassius auratus blastulae embryonic cells using suppression subtractive hybridization. Comp. Biochem. Phys. B 2009, 152, 161–170. [Google Scholar] [CrossRef]
- Qi, D.; Chao, Y.; Wu, R.; Xia, M.; Chen, Q.; Zheng, Z. Transcriptome analysis provides insights into the adaptive responses to hypoxia of a Schizothoracine Fish (Gymnocypris eckloni). Front. Physiol. 2018, 9, 1326. [Google Scholar] [CrossRef] [PubMed]
- Houlihan, D.F.; Carter, C.G.; McCarthy, I.D. Protein synthesis in animals. In Nitrogen Metabolism and Excretion, 1st ed.; Wright, P.J., Walsh, P.A., Eds.; CRC Press: Boca Ratón, FL, USA, 1995; pp. 1–29. [Google Scholar]
- McCarthy, I.D.; Owen, S.F.; Watt, P.W.; Houlihan, D.F. Individuals maintain similar rates of protein synthesis over time on the same plane of nutrition under controlled environmental conditions. PLoS ONE 2016, 11, e0152239. [Google Scholar] [CrossRef]
- Dauer, P.; Sharma, N.S.; Gupta, V.K.; Durden, B.; Hadad, R.; Banerjee, S.; Dudeja, V.; Saluja, A.; Banerjee, S. ER stress sensor, glucose regulatory protein 78 (GRP78) regulates redox status in pancreatic cancer thereby maintaining “stemness”. Cell Death Dis. 2019, 10, 132. [Google Scholar] [CrossRef] [PubMed]
- Park, K.W.; Eun Kim, G.; Morales, R.; Moda, F.; Moreno-Gonzalez, I.; Concha-Marambio, L.; Lee, A.S.; Hetz, C.; Soto, C. The endoplasmic reticulum chaperone grp78/bip modulates prion propagation in vitro and in vivo. Sci. Rep. 2017, 7, 44723. [Google Scholar] [CrossRef] [PubMed]
- Wang, P.; Zhang, N.; Liang, J.; Li, J.; Han, S.; Li, J. Micro-RNA-30a regulates ischemia-induced cell death by targeting heat shock protein HSPA5 in primary cultured cortical neurons and mouse brain after stroke. J. Neurosci. Res. 2015, 93, 1756–1768. [Google Scholar] [CrossRef] [PubMed]
- Pérez-Sánchez, J.; Terova, G.; Simó-Mirabet, P.; Rimoldi, S.; Folkedal, O.; Calduch-Giner, J.A.; Olsen, R.E.; Sitjà-Bobadilla, A. Skin mucus of gilthead sea bream (Sparus aurata L.). Protein mapping and regulation in chronically stressed fish. Front. Phisiol. 2017, 8, 34. [Google Scholar] [CrossRef] [PubMed]
- Gisbert, E.; Andree, K.B.; Quintela, J.C.; Calduch-Giner, J.A.; Ipharraguerre, I.R.; Pérez-Sánchez, J. Olive oil bioactive compounds increase body weight, and improve gut health and integrity in gilthead sea bream (Sparus aurata). Br. J. Nutr. 2017, 117, 351–363. [Google Scholar] [CrossRef]


| N | N-PF | M-HYP | p-Value | |
|---|---|---|---|---|
| Mild-hypoxia conditioning (t0–t45H) | ||||
| Initial body weight (g) | 24.58 ± 0.11 | 24.1 ± 0.10 | 24.19 ± 0.03 | 0.112 |
| Final body weight (g) | 78.69 ± 0.79 b | 66.13 ± 1.41 a | 66.06 ± 0.97 a | <0.001 |
| Feed intake (g DM/fish) | 53.36 ± 0.15 b | 40.77 ± 0.22 a | 40.08 ± 0.84 a | <0.001 |
| Weight gain (%) 1 | 220.30 ± 2.03 b | 174.51 ± 4.50 a | 173.22 ± 3.86 a | <0.001 |
| SGR (%) 2 | 2.59 ± 0.01 b | 2.24 ± 0.04 a | 2.23 ± 0.03 a | <0.001 |
| FCR (%) 3 | 0.98 ± 0.009 | 0.96 ± 0.02 | 0.95 ± 0.008 | 0.285 |
| Normoxia recovery period (t+7N–t+21N) | ||||
| Initial body weight (g) | 98.76 ± 1.20 b | 83.50 ± 0.50 a | 82.00 ± 1.14 a | <0.001 |
| Final body weight (g) | 126.5 ± 1.30 b | 114.7 ± 0.33 a | 111.3 ± 1.81 a | 0.001 |
| Feed intake (g DM/fish) | 37.62 ± 1.28 | 36.57 ± 1.50 | 35.66 ± 0.50 | 0.329 |
| Weight gain (%) 1 | 28.54 ± 0.46 a | 37.22 ± 1.33 b | 35.50 ± 0.85 b | 0.001 |
| SGR (%) 2 | 1.79 ± 0.03 a | 2.26 ± 0.07 b | 2.19 ± 0.04 b | <0.001 |
| FCR (%) 3 | 1.21 ± 0.03 | 1.12 ± 0.02 | 1.14 ± 0.02 | 0.103 |
| N | N-PF | M-HYP | p-Value | |
|---|---|---|---|---|
| Haemoglobin (g/dL) | 8.36 ± 0.38 b | 6.43 ± 0.64 a | 7.88 ± 0.22 b | 0.011 |
| Haematocrit (%) | 34.7 ± 1.24 | 33.7 ± 0.99 | 31.0 ± 1.41 | 0.175 |
| Lactate (mg/dL) | 14.1 ± 0.15 b | 6.32 ± 0.57 a | 4.18 ± 0.77 a | <0.001 |
| Glucose (mg/dL) | 57.1 ± 5.98 | 55.7 ± 2.29 | 56.8 ± 2.35 | 0.493 |
| Triglycerides (mg/dL) | 2.80 ± 0.28 | 4.02 ± 0.34 | 3.02 ± 0.46 | 0.128 |
| Free fatty acids (nmol/µL) | 0.426 ± 0.052 ab | 0.595 ± 0.045 b | 0.388 ± 0.045 a | 0.029 |
| Cortisol (ng/mL) | 24.1 ± 5.43 | 29.3 ± 10.56 | 14.3 ± 4.71 | 0.270 |
| Growth hormone (ng/mL) | 9.19 ± 3.94 | 12.4 ± 5.30 | 13.9 ± 4.87 | 0.752 |
| Insulin-like growth factor-1 (ng/mL) | 69.3 ± 5.74 | 60.5 ± 3.42 | 55.5 ± 3.94 | 0.285 |
| Gh/Igf-1 | 0.13 ± 0.058 a | 0.20 ± 0.081 ab | 0.25 ± 0.041 b | 0.032 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Naya-Català, F.; Martos-Sitcha, J.A.; de las Heras, V.; Simó-Mirabet, P.; Calduch-Giner, J.À.; Pérez-Sánchez, J. Targeting the Mild-Hypoxia Driving Force for Metabolic and Muscle Transcriptional Reprogramming of Gilthead Sea Bream (Sparus aurata) Juveniles. Biology 2021, 10, 416. https://doi.org/10.3390/biology10050416
Naya-Català F, Martos-Sitcha JA, de las Heras V, Simó-Mirabet P, Calduch-Giner JÀ, Pérez-Sánchez J. Targeting the Mild-Hypoxia Driving Force for Metabolic and Muscle Transcriptional Reprogramming of Gilthead Sea Bream (Sparus aurata) Juveniles. Biology. 2021; 10(5):416. https://doi.org/10.3390/biology10050416
Chicago/Turabian StyleNaya-Català, Fernando, Juan A. Martos-Sitcha, Verónica de las Heras, Paula Simó-Mirabet, Josep À. Calduch-Giner, and Jaume Pérez-Sánchez. 2021. "Targeting the Mild-Hypoxia Driving Force for Metabolic and Muscle Transcriptional Reprogramming of Gilthead Sea Bream (Sparus aurata) Juveniles" Biology 10, no. 5: 416. https://doi.org/10.3390/biology10050416
APA StyleNaya-Català, F., Martos-Sitcha, J. A., de las Heras, V., Simó-Mirabet, P., Calduch-Giner, J. À., & Pérez-Sánchez, J. (2021). Targeting the Mild-Hypoxia Driving Force for Metabolic and Muscle Transcriptional Reprogramming of Gilthead Sea Bream (Sparus aurata) Juveniles. Biology, 10(5), 416. https://doi.org/10.3390/biology10050416






