Simple Summary
This review has comprehensively summarized the most recent studies in the last few years about exosomal microRNAs on metastasis of breast cancer (BC), systematically outlined and elucidated the pleiotropic roles that exosomal microRNAs take, and discussed the specific underlying mechanisms related. Besides, we also clearly demonstrate the clinical implications of exosomal microRNAs in various aspects, including early-stage discovery of BC, systematic and targeted therapy, and the selection of anti-cancer chemo-agents. This review clarifies the scope and extent of current studies about the relationship between exosomal microRNAs and metastasis of BC, laying a solid foundation for further laboratory studies. What’s more, it links the theoretical researches and clinical features about exosomal microRNAs on metastasis of BC together, providing novel clues for BC diagnosis, chemotherapeutics and drug efficacy evaluation in clinical, which indicates the great potential of exosomal microRNAs in clinical significance.
Abstract
As a major threat factor for female health, breast cancer (BC) has garnered a lot of attention for its malignancy and diverse molecules participating in its carcinogenesis process. Among these complex carcinogenesis processes, cell proliferation, epithelial-to-mesenchymal transition (EMT), mesenchymal-to-epithelial transition (MET), and angiogenesis are the major causes for the occurrence of metastasis and chemoresistance which account for cancer malignancy. MicroRNAs packaged and secreted in exosomes are termed “exosomal microRNAs (miRNAs)”. Nowadays, more researches have uncovered the roles of exosomal miRNAs played in BC metastasis. In this review, we recapitulated the dual actions of exosomal miRNAs exerted in the aggressiveness of BC by influencing migration, invasion, and distant metastasis. Next, we presented how exosomal miRNAs modify angiogenesis and stemness maintenance. Clinically, several exosomal miRNAs can govern the transformation between drug sensitivity and chemoresistance. Since the balance of the number and type of exosomal miRNAs is disturbed in pathological conditions, they are able to serve as instructive biomarkers for BC diagnosis and prognosis. More efforts are needed to connect the theoretical studies and clinical traits together. This review provides an outline of the pleiotropic impacts of exosomal miRNAs on BC metastasis and their clinical implications, paving the way for future personalized drugs.
1. Introduction
Breast cancer (BC) is one of the most common diseases among women in contemporary society and shows a high mortality rate. In 2018, there are about 2.1 million new cases of BC in the world, accounting for nearly a quarter of female cancer cases. BC is the most commonly diagnosed cancer in the vast majority (83%) of countries, and it is also being the main cause of cancer death in more than 100 countries [1]. As a critical program in BC carcinogenesis, metastasis is attributed to the majority of deaths in people suffering from BC. Metastasis is a complicated and multi-step process, which involves epigenetic and genetic mutations, patterns of dissemination and disaggregation, chaotic and irregular angiogenesis, invasion-metastasis cascades, the epithelia-mesenchymal transition (EMT), and stemness maintenance of cancer stem cells (CSCs). Any treatment targeting molecules in these pathological processes can affect the progress of metastasis [2].
Currently, metastatic disease is regarded to be incurable due to specific characteristics which define it. Some novel traits derived from the metastatic lesions make it more malignant than the original one. This discordance in tumor features of metastatic lesions becomes a great obstacle to the multiple clinical treatments. Metastatic tumor cells generally acquire high genetic instability. Extensive dis-regulation in the genomics repair and transmission processes drives to the high level of genetic mutations. Monogenetic and polygenetic clones are the two major ways in the formation of tumor subpopulations. During the process of cloning, different subpopulations may acquire segregated anatomical distributions and inherit diverse genetic advantages. This molecular discordance can explain the heterogeneity in the biological behavior of metastatic groups. In clinical, the changes in the expression profiles of metastasis groups hide the validated targets for some specialized therapy. And the newly-activated signaling pathways in subpopulations also make the existing medicine less effective. Besides, it is thought that metastases partly evolve from the dissemination of cancerous cells with stemness. While stemness-like properties can impart chemotherapy resistance to cells. What is more, there is another malignant tumor subpopulation, called dormant tumor cells. These cells can stay in a latent state, escape from the initial treatments and awake later. The heterogeneity of metastatic cells. Though some clinical therapies have successfully managed the primary BC lesions, no effective strategies are capable to control relapses and cope with BC metastases [3].
Based on molecular features, BC can be divided into 4 groups, luminal A, luminal B, human epidermal growth factor receptor 2 (HER2), and basal type. Well-established classification contributes to a more accurate diagnosis and the improvement of therapy selection. For patients with luminal A or B type BC, they will bring about a better prognosis after receiving endocrine-based therapy or standard schedule chemotherapy [4]. About HER2-positive patients, anti-HER2 immunotherapy such as targeted antibodies (e.g., trastuzumab/Herceptin) can exert rapidly and sustainedly tumor-inhibitory effects [5]. The clinical features of triple-negative breast cancer (TNBC) appear no expression of estrogen receptors (ER), HER2, and progesterone receptors (PR) on the surface of cancerous cells. The absence of characteristic biomarkers makes endocrine-related therapy not applicable for TNBC therapy. As a novel clinical paradigm for systemic therapy, targeted immunotherapy combined with chemotherapy still shows limited efficacy and deleterious effects on healthy organs in the vicinity. Nanotechnology, an emerging tool, works to artificially design some specific nanoparticles with ideal structures, functions, and properties to satisfy the clinical application. Several products from nanotechnology, such as conjugated dendrimers and modified micelles, possess great prospects in copying with TNBC and can be considered as a new direction to make a breakthrough in BC treatment [6].
Exosomes, with a featured ‘saucer-like’ morphology, are membrane-enclosed vesicles originally made inside the intracellular multivesicular endosomes and multivesicular bodies (MVBs) by invagination and budding [7]. Exosome vesicles contain cytosol and coat the extracellular domain of transferrin receptors at their surface. There are many highly-conserved proteins and lipids in rafts in exosomes with various functions, such as antigen presentation, integrins, and immunoglobulin-family members [8]. Besides, the lumen of exosome has uncovered different nucleic acids, including messenger RNAs (mRNAs), microRNAs (miRNAs), and other non-coding RNAs (ncRNAs). MiRNAs are endogenously small noncoding RNA gene products that commonly regulate mRNA expression post-transcriptionally by repressing translation and/or transcript decay [9]. The biological synthesis of exosomal miRNAs in cells is pictured below in Figure 1. MiRNAs carried in exosomes are not randomly assigned [10]. In addition, their expression levels are not constant in different conditions [11]. More researches have garnered attention for the roles of miRNAs in exosomes.
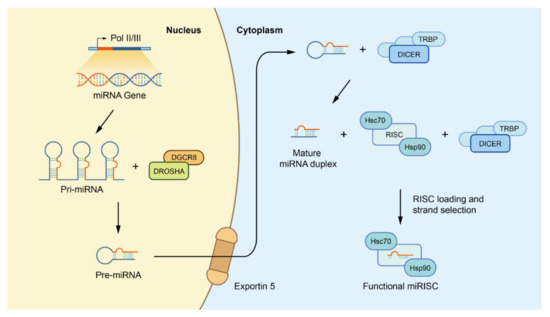
Figure 1.
Canonical biogenesis of miRNAs. Firstly, miRNA genes are transcribed as primary precursors (pri-miRNA) by the action of RNA polymerase and further processed by the Drosha complex, which consists of one ribonuclease called Drosha and two molecules of the RNA-binding protein DGCR8, to form pre-miRNAs. Subsequently, pre miRNAs are exported to the cytoplasm by exportin-5 at the nuclear envelope before being cleaved by the Dicer/TRBP complex to form mature miRNA duplexes. Finally, with the help of the Dicer/TRBP complex and HSC70/HSP90 chaperone proteins, one strand of the miRNA duplex is placed into the RISC to form a functional miRISC for subsequent regulation. Abbreviations: miRNAs: microRNAs; pri-miRNA: primary miRNA; DGCR8: DiGeorge syndrome critical region 8; pre-miRNAs: precursor miRNAs; TRBP: transactivation response RNA binding protein; HSC70: heat shock cognate protein 70; HSP90: heat shock cognate protein 90; RISC: miRNA induced silencing complex.
Commonly, exosomes can be excreted from various cell types, such as smooth muscle cells, endothelial cells, and immunocytes [12,13]. When exosomes circulate in the body fluids, they can be picked up by neighboring cells or distant cells, and subsequently, regulate their biological functions [14]. Several studies have reported that miRNA can take an important part in cell function modification, immunity regulation, and tumorigenesis. For example, miRNAs, as paracrine mediators, are in a relatively high abundance of cardiac fibroblast-derived exosomes, stimulating cardiac remodeling, and inducing cardiomyocyte hypertrophy [15]. Migration and angiogenesis in the human dermal microvascular endothelial cells-1 (HMEC-1) line can be boosted by exosomal miR-214 from HMEC-1 cells nearby [16]. Exosomal miR-21 and miR-29a are able to act as ligands for toll-like receptors (TLRs) and promote the activation of immunocytes [17]. MiR-92 and miR-25-3p contained in plasma exosomes and total blood exosomes of breast cancer patients (BCPs) can induce angiogenesis and epithelial-mesenchymal transition (EMT), and increase the total migration path length of BC cells.
Since the quantity and ingredients of exosomal miRNAs are different between physiological and pathological conditions, its clinical application is also a field worth exploring. Exosomal miRNAs can be used as a marker for disease diagnoses [18], such as glioblastoma [19], chronic obstructive pulmonary disease [20], colorectal cancer [21], prostate cancer [22], and BC [23]. Emerging evidence suggests that tumor-derived (TD) exosomes provide a soil conducive to tumor growth by mediating communication within the tumor microenvironment, i.e., exosomal miR-105 released from the BC cell lines MCF-10A and MDA-MB-231 dissolve endothelial cells identity by weakening Zonula occludens 1 (ZO-1) gene expression which promotes metastases of cancer daughter cells to the lung and brain [24]. Thus, TD-exosomes can be considered as targeted therapies. Specificity, accessibility from the extracellular domain, and stability make exosomes effective markers for tumor diagnosis, treatment, and prognosis [25].
In this study, as shown in Figure 2, we reviewed the latest research progress in this field. We uncovered that exosomal miRNAs not only take part in enhancing cell growth and variance of EMT or MET but also influence the local invasiveness and metastasis of BC cells. Besides, exosomal miRNA can modify tumor immunogenicity, active angiogenic switch, escape programmed apoptosis as well as deliver drug resistance. By consulting numerous kinds of literature, here we discussed the underlying molecular mechanisms involved in these pathological processes, which contribute to the prevention, diagnosis, treatment, and prognosis of BC. How the signaling pathways involved by exosomal miRNAs regulate metastasis of BC cells is the highlight of this review.
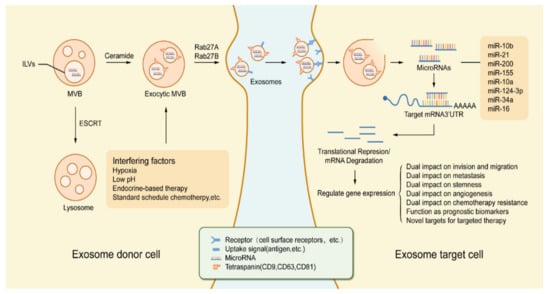
Figure 2.
Tumor regulation process based on exosomal miRNA in the tumor microenvironment. Exosomes are derived from MVB which germinate inward in the form of ILVs. The formation of exosomes as well as the sorting of cargo into lysosomes involves the ESCRT. In addition, exosome production and secretion are also completed by a non-ESCRT dependent process that involves the sphingolipid ceramide and the enzyme neutral sphingomyelinase. Stimulation by various types of chemical, physical factors can induce the secretion of exosomes, such as hypoxia, low pH, endocrine therapy, etc. After the fusion of MVBs with the cell membrane, exosomes are secreted into the extracellular space under the action of Rab GTPases (Rab27A, Rab27B). Exosomes released from exosome donor cells (tumor cells, etc.) will exhibit similar receptor ligands or antigens as their cells of origin. The endocytic process of exosomes occurs with the activation of receptors or bioactive lipid ligands at the cell surface. After exosomes are endocytosed by exosome target cells (endothelial cells, etc.), miRNAs in exosomes are released. Transferred miRNAs are bioactive and can regulate gene expression in exosomal target cells through post-translational regulation of mRNA expression, which in turn leads to mRNA instability or degradation. miRNA-dependent gene regulation can affect various types of processes involved in tumorigenesis and development, and changes in gene expression levels can also be used as a marker for judging tumorigenesis and progression. Abbreviations: MVB: multivesicular body; ILVs: intraluminal vesicles; ESCRT: endosomal sorting complex required for transport; Rab27A: Ras-associated binding 27A; Rab27B: Ras-associated binding 27B.
2. Exosomal microRNAs That Enhance Aggressiveness of BC Cells
2.1. Exosomal microRNAs That Promote Invasion and Migration of BC Cells
2.1.1. miR-21
Early studies have shown that exosomal miR-21 is overexpressed in different types of tumor tissues, including BC [26,27]. It is involved in almost all stages of BC development [28].
The knockdown of exosomal miR-21 can inhibit the invasion and migration of BC cells. On the contrary, the up-regulation of exosomal miR-21 inhibits the expression of the gene of phosphate and tension homology deleted on chromosome ten (PTEN), which suppresses the constitutive activation of downstream components of the phosphatidylinositol 3-kinase (PI3K) pathway, thereby suppressing the expression of the tumor suppressor gene Drg-1 (differentiation-related gene 1) [29].
Besides, several regulatory mechanisms of exosomal miR-21 in BC cells have been proposed by recent studies. They suggested that miR-21 can directly inhibit the expression of the corresponding tumor suppressor by directly targeting the 3’UTR of mammary serine protease inhibitor (maspin), programmed cell death protein 4 (PDCD4), or tumor suppressor gene tropomyosin 1 (TPM1) [30]. Yin et al. found that maspin interacts with the uPA associated with the cell surface and stimulates the internalization of the uPA/uPAR complex [31]. On the other hand, PDCD4 can down-regulate urokinase-type plasminogen activator receptor (uPAR) through its promoter region [32]. uPAR is a promoter of cell invasion and metastasis [33]. The reduction of uPAR level caused by maspin or PDCD4 can significantly inhibit the invasion and migration ability of BC cells. Interestingly, PDCD4 can also inhibit the proliferation and invasion of BC cells by interacting with eukaryotic initiation factor 4A (EIF4A), participating in the down-regulation of mitogen-activated protein kinase kinase kinase kinase 1 (MAP4K1) transcription, and up-regulating p21 to inhibit the transcription of cyclin-dependent kinase 1 (CDK1)/cdc2 [34,35]. For TPM1, Zhu et al. discovered that overexpression of TPM1 inhibits the invasion of BC cells, but the specific mechanism remains to be investigated [30].
2.1.2. miR-10b
Exosomal miR-10b is closely linked to BC size, progression, and degree of invasion [36]. Previous research on BC cells found that exosomal miR-10b can directly inhibit the translation of Homeobox D10 (HOXD10) for promoting the migration of BC [37]. HODX10 is an mRNA encoding a transcriptional repressor, which reduces the expression of some genes involved in extracellular matrix (ECM) remodeling and cell migration, such as rat sarcoma (Ras) homolog gene family member C (RHoC), Membrane-type 1 matrix metalloproteinase (MT1-MMP), a3-integrin, and urokinase plasminogen activator receptor [38]. In addition, studies have confirmed that after inhibiting the expression of TBX5, the up-regulation of exosomal miR-10b levels simultaneously causes the down-regulation of PTEN and another tumor suppressor, Dual-specificity tyrosine phosphorylation-regulated kinase 1A (DYRK1A), which ultimately affects BC invasion and migration [39,40]. Currently, based on the regulatory relationship between sphingolipid ceramide and exosomes, the researchers found that the secretion of exosomal miR-10b is also ceramide-dependent regulation [41]. It is worth noting that exosomal miR-10b can act as a sponge for syndecan-1, and significantly affect the migration and invasion of BC through the miR-10b/syndecan-1 axis. In terms of mechanism, it may be achieved by the activation of miR-10b/syndecan-1 axis that leads to ras homologous oncogenes guanosine triphosphatases (Rho-GTPase)-dependent regulation of cytoskeletal function and down-regulation of E-cadherin expression [42].
2.1.3. miR-1246
MiR-1246, a 25-nt miRNA that is situated on chromosome 2q31.1, originates from U2 small nuclear RNA (RNU2-1) transcript degradation [43]. Studies have shown that exosomal miR-1246 is significantly upregulated in BC [44]. Interestingly, Li et al. found that exosomal miR-1246 is over-expressed in metastatic BC MDA-MB-231 cells, and can also inhibit the expression level of its target gene Cyclin-G2 (CCNG2) to promote BC cell invasion [45]. These findings provide a basis for targeting exosomal miR-1246 to interfere with BC.
2.1.4. miR-373
Exosomal miR-373 has been determined to be functionally related to many biological processes, such as cell proliferation, migration, invasion, and apoptosis of tumors, including BC. Previous studies have found that exosomal miR-373 is highly correlated with BC invasion and migration, especially high-invasive BC [46]. By establishing the MCF-7 cell line, Huang et al. confirmed that exosomal miR-373 promotes cell migration and invasion, at least in part, by limiting CD44 expression directly [47]. In addition, it has recently been reported that exosomal miR-373 may be a plasma-based biomarker that can be used to indicate the lymph node status of BC [48]. These findings provide new ideas for further research on the relationship between exosomal miR-373 and BC aggressiveness.
2.1.5. miR-17-5p
Exosomal miR-17-5p specifically targets HMG-box protein 1 (HBP1)/Wnt/β-catenin signaling cascade to enhance the migratory and invasive ability of BC cell lines. HBP1 serves as a cell-cycle suppressor and oncogene transcriptional suppressor. By controlling the transcriptional actives governed by T-cell factor/lymphoid enhancer-binding factor (TCF/LEF), HBP1 carefully monitors the Wnt/β-catenin signaling network. HBP1-mediated inhibition of exosomal miR-15-5p facilitates the acquisition of BC cells to more malignant phenotypes, as well as invasion and migratory ability [49]. It is worth noting that Lv et al. recently compared the levels of exosomes miR-17-5p in the serum of 83 BC patients and 34 healthy women and found an obvious decrease in the serum of BC patients decreased [50]. These results are consistent with previous studies and strongly suggest that exosomal miR-17-5p can be used as a new diagnostic biomarker for BC.
2.1.6. miR-96
Protein tyrosine phosphatase non-receptor type 9 (PTPN9), an intracellular phosphatase, governs the pro-migratory and pro-invasive functions of several signal molecules, such as epidermal growth factor receptor (EGFR), signal transducer and activator of transcription 3 (STAT3), and human epidermal growth factor receptor-2 (ErbB2). Exosomal miR-96 can specifically recognize and binding to PTPN9 mRNA and cutting off subsequent expression activities, which exerts a promotive effect on non-transform cells for the acquisition of increased motion ability [51].
2.1.7. miR-106b
The content of exosomal miR-106b is particularly abundant in cancerous cells and the plasma of BC patients. BC cells exposed by exosomal miR-106b obtain the increased motion ability. Involved in numerous cellular metabolic activities, restoration of the expression of fucosyltransferase 6 (FUT6) can counteract the oncogenic effects exerted by exosomal miR-106b. Moreover, re-expression of exosomal miR-106b apparently down-regulates the content of FUT6, suggesting that exosomal miR-106b plays the pro-migrative and pro-invasive roles by diminishing the expression of FUT6 [52].
2.2. Exosomal microRNAs That Promote Distant Metastasis of BC Cells
2.2.1. miR-10b
In recent years, exosomal miR-10b has been one of the most thoroughly studied miRNAs. Alongside invasion and migration of BC cells, exosomal miR-10b participates in metastasis of BC as well [39]. Mounting evidence has shown a highly positive correlation between exosomal miR-10b and distant metastasis, as well as lymph node metastasis of BC [53].
Epigenetic signals from the tumor microenvironment regulate the transition of BC cells from dormancy to metastatic growth [54,55]. In addition to exploring crosstalk studies on BC cells themselves, the latest research has investigated the role of exosomal miR-10b in the tumor microenvironment. By establishing an exosome-free culture model of metastatic MDA-MB-231 cells, Ma et al. found that overexpression of miR-10b in MDA-MB-231 cells would lead to an increase in the extracellular level of miR-10b (i.e., exosomal). Exosomal miR-10b can be effectively absorbed by a variety of receptor cells, such as MCF-10A, HEK-239T, and MCF-7 cells [41,56]. Additionally, in the non-metastatic human BC cell line SUM159, the absorption of exosomal miR-10b leads to more pronounced lung metastases and macroscopic peritoneal metastases [56].
Interestingly, after the first identification of the Twist/miR-10b/HOXD10/RHoC signaling pathway, it is demonstrated as a participant when exosomal miR-10b regulates BC metastasis [57,58,59]. Notably, another study found that loss of E-cadherin during BC progression leads to increased expression of exosomal miR-10b. At this point, exosomal miR-10b binds to neurofibromatosis type 1 (NF1) and HODX10 to activate the Rho-associated kinase (ROCK)/c-Jun signaling pathway, thereby promoting the invasion and metastasis of BC cells [60].
2.2.2. miR-503
Xing et al. found that miR-503 is one of the miRNAs with the highest expression in the exosomes of MCF-7 cells with X-inactive specific transcript (XIST) knockdown. They observed that serum levels of exosomal miR-503 in patients with brain metastases from BC are significantly increased. Mechanistically, exosomal tumor cells-derived miR-503 promotes the transformation of microglia from M1 to M2 phenotypes by manipulating STAT3 and NF-κB pathways. Subsequently, activation of these signaling pathways enhanced programmed cell death ligand 1 (PD-L1) expression to suppress local immunity, thus promoting brain metastasis of BC cells [61]. Based on the results above, we suggest exosomal miR-503 is a tumor-promoting exosomal miRNA that endows potential brain metastasis ability to BC cells.
2.2.3. miR-122
Abnormally activated anaerobic glycolysis to reprogram energy metabolism, called the “Warburg effect”, is considered a hallmark of cancer [62]. Quite abundant in all BC cell lines, exosomal miR-122 can be a candidate molecule to reflecting the metastatic phase in the early stage of BC progression. Overexpression of exosomal miR-122 speeds the spontaneous transformation from in situ ductal carcinoma lesions to malignant tumors. Restraining glucose metabolism, exosomal miR-122 inhibits primary lesion growth while functions as a signaling molecule to modify the pre-metastatic microenvironment, and making adequate preparation for further metastatic colonization. After receiving exosomal miR-122, the pyruvate kinase (PKM) involves in glycolysis and glucose transporter 1 (GLUT1), a transporter for glucose uptake is downregulated in niche cells, altering the metabolic rate and energy metabolism. The sufficient energy supply is hijacked by BC cells, building a pro-growth microstructure, and facilitating more metastases occurrence [63].
2.2.4. miR-200
It is proposed that Sec23a facilitates the release of metastasis-suppressive substrates, such as Tinagl1 and Igfbp4. Exosomal miR-200 can directly inhibit the functions of Sec23a to fuel the formation of metastatic subpopulations [64]. Besides, miR-200 is able to downregulate Yes-associated protein 1 (YAP1), which subsequently motivates metastasis efficiently [65].
2.2.5. miR-105
Exosomal miR-105 is specifically enriched in metastatic BC cells, reflecting the progressing of metastasis in early-stage BC. Zonula occludens 1 (ZO-1), a tight junction protein, is the major fundamental molecule composing the intercellular adhesion molecules of epithelial and endothelial cells. Expression of exosomal miR-105 can downregulate the content of ZO-1 in both mRNA and protein levels. Transfected with exosomal miR-105, BC cells show a significant deficiency in ZO-1 and recycle occludin, another functional factor from cell junction. Released from metastatic cells, exosomal miR-105 alters vessel integrity, augments vessel permeability as well as promotes neovessel sprouts, facilitating cancerous cells to seed in anywhere good for further oncogenesis [24].
2.2.6. miR-21
Exosomal miR-21 promotes oncogenic progress in numerous cancers, also motivating metastasis of BC cells. Downregulation of exosomal miR-21 attenuates cell motion and metastatic colonization [66]. As a tumor-suppressor gene, leucine zipper transcription factor-like 1 (LZTFL1) regulates metastasis by blocking β-catenin translocation to the nucleus and activating pro-metastatic gene expressions, such as slug and snail. Targeting LZTFL1, exosomal miR-21 removes the inhibition of LZTFL1 to β-catenin, upregulating EMT marker, and metastatic-dependent functional proteins. MiR-21/LZTFL1/β-catenin/EMT axis effectively fuels metastasis of BC cells [67].
3. Exosomal microRNAs That Attenuate Aggressiveness of BC Cells
3.1. Exosomal microRNAs That Inhibit Invasion and Migration of BC Cells
3.1.1. miR-564
The mitogen-activated protein kinase (MAPK) and PI3K signaling pathways are always hijacked in BC, associated with independent cell growth, EMT, and migration [68]. Exosomal miR-564 impairs cell proliferation, viability, and motility by directly modifying the expression of a cohort of related genes, Akt2, guanine nucleotide-binding protein alpha-12 (GNA12), glycogen synthase 1 (GYS1), and SRF, thereby dually inhibiting MAPK and PI3K pathway and causing irreversible cell-cycle arrest. Acting as an oncogenic suppressor, the expression of exosomal miR-564 effectively blocks the migration and invasion and related pathological process of BC cells [69].
3.1.2. miR-10a
Aberrantly activated PI3K/Akt/mTOR pathway is commonly found in various cancers. BC is no exception. Exosomal miR-10a reduces the expression of the mechanistic target of rapamycin (mTOR), PI3K, and Akt by specifically targeting a key molecule PI3K associated with this pathway. By interfering with the function of this pathway, exosomal miR-10a weakens the migration and motility of BC cells [70].
3.1.3. miR-34c
Metastatic cells selectively downregulate the expression of exosomal miR-34c. After the transfection of exosomal miR-34c, the migration and invasion of BC cell lines are inhibited G-protein-coupled receptor kinase interacting protein-1 (GIT1) exerts a regulatory role in intercellular adhesion and migration. In addition, it also facilitates the stabilization of Paxillin, a fundamental molecule in adhesion. Tao et al. demonstrated that overexpression of exosomal miR-34c can significantly inhibit the invasion and migration [71].
3.1.4. miR-217
As mentioned earlier, miR-217 is considered as a tumor promoter, while some found exosomal miR-217 can inhibit the proliferation and invasion of a variety of cancers, such as pancreatic ductal adenocarcinoma [72], liver cancer [73], and esophageal squamous cell carcinoma [74]. BC is no exception. Interestingly, Zhou et al. discovered that exosomal miR-217 targets krüppel-like factor 5 (KLF5), a powerful carcinogen in both HCC1937 and HCC1806 cell lines, to inhibit the development of TNBC [75]. This supplies the feasibility for the treatment of TNBC through the miR-217/KLF5 axis.
Coincidentally, KLF5 is only highly expressed in HCC1937 and HCC1806 but is low in MCF-7, MDA-MB-231, and SKBR3. Additionally, exosomal miR-217 does not inhibit the expression of DACH1 and PTEN in HCC1937 and HCC1806 cell lines as it did in other cells such as MCF-7 [75]. Therefore, we suggest that the specific role of exosomal miR-217 in BC may be related to cell line specificity.
3.1.5. miR-100
Exosomal miR-100 is a pleiotropic molecule. Wnt/β-catenin signaling pathway is a widely-studied pathway. It takes part in numerous biological processes, including cell proliferation and cell motility, also contributing to tumorigenesis, such as invasion and metastasis. Exosomal miR-100 can repress the expression of pro-invasive and oncogenic factors such as matrix metalloproteinase-7 (MMP-7), cellular-myelocytomatosis viral oncogene (C-myc), and FZD-8. Moreover, it can also upregulate inhibitory molecules such as glycogen synthase kinase 3 (GSK3) belonging to the Wnt/β-catenin signaling pathway to imparting the motion tendency of MDA-MB-231 BC cells [76,77]. Homeobox A1 (HOXA1), an oncogene motivating tumor growth and tumor motility by regulating the expression of downstream pro-migration and pro-invasion genes, cyclin D1 (CCND1), MET, smoothened (SMO), and semaphorin 3C (SEMA3C). Jiang et al. revealed in their research that HOXA1 is blocked by exosomal miR-100 to impair the aggressive phenotypes of BC cell lines [77].
3.1.6. miR-19b-3p, miR-19a-3p, and miR-1226-3p
Aquaporins (AQPs) selectively facilitate the transport of water between different body compartments to maintain homeostasis. AQP3 and AQP5 are quite abundant in triple-negative BC patients. Overexpressed AQP5 in epithelial cells prompts cell detachment and migration, which contributes to invasiveness and metastasis of tumors. AQP5 also regulates the migration of BC. Experiments showed that exosomal miR-19a-3p, miR-19b-3p, and miR-1226-3p are metastatic regulators that exert their roles by regulation of target gene expression, including AQP5 [78].
Exosomal miR-19b-3p blocks the process of transcription by directly binding to AQP5 mRNA. Exosomal miR-19a-3p may bind to AQP5 mRNA at a site different from exosomal miR-19b-3p to inhibit AQP5 expression. Alternatively, it indirectly regulates AQP5 expression by modulating genes that can influence AQP5 expression. For exosomal miR-1226-3p, it decreases the cellular content of AQP5 by motivating the degradation of AQP5 mRNA. So an obvious reduction in migration occurs in MDA-MB-231 cells transfected with these three miRNAs respectively due to the diminished expression of AQP5 [78].
Besides, exosomal miR-19a-3p diminishes the content of Fos-related antigen 1 (FRA-1) which prompts the invasiveness and tumor malignancy of BC cells, by weakening the expression level of the FOS like 1 (FOSL1) gene coding for it. Notably, exosomal miR-1226-3p can reduce mucin 1 expression in BC cells by modifying mucin 1 mRNAs [79]. These exosomal miRNAs provide new ideas for controlling the invasion and metastasis of BC in various ways in the future.
3.1.7. miR-148a and miR-148b-3p
Decreased expression of exosomal miR-148b-3p occurs in various kinds of BC cell lines, which are largely associated with poor prognoses in BC [80]. Tripartite motif-containing 59 (TRIM 59), a major target gene of miR-148b-3p that is highly expressed in BC. Western blot analysis demonstrated that the up-regulation of TRIM 59 expression increased the abundance of EMT-related proteins in MDA-MB-231 cells while inhibiting the levels of apoptosis-related proteins. This reveals the role of TRIM 59 in promoting tumor growth, improving tumor migration and invasion, and playing an anti-apoptotic effect. MiR-148b-3p carried by exosomes derived from human umbilical cord mesenchymal stem cells (HUCMSCs) reduced viability of BC, restrains tumor progression, and weakens tumor malignancy by down-regulating the expression level of TRIM 59 [81]. Therefore, the subsequent oncogenic pathways and the effects in tumorigenesis are almost blocked by exosomal miR-148b-3p [81]. Moreover, previous studies showed that overexpressed exosomal miR-148a takes part in inhibiting tumor motility, including invasion and migration of MCF-7 and MDA-MB-231 BC cells [82]. Interestingly, the latest research also proved that the exosomal miR-148a in BC patients is significantly reduced, along with the adverse outcome of BC [83].
3.1.8. miR-503
Ander varying circumstances, as a pleiotropically-functional factor carried by exosomes from cancerous tissues and endothelial cells, miR-503 may exert opposite effects on the invasiveness of BC cells. As mentioned before, exosomal miR-503 derived from BC cell lines reinforces migration. However, a few studies surprisingly provide new insight into exosomal miR-503 as a metastasis-suppressing exosomal miRNA in BC. Several studies have found exosomal miR-503 from endothelial cells impairs the invasive capacity [61,84]. As a powerful effector, exosomal miR-503 negatively regulates the expression of cyclin D2 (CCND2) and cyclin D3 (CCND3) in both RNA and protein levels. The subsequent impact on BC cells of this interaction is to apparently reduce proliferation rate and diminish invasive capacity [84].
In summary, we cannot conclude whether exosomal miR-503 is a tumor-promoting or suppressing exosomal miRNA; numerous unknown events remain to be discovered to explore the possible explanation for the dual effects of exosomal miR-503 in BC.
3.1.9. miR-17/20
MicroRNA17/20 abundance is reduced in highly invasive BC cell lines and node-positive BC specimens. Serving as tumor-suppressive miRNA, exosomal miR-17/20 blocks irregular cell-cycle by silencing E2F transcription factor 1 (E2F1) expression, as well as causes alteration in the secretion of a series of pro-metastatic cytokines, such as C-X-C motif chemokine ligand 1 (CXCL1), cytokeratin 8 (CK8), and Interleukin-8 (IL-8). By directly binding to IL-8 mRNA, exosomal miR-17/20 impairs the transcription activities of the IL-8 gene. Plasmin, derived from plasminogen, is crucial in the reinforcement of cell motility and metastasis for its role in degrading ECM and is activated by alpha-enolase (alpha-ENO) and CK8 bounce. Exosomal miR-17/20 represses the function of CCND1 to block the activity of CK8 and indirectly reduces alpha-ENO expression. By modifying pro-malignant cytokines excretion and inhibiting plasmin-activator, exosomal miR-17/20 governs BC pre-metastatic behavior [85].
3.2. Exosomal microRNAs That Inhibit Distant Metastasis of BC Cells
3.2.1. miR-429
Exosomal miR-429 is a migration suppressor, belonging to the miR-200 family. As two candidate molecules of exosomal miR-429, zinc finger E-box binding homeobox 1 (ZEB1) and V-crk sarcoma virus CT10 oncogene homolog-like (CRKL) cooperate with other fundamental molecules and govern a series of signal cascades to enhance migratory capacity. Mechanistically, exosomal miR-429 reduces the content of ZEB1 and CRKL at the protein level to interrupt the subsequent signal networks [86]. Another in vitro experimental study uncovered the inhibitory roles of exosomal miR-429 in bone metastasis. Cancerous cells transfected with miR-429 show the decreased expression level of CrkL and matrix metalloproteinases-9 (MMP-9), as well as reduced penetration in the local bone microenvironment and fewer metastatic lesions [87]. These indicate the miR-429/CrkL/MMP-9 axis is capable to alter the tendency of cancerous cells to seed in the bone microenvironment.
3.2.2. miR-124-3p
Compared with adjacent tumor-free tissues, the content of exosomal miR-124-3p is remarkably reduced in former BC tissues [88]. Exosomal miR-124-3p suppresses the EMT processes by upregulating the protein level of E-cadherin and downregulating that of N-cadherin and Vimentin. By directly targeting programmed cell death protein 6 (PDCD6), exosomal miR-124-3p impairs the migratory and motility capacity of BC cells, reducing metastasis [89]. The decline of exosomal miR-124-3p is largely associated with lymph node metastasis and a poorer survival rate [88].
3.2.3. miR-31
Exosomal miR-31 is a pleiotropic molecule taking part in several steps of metastasis and is always specifically diminished or deleted in malignant BC cells [90]. The maintenance of exosomal miR-31 functions facilitates the prevention of normal epithelial cells and tumor cells from obtaining a more aggressive phenotype, such as chemoresistance. By attenuating the expression of a series of pro-metastatic genes, such as frizzled class receptor 3 (Fzd3), c-matrix metalloproteinase-16 (cMMP16), myosin Phosphatase-Rho interacting protein (M-RIP), integrin alpha 5 (ITGA5), hexahydro-1,3,5-trinitro-1,3,5-triazine (RDX), and Ras homolog gene family member A (RhoA), exosomal miR-31 weakens metastatic and metastatic-dependent traits. Gain- and loss-of-function assays dictate that exosomal miR-31 reduces the number of metastatic colonization and restrains the size of the lesion, as well as blocks the early stage of the post-intravasation cascade, including intravasation, intra-capillary viability, and penetration, creating an unfavorable environment for cancer development [91]. The discovery of exosomal miR-31 provides new insights for the treatment and prevention of BC.
3.2.4. miR-124
A number of studies have uncovered the roles played by exosomal miR-124 in the oncogenic process of various malignant cancers, such as cervical cancer, prostate cancer, and BC, demonstrating its tumor-suppressive function [92]. Impairing exosomal miR-124 imparts a more pathological feature of BC cells. Clinically, the low level of exosomal miR-124 contributes to the increased lesion seeding in the bone, rapid appearance of metastasis in bone, and poorer survival free from bone-metastasis [93]. Furthermore, exosomal miR-124 modifies the tumor-microenvironment to weaken the communication between cancerous cells and the bone microenvironment. Interleukin-11 (IL-11), a tumor-motivator, augments the activities of the GP130-Janus kinase network to fuel the tumorigenesis of epithelial tumors, including BC. Excreted by BC cells, IL-11 enhances osteoclastogenesis by facilitating the supplement of osteoclast progenitor cells. Cai et al. have found that exosomal miR-124 exerts its metastasis-inhibitory influence on BC cells by inhibiting IL-11 expression, which significantly suppresses the proliferation and survival of osteoclast progenitor cells [93].
3.2.5. miR-1
Exosomal miR-1 is often found downregulated in BC and associated with worse overall survival. Restoration of the function of BC contributes to the blockage of cell growth, motion, and metastasis. Mechanically, exosomal miR-1 targets Frizzled 7 and TNKS2 to disturb the Wnt/β-catenin signaling cascades and activates Bcl-2, which impairs metastasis and subsequent tumorigenesis [94,95].
3.2.6. miR-193a
Deficiency in exosomal miR-193a activities is common in several cancers [96]. Exosomal miR-193a regulates the cell-cycle cascade induced by epidermal growth factor receptor (EGFR) to dilute cell growth and proliferation. As an oncogenic factor, the methylation of the Wilms’ tumor 1 suppressor gene (WT1) promotor occurs in the early stage of BC. Overexpression of WT1 is positively correlated with the clinicopathological progress of BC. WT1 also takes part in EMT and MET equilibrium. Xie et al. have found that exosomal miR-193a blocks lesion formation and the metastatic tendency of BC cell lines by directly targeting WT1. In the meantime, they also found that ectopic expression of WT1 can partially rescue the metastasis-inhibitory effects exerted by upregulated miR-193a [97]. Therefore, exosomal miR-193a may exhibit biological activity through different proteins in different types of cells.
3.2.7. miR-720
Exosomal miR-720 is attenuated in metastatic BC cell lines, partly demonstrating its tumor-inhibitory identity. By altering the ratio of epithelial markers (β-catenin and E-cadherin) and mesenchymal markers (vimentin, fibronectin, MMP-2, and N-cadherin), exosomal miR-720 impedes the EMT process to repress distant metastasis of BC cells [98]. Twist-related protein 1 (TWIST1), a fundamental molecule involved in various cellular bioactivities, is upregulated in several cancerous cells of the prostate, pancreas, and breast cells. The expression of TWIST1 augments the motility of epithelial cells. Activated by TWIST1, abnormally abundant HER2 appears in 20–30% case of BC, also responsible for more malignant traits and poorer survival rate [93,99]. By mediating the miR-720/TWIST1/HER2 axis, exosomal miR-720 represses the motion tendency of BC cells and metastasis cascade [98].
4. The Regulation of Exosomal microRNAs in the Stemness of BC Cells
CSCs sharing the same characteristics with normal stem cells, such as self-renewal and differentiation, are considered to be situated in the top level of tumor hierarchical organization. CSCs can give rise to phenotypically diverse tumor cell populations or remain undifferentiated with highly-differentiated potential, which imparts tumor heterogeneity and higher malignancy [100]. This subpopulation of tumor cells is the key to perpetuating the tumor. Different subsets of CSCs carrying various biomarkers according to their functions can promote metastasis and tumor recurrence [101].
Some cutting-edge researches have uncovered that exosomal miRNAs exert crucial roles in cancer stemness maintenance [102]. While some effects of exosomal miRNAs in breast cancer stem cells (BCSC) are unclear. Research on the underlying mechanism of exosomal miRNAs provides novel clues for more effective treatments. Next, we review the current understanding of the influence of the interaction between exosomal miRNAs and BC in metastasis and stemness.
Exosomal miRNAs have been demonstrated to involve in regulating cancer progressions, such as stemness and metastasis. By restraining the activity of phosphatase PTEN, exosomal miR-221/222 activates the Akt/NF-κB/COX-2 cascades and imparts stemness-like traits to BC cells [103]. While exosomal miR-143, miR-21, and miR-378e can strongly promote stemness [104]. EMT, a crucial process in tumor invasiveness and metastasis, is a developmental program that facilitates stemness acquisition and maintenance of non-transformed cells and CSCs [105]. Current studies reveal that exosomal miR-200 serves as a tumor suppressor, negatively regulating EMT and stemness by directly activating a series of inhibitory molecules, such as polycomb repressor complex protein B-lymphoma Mo-MLV insertion region 1 (Bmi1) and transcriptional repressor Zeb1/2. TET (Ten eleven translocation) family belongs to DNA demethylase, related to epigenetic modification and cancer cell differentiation. Exosomal miR-22 silences TET/miR-200 axis by inhibiting the TET family which in turn blocks the demethylation of the exosomal miR-200 promoter, contributing to increased EMT and stemness [106].
Exosomal miRNAs can also modify stemness by regulating the expression and function of BC stemness-related genes and the function of related elements. NOTCH1 is an important stemness-regulating gene for mammary stem cells and tumorigenesis. Exosomal miR-34a reduces NOTCH1 expression level to make asymmetric fate definition in BCs and weakens stemness in mammosphere cells [107]. Sox2 and sox9 are oncogenic transcription factors. Sox9 regulates stem cell properties and promotes the transformation from differentiated mammary epithelial cells to mammary stem cells, crucial in BC initiation and malignancy [108]. Sox2 recruits and cooperates with cofactors to constitute a complex binding to multiple genes’ promoters, which causes irregular self-renewal and increased formation of tumor subpopulation [39]. Exosomal miR-140 abundance is reduced in various cancers including BC. Exosomal miR-140 disturbs stem cell renewal and shrinks stem cell population by directly downregulating Sox2/Sox9 expression [39,109].
5. The Regulation of Exosomal microRNAs in the Angiogenesis in BC
Angiogenesis refers to form the neovessel from the preexisting vascular network, which is a highly-active and crucial process in oncogenesis [110]. It is well established that the more pathological angiogenesis occurs, the higher the probability of cancer cells migrating to the circulatory system and achieving distant metastasis. However, in recent years, numerous reviews have summarized and elucidated the mechanism of BC angiogenesis, but only a few have linked BC angiogenesis to exosomal miRNAs. Next, we will review the underlying mechanism of exosomal miRNA taking part in angiogenesis to impair or augment the metastasis of BC.
Exosomal miRNAs that increase the level of any pro-angiogenetic molecules can promote angiogenesis. By establishing an animal model of BC in mice, Kong et al. demonstrated that exosomal miR-155 regulates the von Hippel-Lindau (VHL)/hypoxia-inducible factor (HIF) axis and a cohort of downstream genes, such as interleukin 6 (IL6), vascular endothelial growth factor A (VEGFA), CD44, and pyruvate kinase isozyme type M2 (PKM2), to motivate angiogenetic cascades [111]. Elevated in BC patients, exosomal miR-132 increases the sensitivity of endothelial cells to VEGF by weakening the function of the RAS suppressor, reinforcing angiogenesis [112].
Anti-metastatic exosomal miRNAs regulate the network of angiogenetic-dependent factors to perturb angiogenesis. For example, exosomal miR-16 plays an anti-angiogenetic role by impeding the expression of VEGF [113]. Exosomal miR-503 modifies the tumor microenvironment and blocks the angiogenetic process by simultaneously perturbing the expression of two potent pro-angiogenetic molecules, fibroblast growth factor 2 (FGF2) and VEGFA [114]. After exposure to exosomal miR-100, the formation of human umbilical vein endothelial cells (HUVECs) networks is significantly reduced and the growth of cellular tubes is suppressed. Mechanically, exosomal miR-100 regulates the mTOR/ hypoxia-inducible factor-1 (HIF-1α)/VEGF axis to downregulate the activities of fundamental effectors involved in angiogenesis [115].
6. The Regulation of Exosomal microRNAs in Chemotherapy Resistance in BC
The recognized clinical treatment strategies for BC embrace specific chemotherapy, radiotherapy, and systemically surgical resection. Chemoresistance caused by repetitive and long-range usage of the same kind of drugs is the major obstacle for successful treatment, which accounts for almost 90% of the chemotherapy failures [116]. It is urgent to explore the underlying mechanism in drug resistance acquisition to reduce the recurrence rate. Numerous studies have uncovered the influence of exosomal miRNAs in resistance induction of BC. Here, we will review the signaling pathways targeted by exosomal miRNAs to regulate the drug sensitivity of BC cells. As shown in Table 1, we summarize the exosomal miRNAs and their targets in chemoresistance.

Table 1.
Exosomal microRNAs take part in chemoresistance.
6.1. Topoisomerase Interactive Agents
6.1.1. Doxorubicin (Adriamycin)
Doxorubicin is a commonly used chemotherapeutic drug that shows high activity against a variety of tumors. Early studies proved that doxorubicin can interfere with gene expression through intercalating into DNA double-helix and inhibiting enzymatic activities of topoisomerase II. It can enhance intercellular oxidative stress by promoting the conversion between oxygen and reactive oxygen species (ROS). The excessive stimulation of ROS will attack genomic and mitochondrial DNA, rendering them unstable. This is toxic and harmful for cell survival in doxorubicin-resistance groups [143]. Doxorubicin resistance has traditionally been a hot research topic in recent years. Studies have revealed that 66 miRNAs are increased and 309 miRNAs are decreased [144], which suggests that there is a high correlation between miRNA and doxorubicin resistance. Following, we focus on the concrete impacts of exosomal miRNA on doxorubicin resistance.
As an effective oncogene, Akt is homologous with the retroviral gene v-Akt. Akt and fundamental molecules are involved in its signal cascade to regulate DNA repair and cell proliferation. Highly activated in carcinogenesis, Akt suppresses apoptotic cell death program and improves cell survival by targeting a series of apoptotic effectors [120]. The Akt signaling network is significantly related to chemoresistance. Several studies have indicated that the variance of the Akt pathway by exosome microRNAs influences apoptosis induced by chemo-agents. Mechanically, exosomal miR-221-3p governs the activities of PI3K/Akt/phosphoinositide-3-kinase regulatory subunit 1 (PIK3R1) axis to facilitate the gain of doxorubicin resistance in cancerous cells [118]. Inversely, Exosomal miR-505 indirectly inhibits the Akt3 activities and induces apoptosis to restore doxorubicin sensitivity [120].
As a highly conserved process, autophagy refers to the capture of malfunctioning lipids, proteins, and organelles and carries them to the lysosome for degradation. The influence of autophagy is contentious. It supports cell survival in cytotoxic environments such as hypoxia and starvation. Furthermore, genotypic pressure caused by anti-tumor chemo-agents activates cytoprotective autophagy. However, lack of autophagy disturbs genetic stability and increases oxidative stress, which induces the initiation and progression of tumors [145]. Exosomal microRNAs are able to benefit from autophagy to modify cytotoxic effects induced by drugs and regulate cellular sensitization. Isoliquiritigenin (ISL), a powerful motivator of autophagy-lysosome cascades, is inhibited by exosomal miR-25 to imparts chemoresistance to Doxorubicin [119]. Inversely, by adjusting the interaction between downstream histone deacetylase 1 (HDAC1)/HDAC7/heat shock proteins 70 (Hsp70) K246, exosomal miR-34a inhibits autophagic cell death and weakens chemo-sensitizing potential of BC cells [121,122].
In fact, one exosomal microRNA may exert dual effects by targeting different effectors. The roles of exosomal miR-181a in doxorubicin resistance are dual by targeting different molecules. It increases the sensitivity to doxorubicin by modifying B-cell lymphoma 2 (Bcl-2) [123] and upregulates BCL2-Associated X (Bax) to weaken apoptosis under cytotoxic pressure from anti-cancer drugs [124].
Intriguingly, doxorubicin can suppress the expansion of the primary lesion, but it brings the risk of lung metastasis to some extent. The administration of doxorubicin induces a paracrine signal cascade which modifies the lung microenvironment to establish favorable conditions for future metastasis. Doxorubicin promotes cancerous cells to excrete IL-33 which subsequently induces the release of IL-13 from Th2 cells in the lung. Besides, doxorubicin accelerates the immunoinhibitory cells, myeloid-derived suppressor cells (MDSC), to coat with IL-13 receptors and synthesize exosomal miR-126a. After IL-13 derived from Th2 cells combined with its corresponding receptors on the surface of MDSCs, a vast array of exosomal miR-126a will be excreted and acts on Th2 cells, which, in turn, releases more IL-13 and induces more miR-126a. Exosomal miR-126a targets the S100A9 gene to facilitate the proliferation and survival of MDSC. This positive-feedback network exaggerates the influence of exosomal miR-126a and a cohort of inflammatory cells and molecules in angiogenesis, tumor growth, and metastasis in the lung, making adequate preparations for BC distant metastasis [125]. This finding lays a foundation for future studies to assess whether DOX-induced MDSC exosome miR-126a has an effect on BC cells and contributes to the development of chemoresistance in BC cells.
6.1.2. Mitoxantrone
Mitoxantrone is a powerful topoisomeraseⅡ inhibitor, regarded as a cell-cycle nonspecific chemotherapeutic drug that suppresses the synthesis of genetic materials and blocks cell proliferation in every stage of the cell cycle [146]. Exosomal miR-328 was found to be associated with the cellular content of important breast cancer resistance protein (BCRP/ABCG2). Overexpressed exosomal miR-328 contributes to increased sensitization to Mitoxantrone of MCF-7/MX100 cells by downregulating BCRP/ABCG2 expression [126].
6.2. Platinum Analogs
Cisplatin
Cisplatin can directly bind to DNA, causing cross-linking, which generates double-strand breaks [147]. Numerous miRNAs are found to be dysregulated in these processes for the gain and loss of chemoresistance. Methyl-CpG binding protein 2 (MECP2), a methyl-binding protein involved in DNA transcription activities, is inhibited by exosomal miR-194 and exosomal miR-132 in cisplatin-resistant BC cell lines [127,128]. Besides, some selected candidate exosomal microRNAs can downregulate chemoresistance. Exosomal miR-24 promotes EMT and induces stemness by influencing FIH1 and BimL, enhancing cisplatin-resistance [121]. E2F1 governs G1/S transition and promotes the expression of downstream molecule ataxia-telangiectasia mutated (ATM). By directly perturbing E2F1 functions and indirectly repressing ATM, exosomal miR-302b causes irreversible cell-cycle arrest, deficiency in DNA repair, and promotes apoptosis after cisplatin administration [129].
6.3. Antimicrotubule Agents
Taxane
Taxane, which is a type of tricyclic compound, including paclitaxel (PTX) and docetaxel. The antitumor effects of taxane are generally attributed to inhibition of microtubule dynamics, leading to cell division defects [148].
Increasing studies demonstrate that the roles played by exosomal microRNAs are indispensable in the EMT which is crucial in the acquisition of multidrug resistance (MDR) phenotype [149]. By regulating EMT-associated molecules, such as Twinfilin 1 (TWF1) and Interleukin-11 (IL-11), exosomal miR-30c causes acquired PTX resistance [130]. In contrast, semaphorin 4C (Sema4C), a natural inducer of EMT, is inhibited by exosomal miR-125b, which reverse EMT phenotypes and augments sensitivity to PTX [132]. Re-expression of miR-200 family suppresses EMT by regulating ZEB1/ZEB2/E-cadherin, restoring sensitivity to microtubule-modulating drugs [131,133].
One of the hallmarks of cancer is the successful escape from programmed cell death [62]. Apoptosis-related molecules, such as BCL2 and BCL2 homologous antagonist/killer (BAK1) are the major effectors involved in this process, as well as take part in the exosomal microRNAs-mediated chemoresistance. By targeting BAK1, exosomal miR-125 interfering with and weakening PTX-caused apoptosis and cellular toxicity [131]. Inversely, the exosomal miR-16/BCL2 axis leads to an increased apoptotic program and imparts docetaxel sensitivity to cancerous cells [131].
6.4. Hormonal Agents
6.4.1. Tamoxifen
Tamoxifen, an antagonist of the ER, suppresses estrogen-dependent growth [150]. As a kind of phosphatase, PTEN is a crucial inhibitor of PI3K, which precisely governs the PI3K/Akt/mTOR signal way. Loss or diminution of PTEN activities favors oncogenic progress and chemoresistance [151]. Exosomal miR-301 targets PTEN to cause the loss of response to tamoxifen therapy [134]. Exosomal miR-101 inhibits PTEN and activates Akt by targeting membrane-associated guanylate kinase (MAGI-2) to confer tamoxifen resistance to BC cells [135].
The balance between cell cycle arrest and cell death is usually inconstant in cancer treatment. Exosomal microRNAs can regulate the bioactivities of E2F, cyclins, and CDKs to coordinate cell cycle pace or make cell cycle chaotic to modify chemoresistance. MiR-320a modifies the cAMP-regulated phosphoprotein (ARPP-19)/estrogen-related receptor gamma (ERRγ) pathways to reduce the content of CCND1 and c-Myc, which re-sensitizes resistant cells to tamoxifen therapy [136].
Autophagy is an essential process to maintain homeostasis and is closely related to chemo-resistance. By directly inhibiting uncoupling protein 2 (UCP2)-mediated autophagy, exosomal miR-214 restores the response of cancerous cells to tamoxifen [137]. In addition, exosomal miR-415a weakens chemo-resistant potential to tamoxifen by adjusting the interaction of ERα and 14-3-3 protein zeta (14-3-3ζ), as well as inhibiting autophagy [138].
6.4.2. Fulvestrant
Fulvestrant, by selectively reducing the estrogen receptor, and inhibiting aromatase, represses the synthesis of estrogen, sufficiently improving the survival rate of ER-positive BC patients [152]. Loss of sensitivity to tamoxifen happens in almost 50% of ER-positive BC patients [153]. Exosomal miR-221/222 is a pleiotropic and powerful oncogenic molecule, elevated after fulvestrant treatment. By dysfunction of transforming growth factor-beta (TGF-β) and β-Catenin and their downstream signaling network, exosomal miR-221/222 imparts fulvestrant resistance to BC cell lines [139]. Enhancer of zeste homolog 2 (EZH2), a tri-methylase for histone H3 lysine 27 to turn the chromosomes into a translation-inhibitory state, is bound by exosomal miR-101 to bring fulvestrant resistance to sensitive BC cells [131,135].
6.5. Monoclonal Antibodies
Trastuzumab
Trastuzumab suppresses irregular cell growth of BC by combining with HER2 whose gene sequence is amplified in 20% to 25% of BC patients [154].
The interaction between growth factors and their corresponding receptors is critical in metabolic activities, such as cell viability, inter- or intra-cellular communication, and response to therapy [155]. Transforming growth factor (TGF), EGFR and insulin-like growth factor 1 receptor (IGF1R) are the key molecules in these cellular processes. While exosomal miR-205-5p can sensitize BC cells to trastuzumab by targeting ERBB2 and regulating the P63/EGFR axis [140].
Besides, exosomal microRNAs can target some novel factors to influence chemoresistance. Exosomal miR-16 inhibits the formation of trastuzumab-resistant BC cell lines in vivo and vitro. Mechanically, miR-16 modifies chromatin assembly and regulates far upstream element-binding protein 1 (FUBP1) and cyclin J to enhance the cytotoxic effects of trastuzumab [141].
6.6. Others
Verapamil
Verapamil is a suppressor of the P-glycoprotein (P-gp), a transporter protein pumping anti-cancer chemo-agents out of cells [156]. Transfection of let-7 can decrease the sensitivity of verapamil. Exosomal let-7 can regulate the functions of RAS, estrogen receptor 1 (ESR1), caspase-3 (CASP3), and high mobility group AT-hook 2 (HMGA2), influencing cell-cycle, the transformation between EMT and MET, and hormone receptor expression [131].
In conclusion, the acquisition of drug chemoresistance by exosomal miRNAs can be generalized in the following ways. First, grab some critical signaling pathways to support cancer cellular independent growth. Second, promote EMT and stimulate the formation and expansion of the CSC population to dictate a more aggressive phenotype to cancer cells. Third, facilitate cell survival by the imbalance of autophagy/apoptosis and upregulation of anti-apoptotic molecules. Finally, yet importantly, dis-regulate drug transport to enhance the elimination of cytotoxic drugs from cells.
7. Exosomal microRNAs That Function as Prognostic Biomarkers for BC Patients
7.1. Exosomal microRNAs for Early-Stage Discovery of BC
Exosomal miRNAs work as effective instructors to tell us in which step is the oncogenic process going on and help doctors distinguish what kinds of BC are those states belonging to, which facilitates doctors to take further steps to copy with BC.
Exosomal miRNAs can predict the early occurrence of BC. Exosomal miR-221/222 almost penetrate every step of the oncogenesis of BC, which makes exosomal miR-221 an informative indicator for predictive BC origin, progress and treatment effects [157]. The increased concentration of miR-409-3p, miR-801, miR-127-3p, miR-652, and miR-148b in serum are found in stages I and II of BC, indicating their roles in detecting cancer early origin [158]. After amplification and quantitative detection of exosomal miR-21 by real-time PCR, the serum changes of exosomal miR-21 in 89 BC patients and 55 healthy controls were measured. The obvious increase of exosomal miR-21 in BC patients suggests its role in serving as biomarkers for early-phase BC, with 87.3% specificity and 87.6% sensitivity [159].
Besides, exosomal miRNAs are of great significance in predicting TNM staging, deterioration, and BC types with worse prognoses. The content of miR-197, miR-29b-2, miR-205, and miR-155 correspond to the lymph node involved in lesion metastasis (N3 versus N2) and tumor size (T3 versus T2). The occurrence of exosomal miR-205 and exosomal miR-155 suggests lesion metastases in the distance [160]. The elevation of exosomal miR-373 in TNBC is more significant than that in the luminal subtype whose presence demonstrates the hormone receptor-negative and triple-negative types of BC [46,161].
7.2. Exosomal microRNAs for Treatment Assessment of BC
Numerous studies demonstrate that the level changes of exosomal miRNAs before and after treatment can serve as the value index to reflect the therapy effect. Endothelial cells release exosomal miR-503 to regulate cell growth and migration of BC cell lines in vitro, reversing the aggressive phenotype of BC, which becomes elevated in patients after neoadjuvant treatment [162]. The anti-cancer drug can modify the quality and quantity of exosomal miRNAs, which informs us of the alteration in cancer status. Four overexpressed exosomal miRNAs, miR-376c, miR-27a, miR-155, and miR-376a, all turn back to normal concentration after neoadjuvant therapy, for further monitoring treatment progression [84]. After the administration of Trastuzumab, the concentration of exosomal miR-21 in HER2-positive patients is decreased, which suggests exosomal miR-21 can work as a Trastuzumab treatment assessment [163]. Elevated exosomal miR-454 level and reduced exosomal miR-374a/b indicate poor disease-free survival. Similarly, the increased content of some exosomal miRNAs (miR-210, miR-21, miR-454, miR-27a/b) and reduced expression of exosomal miR-155 are all typical markers for poor overall poor survival [164].
8. Exosomal microRNAs as Novel Targets for Targeted Therapy
The roles of exosomal miRNAs in different stages of BC carcinogenesis have been reviewed in detail. We should combine the research value of exosomal miRNAs with their clinical value, making the research value better serve the clinical treatment. After clearly identifying the composition of miRNAs in different phases of BC, as shown in Figure 3, we can target specific miRNAs to treat various types of BC, timely change anti-cancer drugs to cope with chemoresistance, and monitor the real-time treatment response.
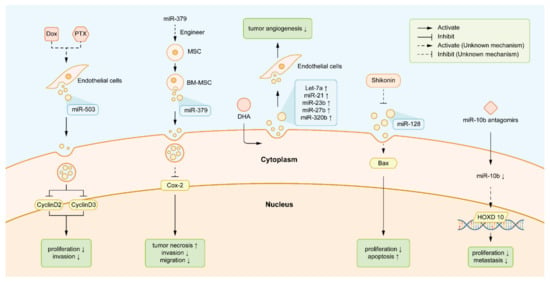
Figure 3.
Exosomal miRNAs as novel targets for targeted therapy. The text legends from left to right are: (a) DOX and PTX induced endothelial cells to deliver more exosomal miR-503. The increased miR-503 interrupted the expression of CCND2 and CCND3, inhibiting BC cell proliferation and invasion consequently. (b) Engineering MSCs carried more exosomal miR-379 and released it into the tumor micro-environment by BM-MSCs, which interfered with the migration capacity of BC and induced the apparent tumor lesion necrosis. (c) DHA induced the elevated level of some exosomal miRNAs with negative effects on angiogenesis and tumorigenesis, such as let-7, miR-21, miR-23b, miR-27b, and miR-320b. (d) Shikonin inhibited the secretion of exosomal miR-128 in breast cancer cells, thus lifting its inhibition to the expression of the apoptotic gene Bax. Shikonin modified the interaction between exosomal miR-128 and Bax, subsequently promoting apoptosis. (e) MiR-10b antagomirs directly attenuated the bioactivities of exosomal miR-10b, which subsequently promoted the translation of HOXD10 and suppresses the migration of BC. Abbreviations: DOX: doxorubicin. PTX: paclitaxel. CCND2: cyclin D2. CCND3: cyclin D3. BC: breast cancer. MSCs: mesenchymal stem cells. BM-MSCs: bone marrow-derived mesenchymal stem cells. DHA: docosa-hexaenoic acid. HOXD10: Homeobox D10.
Drugs that elevate the tumor-suppressive exosomal miRNAs can moderate the state of illness. Exosomal miR-503 is a powerful tumor- suppressor. After administration of neoadjuvant treatments, such as epirubicin and PTX, more exosomal miR-503 released from endothelial cells attenuates the aggressive traits of tumor [162]. Engineering mesenchymal stem cells (MSCs) carry more exosomal miR-379 and release the elevated level of miR-379, which contributes to the reduction in cell migration capacity and apparent BC tumor lesion necrosis [165]. Under the assistance of the Rab GTPase, Rab27A, docosahexaenoic acid (DHA) induces the elevated level of some exosomal miRNAs with anti-angiogenetic and tumor-inhibitory roles, such as let-7, miR-21, miR-23b, miR-27b, and miR-320b [166]. Sinomenine exerts its anti-tumor roles by regulating the miR-29/PDCD-4 axis, which subsequently suppresses cell viability capacity [167].
Simultaneously, the diminishment of tumor-promoting miRNAs is also the target task of anti-tumor medicine. Shikonin, an effective anti-tumor drug, inhibits the excretion of pro-tumorigenic exosomal miR-128 which facilitates the escape of BC cells from apoptosis [168]. Matrine decreases cell growth and enhances apoptosis by targeting the miR-21/PTEN/Akt pathway and downstream tumor-suppressive molecules [169]. MiR-10b antagomirs, an artificially-engineered anti-miRNA for systemic treatment, attenuates the activities of exosomal miR-10b and blocks the metastasis of BC cells to surround organs [56,59]. With superior efficacy in cancer treatment, doxycycline exerts powerful inhibitory effects on the PAR1/Akt/NF-κB/miR-17/E-cadherin pathway, which reverses aggressive phenotype and represses the motility of cancerous cells [170].
9. Discussion
Metastasis is the major cause of poor overall survival in BC patients. Distant metastatic lesions occur in almost half of BC patients administrated with hormone treatments and chemotherapy. Only 20% of the five-year survival rate happens in these patients [171]. The discovery and deep study of exosomal miRNA deepen an understanding of the potential mechanism of BC oncogenic progress. It has been indicated that exosomal miRNAs almost penetrate every step of various biological processes and takes indispensable roles [172]. Though the exploration of BC is quite advanced, only a small proportion of studies focus on the relationship between exosomal miRNA and BC metastasis in the last 5 years. In Table 2, we list the underlying mechanisms of different processes that exosomal miRNAs are involved in to contribute to metastasis and metastatic-related processes.

Table 2.
Different roles of exosomal microRNAs playing in BC progress.
In this study, we have reviewed the latest findings of the roles played by exosomal miRNAs in BC metastasis and metastatic-related processes, including, migration, invasion, distal metastasis, stemness, and angiogenesis.
- MiRNAs involve controlling the aggressiveness of BC cells (such as miR-21, miR-10b, miR-1246, miR-373, and miR-17-5p).
- MiRNAs modify the stemness properties of BC cells (such as miR-22, miR-221/222, and miR-143).
- MiRNAs alter the angiogenetic process of BC (such as miR-155, miR-132, and miR-16).
In the next part, we review how exosomal miRNAs verify the sensitivity of BC cell lines to chemotherapy, and their instructive functions as biomarkers for diagnosis and prognosis (such as miR-221/222, miR-195, miR-1246, miR-484, and miR-182).
Demonstrated by different studies and conclusions, we tried to conclude some exosomal miRNAs carried dual effects on BC metastasis and metastasis-related processes. For example, different studies suggested miR-200 family (miR-200a, miR-200b, miR-200c, miR-429, miR-141) takes opposite roles in BC. The materials and methods employed in these experiments were different. Minh et al. transferred extracellular vesicles with miR-200 in different TNBC cell lines, 4TO7 and 4T1, observing the alteration of motion behavior of recipient cells [173]. Zhi-bin et al. used MDA-MB-231 cells for building a bone metastasis model, thereby uncovering the key modulator in the motion of BC cells to the bone micro-environment was miR-429/ZAB1/CRKL signaling network [86]. It can be deduced that the most likely reason for the discrepancy was the tumor microenvironment provided by different cell lines or inner interaction between miRNAs. More experiments that are comprehensive need to further investigate how the miR-200 family regulates the oncogenesis of BC.
At present, studies on whether and how exosomal miRNAs play a role in the treatment of BC are relatively abundant, and some of which, as we discussed earlier, give us a clearer understanding of the molecular mechanism of chemoresistance. In terms of radiotherapy, Tang et al. found that radiation-induced exosomal miR-208a enhanced the radioresistance of lung cancer cells, which first revealed the relationship between exosomal miRNAs and radiosensitivity [174]. Interestingly, recent experiments have confirmed that downregulation of miRNA-26b-5p enhances radiosensitization in lung adenocarcinoma cells [175]. Previous experimental results suggest that different exosomal miRNAs may exert opposite effects in different types of cancer cells. Unfortunately, as of this writing, no studies have directly elucidated the relationship between exosomal miRNAs and radiosensitivity of BC cells. We believe that this will be a promising area for future research.
As a highly conserved process, autophagy refers to assemble dysfunctional substances, such as damaged organelles and cytoplasmic proteins into bilayer autophagosomes. And after processing in lysosomes, components of autophagosomes are chosen to be degraded or recycled, which maintains cellular homeostasis [176]. Roles of autophagy in tumorigenesis are dual. At the beginning of metastasis, increased autophagy counteracts deleterious situations such as oxidative cytotoxicity and metabolic pressure, assisting the survival of cells and speeding up metastasis [177]. However, autophagy promotes inflammatory reactions and restrains the formation and expansion of primary lesions [178]. Some studies demonstrated the involvement of exosomal miRNAs in autophagy happening in BC. Biological activities of exosomal miR-92b induce autophagy in BC cells by downregulating the expression of EZH2 at mRNA level [179]. Instead, as a tumor-suppressive miRNA, exosomal miR-107 inhibits autophagy by mediating HMGB1 expression [180].
It was uncovered that autophagy can fuel tumor progress by weakening the negative influence of anoikis on cancerous cells [181]. Anoikis, as an indispensable tumor-inhibitory process, induces programmed cell death when sensing the detachment of cells from the native ECM, which is considered as an effective repressor in anchorage-free growth and abnormal detachment. Anoikis resistance refers to adherent-free growth and spontaneous EMT which are pivotal in facilitating distant metastasis and tumorigenesis. Elevated anoikis resistance imparts more aggressive traits to metastatic cells [182]. Erin N Howe et al. revealed that as a receptor tyrosine kinase situated in the cell surface, TrkB imparts increased anoikis resistance to BC cells after activation by its ligands NTF3. By directly targeting NTF3, exosomal miR-200 restores the anoikis sensitivity of cancerous cells of TNBCs [183]. However, at the time of writing, only a few studies have detailedly discussed the relationship of exosomal miRNA and anoikis in BC, which points out the direction for the next research.
BC cells are divided into five subtypes, HER2-overexpressing, basal-like, normal-like, luminal A, and luminal B. Due to their different clinicopathological features and prognostics, it is important in identifying candidate miRNAs to distinguish several subtypes for assessing the targeted clinical treatments. After analyzing clinical cases, exosomal miR-21 was found specifically elevated in several cancers, which effectively instructs TNBC [164]. Besides, it was discovered that exosomal miR-373, speeding up the tumorigenic processes, is significantly elevated in TNBC and ER/PR-negative patients compared with receptor-positive patients [184]. The content of exosomal let-7a, miR-1246, miR-10b, miR-21, and miR-122 all increase to varying degrees, which tells healthy patients apart from BC patients [161,185]. As consulting numerous literature, many studies have focused on how to distinguish healthy groups from BC patients, and tell TNBC apart from other types. Only a few studies clearly indicated the content difference between these five subtypes of BC and evidence to discriminate them under the assistance of exosomal miRNAs.
This comprehensive review provides a systematic overview to improve our understanding of the mechanisms behind the origin, maintenance, and progression of tumors. More importantly, new insights have helped to identify diagnostic, prognostic, and predictive markers at the molecular and cellular levels, paving the way for new therapeutic approaches.
Author Contributions
All authors contributed to the study conception and design. Material preparation was performed by L.-B.T. and S.-X.M. Data collection and analysis were performed by L.-B.T., S.-X.M., Z.-H.C., and Q.-Y.H. The first draft of the manuscript was written by L.-B.T., S.-X.M., and L.-Y.W. Q.-Y.H., R.-C.Z., Y.W. and L.-X.X. critically revised the work. All authors commented on previous versions of the manuscript. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the National Natural Science Foundation of China (No. 31860317) and the National Natural Science Foundation of Jiangxi province (No.20202BAB206056), as well as the postgraduates innovation special fund project of Nanchang University (No. YC2020-S049) and the college students innovation training program of Nanchang University (No. S202010403034).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
The data presented in this study are available on request from the corresponding author.
Acknowledgments
We would like to thank the technical support by Si-Di Lv, and the support from our families and friends.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Bray, F.; Ferlay, J.; Soerjomataram, I.; Siegel, R.L.; Torre, L.A.; Jemal, A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. Cancer J. Clin. 2018, 68, 394–424. [Google Scholar] [CrossRef] [PubMed]
- Scully, O.J.; Bay, B.H.; Yip, G.; Yu, Y. Breast cancer metastasis. Cancer Genom. Proteom. 2012, 9, 311–320. [Google Scholar]
- Marino, N.; Woditschka, S.; Reed, L.T.; Nakayama, J.; Mayer, M.; Wetzel, M.; Steeg, P.S. Breast cancer metastasis: Issues for the personalization of its prevention and treatment. Am. J. Pathol. 2013, 183, 1084–1095. [Google Scholar] [CrossRef] [PubMed]
- Qiu, A.F.; Miao, Z.L.; Ge, G.K.; Wang, C.B.; Bian, J.; Ma, H.Y.; Xu, Q. Response and prognosis of neoadjuvant dose-dense or standard schedule chemotherapy with anthracyclines and taxanes for Luminal B breast cancer. Zhonghua Yi Xue Za Zhi 2017, 97, 3466–3470. [Google Scholar] [CrossRef] [PubMed]
- Griguolo, G.; Pascual, T.; Dieci, M.V.; Guarneri, V.; Prat, A. Interaction of host immunity with HER2-targeted treatment and tumor heterogeneity in HER2-positive breast cancer. J. Immunother. Cancer 2019, 7, 90. [Google Scholar] [CrossRef]
- Thakur, V.; Kutty, R.V. Recent advances in nanotheranostics for triple negative breast cancer treatment. J. Exp. Clin. Cancer Res. 2019, 38, 430. [Google Scholar] [CrossRef]
- Colombo, M.; Raposo, G.; Théry, C. Biogenesis, secretion, and intercellular interactions of exosomes and other extracellular vesicles. Annu. Rev. Cell Dev. Biol. 2014, 30, 255–289. [Google Scholar] [CrossRef]
- Théry, C.; Zitvogel, L.; Amigorena, S. Exosomes: Composition, biogenesis and function. Nat. Rev. Immunol. 2002, 2, 569–579. [Google Scholar] [CrossRef] [PubMed]
- Lu, T.X.; Rothenberg, M.E. MicroRNA. J. Allergy Clin. Immunol. 2018, 141, 1202–1207. [Google Scholar] [CrossRef] [PubMed]
- Guduric-Fuchs, J.; O’Connor, A.; Camp, B.; O’Neill, C.L.; Medina, R.J.; Simpson, D.A. Selective extracellular vesicle-mediated export of an overlapping set of microRNAs from multiple cell types. BMC Genom. 2012, 13, 357. [Google Scholar] [CrossRef]
- Skog, J.; Würdinger, T.; van Rijn, S.; Meijer, D.H.; Gainche, L.; Curry, W.T., Jr.; Carter, B.S.; Krichevsky, A.M.; Breakefield, X.O. Glioblastoma microvesicles transport RNA and proteins that promote tumour growth and provide diagnostic biomarkers. Nat. Cell Biol. 2008, 10, 1470–1476. [Google Scholar] [CrossRef] [PubMed]
- Liao, J.; Liu, R.; Yin, L.; Pu, Y. Expression profiling of exosomal miRNAs derived from human esophageal cancer cells by Solexa high-throughput sequencing. Int. J. Mol. Sci. 2014, 15, 15530–15551. [Google Scholar] [CrossRef] [PubMed]
- Wieckowski, E.; Whiteside, T.L. Human tumor-derived vs dendritic cell-derived exosomes have distinct biologic roles and molecular profiles. Immunol. Res. 2006, 36, 247–254. [Google Scholar] [CrossRef]
- Zhang, J.; Li, S.; Li, L.; Li, M.; Guo, C.; Yao, J.; Mi, S. Exosome and exosomal microRNA: Trafficking, sorting, and function. Genom. Proteom. Bioinform. 2015, 13, 17–24. [Google Scholar] [CrossRef]
- Bang, C.; Batkai, S.; Dangwal, S.; Gupta, S.K.; Foinquinos, A.; Holzmann, A.; Just, A.; Remke, J.; Zimmer, K.; Zeug, A.; et al. Cardiac fibroblast-derived microRNA passenger strand-enriched exosomes mediate cardiomyocyte hypertrophy. J. Clin. Investig. 2014, 124, 2136–2146. [Google Scholar] [CrossRef] [PubMed]
- Van Balkom, B.W.; de Jong, O.G.; Smits, M.; Brummelman, J.; den Ouden, K.; de Bree, P.M.; van Eijndhoven, M.A.; Pegtel, D.M.; Stoorvogel, W.; Würdinger, T.; et al. Endothelial cells require miR-214 to secrete exosomes that suppress senescence and induce angiogenesis in human and mouse endothelial cells. Blood 2013, 121, 3997–4006. [Google Scholar] [CrossRef]
- Fabbri, M.; Paone, A.; Calore, F.; Galli, R.; Gaudio, E.; Santhanam, R.; Lovat, F.; Fadda, P.; Mao, C.; Nuovo, G.J.; et al. MicroRNAs bind to Toll-like receptors to induce prometastatic inflammatory response. Proc. Natl. Acad. Sci. USA 2012, 109, 2110–2116. [Google Scholar] [CrossRef]
- Sheridan, C. Exosome cancer diagnostic reaches market. Nat. Biotechnol. 2016, 34, 359–360. [Google Scholar] [CrossRef]
- Saadatpour, L.; Fadaee, E.; Fadaei, S.; Nassiri Mansour, R.; Mohammadi, M.; Mousavi, S.M.; Goodarzi, M.; Verdi, J.; Mirzaei, H. Glioblastoma: Exosome and microRNA as novel diagnosis biomarkers. Cancer Gene Ther. 2016, 23, 415–418. [Google Scholar] [CrossRef]
- Salimian, J.; Mirzaei, H.; Moridikia, A.; Harchegani, A.B.; Sahebkar, A.; Salehi, H. Chronic obstructive pulmonary disease: MicroRNAs and exosomes as new diagnostic and therapeutic biomarkers. J. Res. Med Sci. 2018, 23, 27. [Google Scholar] [CrossRef]
- Ogata-Kawata, H.; Izumiya, M.; Kurioka, D.; Honma, Y.; Yamada, Y.; Furuta, K.; Gunji, T.; Ohta, H.; Okamoto, H.; Sonoda, H.; et al. Circulating exosomal microRNAs as biomarkers of colon cancer. PLoS ONE 2014, 9, e92921. [Google Scholar] [CrossRef]
- Huang, X.; Yuan, T.; Tschannen, M.; Sun, Z.; Jacob, H.; Du, M.; Liang, M.; Dittmar, R.L.; Liu, Y.; Liang, M.; et al. Characterization of human plasma-derived exosomal RNAs by deep sequencing. BMC Genom. 2013, 14, 319. [Google Scholar] [CrossRef]
- Sempere, L.F.; Keto, J.; Fabbri, M. Exosomal MicroRNAs in Breast Cancer towards Diagnostic and Therapeutic Applications. Cancers 2017, 9, 71. [Google Scholar] [CrossRef] [PubMed]
- Zhou, W.; Fong, M.Y.; Min, Y.; Somlo, G.; Liu, L.; Palomares, M.R.; Yu, Y.; Chow, A.; O’Connor, S.T.; Chin, A.R.; et al. Cancer-secreted miR-105 destroys vascular endothelial barriers to promote metastasis. Cancer Cell 2014, 25, 501–515. [Google Scholar] [CrossRef] [PubMed]
- Zhao, L.; Liu, W.; Xiao, J.; Cao, B. The role of exosomes and “exosomal shuttle microRNA” in tumorigenesis and drug resistance. Cancer Lett. 2015, 356, 339–346. [Google Scholar] [CrossRef] [PubMed]
- Shi, J. Considering Exosomal miR-21 as a Biomarker for Cancer. J. Clin. Med. 2016, 5, 42. [Google Scholar] [CrossRef]
- Volinia, S.; Calin, G.A.; Liu, C.G.; Arnbs, S.; Cimmino, A.; Petrocca, F.; Visone, R.; Iorio, M.; Roldo, C.; Ferracin, M. A microRNA expression signature of human solid tumors defines cancer gene targets. Proc. Natl. Acad. Sci. USA 2006, 103, 2257–2261. [Google Scholar] [CrossRef] [PubMed]
- Feng, Y.H.; Tsao, C.-J. Emerging role of microRNA-21 in cancer. Biomed. Rep. 2016, 5, 395–402. [Google Scholar] [CrossRef] [PubMed]
- Bandyopadhyay, S.; Pai, S.K.; Hirota, S.; Hosobe, S.; Tsukada, T.; Miura, K.; Takano, Y.; Saito, K.; Commes, T.; Piquemal, D.; et al. PTEN up-regulates the tumor metastasis suppressor gene Drg-1 in prostate and breast cancer. Cancer Res. 2004, 64, 7655–7660. [Google Scholar] [CrossRef] [PubMed]
- Zhu, S.; Wu, H.; Wu, F.; Nie, D.; Sheng, S.; Mo, Y.Y. MicroRNA-21 targets tumor suppressor genes in invasion and metastasis. Cell Res. 2008, 18, 350–359. [Google Scholar] [CrossRef] [PubMed]
- Yin, S.; Lockett, J.; Meng, Y.; Biliran, H., Jr.; Blouse, G.E.; Li, X.; Reddy, N.; Zhao, Z.; Lin, X.; Anagli, J.; et al. Maspin retards cell detachment via a novel interaction with the urokinase-type plasminogen activator/urokinase-type plasminogen activator receptor system. Cancer Res. 2006, 66, 4173–4181. [Google Scholar] [CrossRef]
- Leupold, J.H.; Yang, H.S.; Colburn, N.H.; Asangani, I.; Post, S.; Allgayer, H. Tumor suppressor Pdcd4 inhibits invasion/intravasation and regulates urokinase receptor (u-PAR) gene expression via Sp-transcription factors. Oncogene 2007, 26, 4550–4562. [Google Scholar] [CrossRef]
- Stephens, R.W.; Nielsen, H.J.; Christensen, I.J.; Thorlacius-Ussing, O.; Sørensen, S.; Danø, K.; Brunner, N. Plasma urokinase receptor levels in patients with colorectal cancer: Relationship to prognosis. J. Natl. Cancer Inst. 1999, 91, 869–874. [Google Scholar] [CrossRef] [PubMed]
- Yang, H.S.; Jansen, A.P.; Komar, A.A.; Zheng, X.; Merrick, W.C.; Costes, S.; Lockett, S.J.; Sonenberg, N.; Colburn, N.H. The transformation suppressor Pdcd4 is a novel eukaryotic translation initiation factor 4A binding protein that inhibits translation. Mol. Cell. Biol. 2003, 23, 26–37. [Google Scholar] [CrossRef] [PubMed]
- Göke, R.; Barth, P.; Schmidt, A.; Samans, B.; Lankat-Buttgereit, B. Programmed cell death protein 4 suppresses CDK1/cdc2 via induction of p21(Waf1/Cip1). Am. J. Physiol. Cell Physiol. 2004, 287, 1541–1546. [Google Scholar] [CrossRef]
- Min, W.; Wang, B.; Li, J.; Han, J.; Zhao, Y.; Su, W.; Dai, Z.; Wang, X.; Ma, Q. The expression and significance of five types of miRNAs in breast cancer. Med. Sci. Monit. Basic Res. 2014, 20, 97–104. [Google Scholar] [CrossRef] [PubMed]
- Liang, A.-L.; Zhang, T.-T.; Zhou, N.; Wu, C.Y.; Lin, M.-H.; Liu, Y.-J. MiRNA-10b sponge: An anti-breast cancer study in vitro. Oncol. Rep. 2016, 35, 1950–1958. [Google Scholar] [CrossRef] [PubMed]
- Betel, D.; Koppal, A.; Agius, P.; Sander, C.; Leslie, C. Comprehensive modeling of microRNA targets predicts functional non-conserved and non-canonical sites. Genome Biol. 2010, 11, R90. [Google Scholar] [CrossRef]
- Kim, J.; Siverly, A.N.; Chen, D.; Wang, M.; Yuan, Y.; Wang, Y.; Lee, H.; Zhang, J.; Muller, W.J.; Liang, H.; et al. Ablation of miR-10b Suppresses Oncogene-Induced Mammary Tumorigenesis and Metastasis and Reactivates Tumor-Suppressive Pathways. Cancer Res. 2016, 76, 6424–6435. [Google Scholar] [CrossRef] [PubMed]
- Alimonti, A.; Carracedo, A.; Clohessy, J.G.; Trotman, L.C.; Nardella, C.; Egia, A.; Salmena, L.; Sampieri, K.; Haveman, W.J.; Brogi, E.; et al. Subtle variations in Pten dose determine cancer susceptibility. Nat. Genet. 2010, 42, 454–458. [Google Scholar] [CrossRef]
- Singh, R.; Pochampally, R.; Watabe, K.; Lu, Z.; Mo, Y.-Y. Exosome-mediated transfer of miR-10b promotes cell invasion in breast cancer. Mol. Cancer 2014, 13, 256. [Google Scholar] [CrossRef] [PubMed]
- Ibrahim, S.A.; Yip, G.W.; Stock, C.; Pan, J.-W.; Neubauer, C.; Poeter, M.; Pupjalis, D.; Koo, C.Y.; Kelsch, R.; Schüle, R.; et al. Targeting of syndecan-1 by microRNA miR-10b promotes breast cancer cell motility and invasiveness via a Rho-GTPase- and E-cadherin-dependent mechanism. Int. J. Cancer 2012, 131, E884–E896. [Google Scholar] [CrossRef]
- Xu, Y.-F.; Hannafon, B.N.; Khatri, U.; Gin, A.; Ding, W.-Q. The origin of exosomal miR-1246 in human cancer cells. RNA Biol. 2019, 16, 770–784. [Google Scholar] [CrossRef]
- Fu, L.; Li, Z.; Zhu, J.; Wang, P.; Fan, G.; Dai, Y.; Zheng, Z.; Liu, Y. Serum expression levels of microRNA-382-3p, -598-3p, -1246 and -184 in breast cancer patients. Oncol. Lett. 2016, 12, 269–274. [Google Scholar] [CrossRef] [PubMed]
- Li, X.J.; Ren, Z.J.; Tang, J.H.; Yu, Q. Exosomal MicroRNA MiR-1246 Promotes Cell Proliferation, Invasion and Drug Resistance by Targeting CCNG2 in Breast Cancer. Cell. Physiol. Biochem. 2017, 44, 1741–1748. [Google Scholar] [CrossRef]
- Eichelser, C.; Stückrath, I.; Müller, V.; Milde-Langosch, K.; Wikman, H.; Pantel, K.; Schwarzenbach, H. Increased serum levels of circulating exosomal microRNA-373 in receptor-negative breast cancer patients. Oncotarget 2014, 5, 9650–9663. [Google Scholar] [CrossRef] [PubMed]
- Huang, Q.; Gumireddy, K.; Schrier, M.; le Sage, C.; Nagel, R.; Nair, S.; Egan, D.A.; Li, A.; Huang, G.; Klein-Szanto, A.J.; et al. The microRNAs miR-373 and miR-520c promote tumour invasion and metastasis. Nat. Cell Biol. 2008, 10, 202–210. [Google Scholar] [CrossRef]
- Chen, W.; Cai, F.; Zhang, B.; Barekati, Z.; Zhong, X.Y. The level of circulating miRNA-10b and miRNA-373 in detecting lymph node metastasis of breast cancer: Potential biomarkers. Tumour Biol. 2013, 34, 455–462. [Google Scholar] [CrossRef]
- Li, H.; Bian, C.; Liao, L.; Li, J.; Zhao, R.C. miR-17-5p promotes human breast cancer cell migration and invasion through suppression of HBP1. Breast Cancer Res. Treat. 2010, 126, 565–575. [Google Scholar] [CrossRef]
- Lv, S.; Wang, Y.; Xu, W.; Dong, X. Serum Exosomal miR-17-5p as a Promising Biomarker Diagnostic Biomarker for Breast Cancer. Clin. Lab. 2020, 66. [Google Scholar] [CrossRef]
- Hong, Y.; Liang, H.; Rehman, U.-U.; Wang, Y.; Zhang, W.; Zhou, Y.; Chen, S.A.; Yu, M.; Cui, S.; Liu, M.; et al. miR-96 promotes cell proliferation, migration and invasion by targeting PTPN9 in breast cancer. Sci. Rep. 2016, 6. [Google Scholar] [CrossRef]
- Li, N.; Liu, Y.; Miao, Y.; Zhao, L.; Zhou, H.; Jia, L. MicroRNA-106b targets FUT6 to promote cell migration, invasion, and proliferation in human breast cancer. IUBMB Life 2016, 68, 764–775. [Google Scholar] [CrossRef] [PubMed]
- Ahmad, A.; Sethi, S.; Chen, W.; Ali-Fehmi, R.; Mittal, S.; Sarkar, F.H. Up-regulation of microRNA-10b is associated with the development of breast cancer brain metastasis. Am. J. Transl. Res. 2014, 6, 384–390. [Google Scholar] [PubMed]
- Barkan, D.; Kleinman, H.; Simmons, J.L.; Asmussen, H.; Kamaraju, A.K.; Hoenorhoff, M.J.; Liu, Z.-Y.; Costes, S.V.; Cho, E.H.; Lockett, S.; et al. Inhibition of Metastatic Outgrowth from Single Dormant Tumor Cells by Targeting the Cytoskeleton. Cancer Res. 2008, 68, 6241–6250. [Google Scholar] [CrossRef] [PubMed]
- Rana, S.; Malinowska, K.; Zöller, M. Exosomal tumor microRNA modulates premetastatic organ cells. Neoplasia 2013, 15, 281–295. [Google Scholar] [CrossRef] [PubMed]
- Ma, L.; Teruya-Feldstein, J.; Weinberg, R.A. Tumour invasion and metastasis initiated by microRNA-10b in breast cancer. Nature 2007, 449, 682–688. [Google Scholar] [CrossRef]
- Ma, L. Role of miR-10b in breast cancer metastasis. Breast Cancer Res. 2010, 12, 210. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Zhao, J.; Zhang, P.-Y.; Zhang, Y.; Sun, S.-Y.; Yu, S.-Y.; Xi, Q.-S. MicroRNA-10b targets E-cadherin and modulates breast cancer metastasis. Med. Sci. Monit. 2012, 18, BR299–BR308. [Google Scholar] [CrossRef]
- Ma, L.; Reinhardt, F.; Pan, E.; Soutschek, J.; Bhat, B.; Marcusson, E.G.; Teruya-Feldstein, J.; Bell, G.W.; Weinberg, R.A. Therapeutic silencing of miR-10b inhibits metastasis in a mouse mammary tumor model. Nat. Biotechnol. 2010, 28, 341–347. [Google Scholar] [CrossRef]
- Knirsh, R.; Ben-Dror, I.; Modai, S.; Shomron, N.; Vardimon, L. MicroRNA 10b promotes abnormal expression of the proto-oncogene c-Jun in metastatic breast cancer cells. Oncotarget 2016, 7, 59932–59944. [Google Scholar] [CrossRef][Green Version]
- Xing, F.; Liu, Y.; Wu, S.-Y.; Wu, K.; Sharma, S.; Mo, Y.-Y.; Feng, J.; Sanders, S.; Jin, G.; Singh, R.; et al. Loss of XIST in Breast Cancer Activates MSN-c-Met and Reprograms Microglia via Exosomal miRNA to Promote Brain Metastasis. Cancer Res. 2018, 78, 4316–4330. [Google Scholar] [CrossRef]
- Hanahan, D.; Weinberg, R.A. Hallmarks of cancer: The next generation. Cell 2011, 144, 646–674. [Google Scholar] [CrossRef]
- Fong, M.Y.; Zhou, W.; Liu, L.; Alontaga, A.Y.; Chandra, M.; Ashby, J.; Chow, A.; O’Connor, S.T.; Li, S.; Chin, A.R.; et al. Breast-cancer-secreted miR-122 reprograms glucose metabolism in premetastatic niche to promote metastasis. Nat. Cell Biol. 2015, 17, 183–194. [Google Scholar] [CrossRef]
- Korpal, M.; Ell, B.J.; Buffa, F.M.; Ibrahim, T.; Blanco, M.A.; Celià-Terrassa, T.; Mercatali, L.; Khan, Z.; Goodarzi, H.; Hua, Y.; et al. Direct targeting of Sec23a by miR-200s influences cancer cell secretome and promotes metastatic colonization. Nat. Med. 2011, 17, 1101–1108. [Google Scholar] [CrossRef]
- Yu, S.J.; Hu, J.Y.; Kuang, X.Y.; Luo, J.M.; Hou, Y.F.; Di, G.H.; Wu, J.; Shen, Z.Z.; Song, H.Y.; Shao, Z.M. MicroRNA-200a promotes anoikis resistance and metastasis by targeting YAP1 in human breast cancer. Clin. Cancer Res. 2013, 19, 1389–1399. [Google Scholar] [CrossRef] [PubMed]
- Yan, L.X.; Huang, X.; Shao, Q.; Huang, M.-Y.; Deng, L.; Wu, Q.-L.; Zeng, Y.-X.; Shao, J.-Y. MicroRNA miR-21 overexpression in human breast cancer is associated with advanced clinical stage, lymph node metastasis and patient poor prognosis. RNA 2008, 14, 2348–2360. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Tan, Z.; Hu, H.; Liu, H.; Wu, T.; Zheng, C.; Wang, X.; Luo, Z.; Wang, J.; Liu, S.; et al. microRNA-21 promotes breast cancer proliferation and metastasis by targeting LZTFL1. BMC Cancer 2019, 19, 738. [Google Scholar] [CrossRef] [PubMed]
- Adeyinka, A.; Nui, Y.; Cherlet, T.; Snell, L.; Watson, P.H.; Murphy, L.C. Activated mitogen-activated protein kinase expression during human breast tumorigenesis and breast cancer progression. Clin. Cancer Res. 2002, 8, 1747–1753. [Google Scholar]
- Mutlu, M.; Saatci, Ö.; Ansari, S.A.; Yurdusev, E.; Shehwana, H.; Konu, Ö.; Raza, U.; Şahin, Ö. miR-564 acts as a dual inhibitor of PI3K and MAPK signaling networks and inhibits proliferation and invasion in breast cancer. Sci. Rep. 2016, 6. [Google Scholar] [CrossRef]
- Ke, K.; Lou, T. MicroRNA-10a suppresses breast cancer progression via PI3K/Akt/mTOR pathway. Oncol. Lett. 2017, 14. [Google Scholar] [CrossRef]
- Tao, W.-Y.; Wang, C.-Y.; Sun, Y.-H.; Su, Y.-H.; Pang, D.; Zhang, G.-Q. MicroRNA-34c Suppresses Breast Cancer Migration and Invasion by Targeting GIT1. J. Cancer 2016, 7, 1653–1662. [Google Scholar] [CrossRef] [PubMed]
- Zhao, W.G.; Yu, S.N.; Lu, Z.H.; Ma, Y.H.; Gu, Y.M.; Chen, J. The miR-217 microRNA functions as a potential tumor suppressor in pancreatic ductal adenocarcinoma by targeting KRAS. Carcinogenesis 2010, 31, 1726–1733. [Google Scholar] [CrossRef] [PubMed]
- Su, J.; Wang, Q.; Liu, Y.; Zhong, M. miR-217 inhibits invasion of hepatocellular carcinoma cells through direct suppression of E2F3. Mol. Cell. Biochem. 2014, 392, 289–296. [Google Scholar] [CrossRef]
- Wang, X.; Li, M.; Wang, Z.; Han, S.; Tang, X.; Ge, Y.; Zhou, L.; Zhou, C.; Yuan, Q.; Yang, M. Silencing of long noncoding RNA MALAT1 by miR-101 and miR-217 inhibits proliferation, migration, and invasion of esophageal squamous cell carcinoma cells. J. Biol. Chem. 2015, 290, 3925–3935. [Google Scholar] [CrossRef]
- Zhou, W.; Song, F.; Wu, Q.; Liu, R.; Wang, L.; Liu, C.; Peng, Y.; Mao, S.; Feng, J.; Chen, C. miR-217 inhibits triple-negative breast cancer cell growth, migration, and invasion through targeting KLF5. PLoS ONE 2017, 12, e0176395. [Google Scholar] [CrossRef] [PubMed]
- MacDonald, B.T.; Tamai, K.; He, X. Wnt/beta-catenin signaling: Components, mechanisms, and diseases. Dev. Cell 2009, 17, 9–26. [Google Scholar] [CrossRef] [PubMed]
- Jiang, Q.; He, M.; Guan, S.; Ma, M.; Wu, H.; Yu, Z.; Jiang, L.; Wang, Y.; Zong, X.; Jin, F.; et al. MicroRNA-100 suppresses the migration and invasion of breast cancer cells by targeting FZD-8 and inhibiting Wnt/beta-catenin signaling pathway. Tumour Biol. 2016, 37, 5001–5011. [Google Scholar] [CrossRef] [PubMed]
- Park, E.J.; Jung, H.J.; Choi, H.J.; Jang, H.J.; Park, H.J.; Nejsum, L.N.; Kwon, T.H. Exosomes co-expressing AQP5-targeting miRNAs and IL-4 receptor-binding peptide inhibit the migration of human breast cancer cells. FASEB J. 2020, 34, 3379–3398. [Google Scholar] [CrossRef]
- Jin, C.; Rajabi, H.; Kufe, D. miR-1226 targets expression of the mucin 1 oncoprotein and induces cell death. Int. J. Oncol. 2010, 37, 61–69. [Google Scholar] [CrossRef]
- Shah, S.P.; Roth, A.; Goya, R.; Oloumi, A.; Ha, G.; Zhao, Y.; Turashvili, G.; Ding, J.; Tse, K.; Haffari, G.; et al. The clonal and mutational evolution spectrum of primary triple-negative breast cancers. Nature 2012, 486, 395–399. [Google Scholar] [CrossRef]
- Yuan, L.; Liu, Y.; Qu, Y.; Liu, L.; Li, H. Exosomes Derived From MicroRNA-148b-3p-Overexpressing Human Umbilical Cord Mesenchymal Stem Cells Restrain Breast Cancer Progression. Front. Oncol. 2019, 9, 1076. [Google Scholar] [CrossRef] [PubMed]
- Jiang, Q.; He, M.; Ma, M.T.; Wu, H.Z.; Yu, Z.J.; Guan, S.; Jiang, L.Y.; Wang, Y.; Zheng, D.D.; Jin, F.; et al. MicroRNA-148a inhibits breast cancer migration and invasion by directly targeting WNT-1. Oncol. Rep. 2015, 35, 1425–1432. [Google Scholar] [CrossRef] [PubMed]
- Li, D.; Wang, J.; Ma, L.J.; Yang, H.B.; Jing, J.F.; Jia, M.M.; Zhang, X.J.; Guo, F.; Gao, J.N. Identification of serum exosomal miR-148a as a novel prognostic biomarker for breast cancer. Eur. Rev. Med Pharmacol. Sci. 2020, 24, 7303–7309. [Google Scholar] [CrossRef] [PubMed]
- Bovy, N.; Blomme, B.; Frères, P.; Dederen, S.; Nivelles, O.; Lion, M.; Carnet, O.; Martial, J.A.; Noël, A.; Thiry, M.; et al. Endothelial exosomes contribute to the antitumor response during breast cancer neoadjuvant chemotherapy via microRNA transfer. Oncotarget 2015, 6, 10253–10266. [Google Scholar] [CrossRef] [PubMed]
- Yu, Z.; Willmarth, N.; Zhou, J.; Katiyar, S.; Wang, M.; Liu, Y.; McCue, P.A.; Quong, A.A.; Lisanti, M.P.; Pestell, R.G. microRNA 17/20 inhibits cellular invasion and tumor metastasis in breast cancer by heterotypic signaling. Proc. Natl. Acad. Sci. USA 2010, 107, 8231–8236. [Google Scholar] [CrossRef]
- Ye, Z.-B.; Ma, G.; Zhao, Y.-H.; Xiao, Y.; Zhan, Y.; Jing, C.; Gao, K.A.I.; Liu, Z.-H.; Yu, S.-J. miR-429 inhibits migration and invasion of breast cancer cells in vitro. Int. J. Oncol. 2015, 46, 531–538. [Google Scholar] [CrossRef]
- Zhang, X.; Yu, X.; Zhao, Z.; Yuan, Z.; Ma, P.; Ye, Z.; Guo, L.; Xu, S.; Xu, L.; Liu, T.; et al. MicroRNA-429 inhibits bone metastasis in breast cancer by regulating CrkL and MMP-9. Bone 2020, 130, 115139. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Luo, J.; Wang, B.; Wang, D.; Xie, X.; Yuan, L.; Guo, J.; Xi, S.; Gao, J.; Lin, X.; et al. Microrna-124 targets flotillin-1 to regulate proliferation and migration in breast cancer. Mol. Cancer 2013, 12, 163. [Google Scholar] [CrossRef]
- Zhang, L.; Chen, X.; Liu, B.; Han, J. MicroRNA-124-3p directly targets PDCD6 to inhibit metastasis in breast cancer. Oncol. Lett. 2017, 15, 984–990. [Google Scholar] [CrossRef] [PubMed]
- Calin, G.A.; Croce, C.M. MicroRNA signatures in human cancers. Nat. Rev. Cancer 2006, 6, 857–866. [Google Scholar] [CrossRef]
- Valastyan, S.; Reinhardt, F.; Benaich, N.; Calogrias, D.; Szász, A.M.; Wang, Z.C.; Brock, J.E.; Richardson, A.L.; Weinberg, R.A. A pleiotropically acting microRNA, miR-31, inhibits breast cancer metastasis. Cell 2009, 137, 1032–1046. [Google Scholar] [CrossRef]
- Lv, X.B.; Jiao, Y.; Qing, Y.; Hu, H.; Cui, X.; Lin, T.; Song, E.; Yu, F. miR-124 suppresses multiple steps of breast cancer metastasis by targeting a cohort of pro-metastatic genes in vitro. Chin. J. Cancer 2011, 30, 821–830. [Google Scholar] [CrossRef] [PubMed]
- Cai, W.L.; Huang, W.D.; Li, B.; Chen, T.R.; Li, Z.X.; Zhao, C.L.; Li, H.Y.; Wu, Y.M.; Yan, W.A.-O.; Xiao, J.R. microRNA-124 inhibits bone metastasis of breast cancer by repressing Interleukin-11. Mol. Cancer 2018, 17, 9. [Google Scholar] [CrossRef] [PubMed]
- Liu, T.; Hu, K.; Zhao, Z.; Chen, G.; Ou, X.; Zhang, H.; Zhang, X.; Wei, X.; Wang, D.; Cui, M.; et al. MicroRNA-1 down-regulates proliferation and migration of breast cancer stem cells by inhibiting the Wnt/β-catenin pathway. Oncotarget 2015, 6, 41638–41649. [Google Scholar] [CrossRef]
- Peng, J.; Yuan, C.; Wu, Z.; Wang, Y.; Yin, W.; Lin, Y.; Zhou, L.; Lu, J. Upregulation of microRNA-1 inhibits proliferation and metastasis of breast cancer. Mol. Med. Rep. 2020, 22, 454–464. [Google Scholar] [CrossRef]
- Kwon, J.E.; Kim, B.; Kwak, S.-Y.; Bae, I.-H.; Han, Y.H. Ionizing radiation-inducible microRNA miR-193a-3p induces apoptosis by directly targeting Mcl-1. Apoptosis 2013, 18, 896–909. [Google Scholar] [CrossRef]
- Xie, F.; Hosany, S.; Zhong, S.; Jiang, Y.; Zhang, F.; Lin, L.; Wang, X.; Gao, S.; Hu, X. MicroRNA-193a inhibits breast cancer proliferation and metastasis by downregulating WT1. PLoS ONE 2017, 12, e0185565. [Google Scholar] [CrossRef]
- Li, L.Z.; Zhang, C.Z.; Liu, L.-L.; Yi, C.; Lu, S.-X.; Zhou, X.; Zhang, Z.-J.; Peng, Y.-H.; Yang, Y.-Z.; Yun, J.-P. miR-720 inhibits tumor invasion and migration in breast cancer by targeting TWIST1. Carcinogenesis 2013, 35, 469–478. [Google Scholar] [CrossRef]
- Yang, J.; Mani, S.A.; Donaher, J.L.; Ramaswamy, S.; Itzykson, R.A.; Come, C.; Savagner, P.; Gitelman, I.; Richardson, A.; Weinberg, R.A. Twist, a master regulator of morphogenesis, plays an essential role in tumor metastasis. Cell 2004, 117, 927–939. [Google Scholar] [CrossRef] [PubMed]
- Vlashi, E.; Pajonk, F. Cancer stem cells, cancer cell plasticity and radiation therapy. Semin. Cancer Biol. 2015, 31, 28–35. [Google Scholar] [CrossRef] [PubMed]
- Beck, B.; Blanpain, C. Unravelling cancer stem cell potential. Nat. Rev. Cancer 2013, 13, 727–738. [Google Scholar] [CrossRef]
- Hu, J.L.; Wang, W.; Lan, X.L.; Zeng, Z.C.; Liang, Y.S.; Yan, Y.R.; Song, F.Y.; Wang, F.F.; Zhu, X.H.; Liao, W.J.; et al. CAFs secreted exosomes promote metastasis and chemotherapy resistance by enhancing cell stemness and epithelial-mesenchymal transition in colorectal cancer. Mol. Cancer 2019, 18, 91. [Google Scholar] [CrossRef] [PubMed]
- Li, B.; Lu, Y.; Yu, L.; Han, X.; Wang, H.; Mao, J.; Shen, J.; Wang, B.; Tang, J.; Li, C.; et al. miR-221/222 promote cancer stem-like cell properties and tumor growth of breast cancer via targeting PTEN and sustained Akt/NF-κB/COX-2 activation. Chem. Biol. Interact. 2017, 277, 33–42. [Google Scholar] [CrossRef] [PubMed]
- Donnarumma, E.; Fiore, D.; Nappa, M.; Roscigno, G.; Adamo, A.; Iaboni, M.; Russo, V.; Affinito, A.; Puoti, I.; Quintavalle, C.; et al. Cancer-associated fibroblasts release exosomal microRNAs that dictate an aggressive phenotype in breast cancer. Oncotarget 2017, 8, 19592–19608. [Google Scholar] [CrossRef]
- Chaffer, C.L.; Weinberg, R.A. A perspective on cancer cell metastasis. Science 2011, 331, 1559–1564. [Google Scholar] [CrossRef] [PubMed]
- Song, S.J.; Poliseno, L.; Song, M.S.; Ala, U.; Webster, K.; Ng, C.; Beringer, G.; Brikbak, N.J.; Yuan, X.; Cantley, L.C.; et al. MicroRNA-antagonism regulates breast cancer stemness and metastasis via TET-family-dependent chromatin remodeling. Cell 2013, 154, 311–324. [Google Scholar] [CrossRef] [PubMed]
- Park, E.Y.; Chang, E.; Lee, E.J.; Lee, H.-W.; Kang, H.-G.; Chun, K.-H.; Woo, Y.M.; Kong, H.K.; Ko, J.Y.; Suzuki, H.; et al. Targeting of miR34a–NOTCH1 Axis Reduced Breast Cancer Stemness and Chemoresistance. Cancer Res. 2014, 74, 7573–7582. [Google Scholar] [CrossRef] [PubMed]
- Guo, W.; Keckesova, Z.; Donaher, J.L.; Shibue, T.; Tischler, V.; Reinhardt, F.; Itzkovitz, S.; Noske, A.; Zürrer-Härdi, U.; Bell, G.; et al. Slug and Sox9 cooperatively determine the mammary stem cell state. Cell 2012, 148, 1015–1028. [Google Scholar] [CrossRef]
- Wolfson, B.; Eades, G.; Zhou, Q. Roles of microRNA-140 in stem cell-associated early stage breast cancer. World J. Stem Cells 2014, 6. [Google Scholar] [CrossRef] [PubMed]
- Viallard, C.; Larrivée, B. Tumor angiogenesis and vascular normalization: Alternative therapeutic targets. Angiogenesis 2017, 20, 409–426. [Google Scholar] [CrossRef]
- Kong, W.; He, L.; Richards, E.J.; Challa, S.; Xu, C.X.; Permuth-Wey, J.; Lancaster, J.M.; Coppola, D.; Sellers, T.A.; Djeu, J.Y.; et al. Upregulation of miRNA-155 promotes tumour angiogenesis by targeting VHL and is associated with poor prognosis and triple-negative breast cancer. Oncogene 2013, 33, 679–689. [Google Scholar] [CrossRef]
- Kontomanolis, E.; Mitrakas, A.; Giatromanolaki, A.; Kareli, D.; Panteliadou, M.; Pouliliou, S.; Koukourakis, M.I. A pilot study on plasma levels of micro-RNAs involved in angiogenesis and vascular maturation in patients with breast cancer. Med. Oncol. 2017, 34, 20. [Google Scholar] [CrossRef]
- Lee, J.K.; Park, S.R.; Jung, B.K.; Jeon, Y.K.; Lee, Y.S.; Kim, M.K.; Kim, Y.G.; Jang, J.Y.; Kim, C.W. Exosomes derived from mesenchymal stem cells suppress angiogenesis by down-regulating VEGF expression in breast cancer cells. PLoS ONE 2013, 8, e84256. [Google Scholar] [CrossRef]
- Zhou, B.; Ma, R.; Si, W.; Li, S.; Xu, Y.; Tu, X.; Wang, Q. MicroRNA-503 targets FGF2 and VEGFA and inhibits tumor angiogenesis and growth. Cancer Lett. 2013, 333, 159–169. [Google Scholar] [CrossRef]
- Pakravan, K.; Babashah, S.; Sadeghizadeh, M.; Mowla, S.J.; Mossahebi-Mohammadi, M.; Ataei, F.; Dana, N.; Javan, M. MicroRNA-100 shuttled by mesenchymal stem cell-derived exosomes suppresses in vitro angiogenesis through modulating the mTOR/HIF-1alpha/VEGF signaling axis in breast cancer cells. Cell Oncol. 2017, 40, 457–470. [Google Scholar] [CrossRef]
- Pasquier, J.; Magal, P.; Boulangé-Lecomte, C.; Webb, G.; Le Foll, F. Consequences of cell-to-cell P-glycoprotein transfer on acquired multidrug resistance in breast cancer: A cell population dynamics model. Biol. Direct 2011, 6, 5. [Google Scholar] [CrossRef]
- Chen, Z.; Pan, T.; Jiang, D.; Jin, L.; Geng, Y.; Feng, X.; Shen, A.; Zhang, L. The lncRNA-GAS5/miR-221-3p/DKK2 Axis Modulates ABCB1-Mediated Adriamycin Resistance of Breast Cancer via the Wnt/β-Catenin Signaling Pathway. Mol. Ther. Nucleic Acids 2020, 19, 1434–1448. [Google Scholar] [CrossRef] [PubMed]
- Pan, X.; Hong, X.; Lai, J.; Cheng, L.; Cheng, Y.; Yao, M.; Wang, R.; Hu, N. Exosomal MicroRNA-221-3p Confers Adriamycin Resistance in Breast Cancer Cells by Targeting PIK3R1. Front. Oncol. 2020, 10, 441. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Wang, N.; Liu, P.; Chen, Q.; Situ, H.; Xie, T.; Zhang, J.; Peng, C.; Lin, Y.; Chen, J. MicroRNA-25 regulates chemoresistance-associated autophagy in breast cancer cells, a process modulated by the natural autophagy inducer isoliquiritigenin. Oncotarget 2014, 5, 7013–7026. [Google Scholar] [CrossRef]
- Yamamoto, Y.; Yoshioka, Y.; Minoura, K.; Takahashi, R.-U.; Takeshita, F.; Taya, T.; Horii, R.; Fukuoka, Y.; Kato, T.; Kosaka, N.; et al. An integrative genomic analysis revealed the relevance of microRNA and gene expression for drug-resistance in human breast cancer cells. Mol. Cancer 2011, 10, 135. [Google Scholar] [CrossRef] [PubMed]
- Hu, W.; Tan, C.; He, Y.; Zhang, G.; Xu, Y.; Tang, J. Functional miRNAs in breast cancer drug resistance. OncoTargets Ther. 2018, 11, 1529–1541. [Google Scholar] [CrossRef]
- Wu, M.Y.; Fu, J.; Xiao, X.; Wu, J.; Wu, R.C. MiR-34a regulates therapy resistance by targeting HDAC1 and HDAC7 in breast cancer. Cancer Lett. 2014, 354, 311–319. [Google Scholar] [CrossRef]
- Zhu, Y.; Wu, J.; Li, S.; Ma, R.; Cao, H.; Ji, M.; Jing, C.; Tang, J. The function role of miR-181a in chemosensitivity to adriamycin by targeting Bcl-2 in low-invasive breast cancer cells. Cell. Physiol. Biochem. 2013, 32, 1225–1237. [Google Scholar] [CrossRef] [PubMed]
- Clarke, R.; Skaar, T.C.; Bouker, K.B.; Davis, N.; Lee, Y.; Welch, J.N.; Leonessa, F. Molecular and pharmacological aspects of antiestrogen resistance. J. Steroid Biochem. Mol. Biol. 2001, 76, 71–84. [Google Scholar] [CrossRef]
- Deng, Z.; Rong, Y.; Teng, Y.; Zhuang, X.; Samykutty, A.; Mu, J.; Zhang, L.; Cao, P.; Yan, J.; Miller, D.; et al. Exosomes miR-126a released from MDSC induced by DOX treatment promotes lung metastasis. Oncogene 2017, 36, 639–651. [Google Scholar] [CrossRef] [PubMed]
- Pan, Y.Z.; Morris, M.E.; Yu, A.M. MicroRNA-328 negatively regulates the expression of breast cancer resistance protein (BCRP/ABCG2) in human cancer cells. Mol. Pharmacol. 2009, 75, 1374–1379. [Google Scholar] [CrossRef]
- Pogribny, I.P.; Filkowski, J.N.; Tryndyak, V.P.; Golubov, A.; Shpyleva, S.I.; Kovalchuk, O. Alterations of microRNAs and their targets are associated with acquired resistance of MCF-7 breast cancer cells to cisplatin. Int. J. Cancer 2010, 127, 1785–1794. [Google Scholar] [CrossRef] [PubMed]
- Cataldo, A.; Cheung, D.G.; Balsari, A.; Tagliabue, E.; Coppola, V.; Iorio, M.V.; Palmieri, D.; Croce, C.M. miR-302b enhances breast cancer cell sensitivity to cisplatin by regulating E2F1 and the cellular DNA damage response. Oncotarget 2016, 7, 786–797. [Google Scholar] [CrossRef]
- Bockhorn, J.; Dalton, R.; Nwachukwu, C.; Huang, S.; Prat, A.; Yee, K.; Chang, Y.-F.; Huo, D.; Wen, Y.; Swanson, K.E.; et al. MicroRNA-30c inhibits human breast tumour chemotherapy resistance by regulating TWF1 and IL-11. Nat. Commun. 2013, 4, 1393. [Google Scholar] [CrossRef]
- Kutanzi, K.R.; Yurchenko, O.V.; Beland, F.A.; Checkhun, V.F.; Pogribny, I.P. MicroRNA-mediated drug resistance in breast cancer. Clin. Epigenetics 2011, 2, 171–185. [Google Scholar] [CrossRef]
- Yang, Q.; Wang, Y.; Lu, X.; Zhao, Z.; Zhu, L.; Chen, S.; Wu, Q.; Chen, C.; Wang, Z. MiR-125b regulates epithelial-mesenchymal transition via targeting Sema4C in paclitaxel-resistant breast cancer cells. Oncotarget 2015, 6, 3268–3279. [Google Scholar] [CrossRef]
- Cochrane, D.R.; Spoelstra, N.S.; Howe, E.N.; Nordeen, S.K.; Richer, J.K. MicroRNA-200c mitigates invasiveness and restores sensitivity to microtubule-targeting chemotherapeutic agents. Mol. Cancer Ther. 2009, 8, 1055–1066. [Google Scholar] [CrossRef]
- Kastl, L.; Brown, I.; Schofield, A.C. miRNA-34a is associated with docetaxel resistance in human breast cancer cells. Breast Cancer Res. Treat. 2011, 131, 445–454. [Google Scholar] [CrossRef] [PubMed]
- Shi, W.; Gerster, K.; Alajez, N.M.; Tsang, J.; Waldron, L.; Pintilie, M.; Hui, A.B.; Sykes, J.; P’ng, C.; Miller, N.; et al. MicroRNA-301 mediates proliferation and invasion in human breast cancer. Cancer Res. 2011, 71, 2926–2937. [Google Scholar] [CrossRef] [PubMed]
- Sachdeva, M.; Wu, H.; Ru, P.; Hwang, L.; Trieu, V.; Mo, Y.Y. MicroRNA-101-mediated Akt activation and estrogen-independent growth. Oncogene 2010, 30, 822–831. [Google Scholar] [CrossRef] [PubMed]
- Lü, M.; Ding, K.; Zhang, G.; Yin, M.; Yao, G.; Tian, H.; Lian, J.; Liu, L.; Liang, M.; Zhu, T.; et al. MicroRNA-320a sensitizes tamoxifen-resistant breast cancer cells to tamoxifen by targeting ARPP-19 and ERRγ. Sci. Rep. 2015, 5, 8735. [Google Scholar] [CrossRef]
- Yu, X.; Luo, A.; Liu, Y.; Wang, S.; Li, Y.; Shi, W.; Liu, Z.; Qu, X. MiR-214 increases the sensitivity of breast cancer cells to tamoxifen and fulvestrant through inhibition of autophagy. Mol. Cancer 2015, 14, 208. [Google Scholar] [CrossRef]
- Liu, Z.R.; Song, Y.; Wan, L.H.; Zhang, Y.Y.; Zhou, L.M. Over-expression of miR-451a can enhance the sensitivity of breast cancer cells to tamoxifen by regulating 14-3-3ζ, estrogen receptor α, and autophagy. Life Sci. 2016, 149, 104–113. [Google Scholar] [CrossRef]
- Rao, X.; Di Leva, G.; Li, M.; Fang, F.; Devlin, C.; Hartman-Frey, C.; Burow, M.E.; Ivan, M.; Croce, C.M.; Nephew, K.P. MicroRNA-221/222 confers breast cancer fulvestrant resistance by regulating multiple signaling pathways. Oncogene 2010, 30, 1082–1097. [Google Scholar] [CrossRef]
- De Cola, A.; Volpe, S.; Budani, M.C.; Ferracin, M.A.-O.; Lattanzio, R.; Turdo, A.; D’Agostino, D.; Capone, E.; Stassi, G.; Todaro, M.; et al. miR-205-5p-mediated downregulation of ErbB/HER receptors in breast cancer stem cells results in targeted therapy resistance. Cell Death Dis. 2015, 6, e1823. [Google Scholar] [CrossRef]
- Venturutti, L.; Cordo Russo, R.I.; Rivas, M.A.; Mercogliano, M.F.; Izzo, F.; Oakley, R.H.; Pereyra, M.G.; De Martino, M.; Proietti, C.J.; Yankilevich, P.; et al. MiR-16 mediates trastuzumab and lapatinib response in ErbB-2-positive breast and gastric cancer via its novel targets CCNJ and FUBP1. Oncogene 2016, 35, 6189–6202. [Google Scholar] [CrossRef]
- Guo, L.; Cheng, X.; Chen, H.; Chen, C.; Xie, S.; Zhao, M.; Liu, D.; Deng, Q.; Liu, Y.; Wang, X.; et al. Induction of breast cancer stem cells by M1 macrophages through Lin-28B-let-7-HMGA2 axis. Cancer Lett. 2019, 452, 213–225. [Google Scholar] [CrossRef] [PubMed]
- Van der Zanden, S.Y.; Qiao, X.; Neefjes, J. New insights into the activities and toxicities of the old anti-cancer drug doxorubicin. FEBS J. 2020. [Google Scholar] [CrossRef] [PubMed]
- Chen, W.X.; Xu, L.Y.; Qian, Q.; He, X.; Peng, W.T.; Zhu, Y.L.; Cheng, L. Analysis of miRNA signature differentially expressed in exosomes from adriamycin-resistant and parental human breast cancer cells. Biosci. Rep. 2018, 38, BSR20181090. [Google Scholar] [CrossRef]
- Soni, M.; Patel, Y.; Markoutsa, E.; Jie, C.; Liu, S.; Xu, P.; Chen, H. Autophagy, Cell Viability, and Chemoresistance Are Regulated By miR-489 in Breast Cancer. Mol. Cancer Res. 2018, 16, 1348–1360. [Google Scholar] [CrossRef]
- Evison, B.J.; Sleebs, B.E.; Watson, K.G.; Phillips, D.R.; Cutts, S.M. Mitoxantrone, More than Just Another Topoisomerase II Poison. Med. Res. Rev. 2015, 36, 248–299. [Google Scholar] [CrossRef] [PubMed]
- Dickson, M.A.; Carvajal, R.D.; Merrill, A.H., Jr.; Gonen, M.; Cane, L.M.; Schwartz, G.K. A phase I clinical trial of safingol in combination with cisplatin in advanced solid tumors. Clin. Cancer Res. 2011, 17, 2484–2492. [Google Scholar] [CrossRef]
- Willson, M.L.; Burke, L.; Ferguson, T.; Ghersi, D.; Nowak, A.K.; Wilcken, N. Taxanes for adjuvant treatment of early breast cancer. Cochrane Database Syst. Rev. 2019, 9, CD004421. [Google Scholar] [CrossRef]
- Shang, Y.; Cai, X.; Fan, D. Roles of epithelial-mesenchymal transition in cancer drug resistance. Curr. Cancer Drug Targets 2013, 13, 915–929. [Google Scholar] [CrossRef]
- Cui, J.; Yang, Y.; Li, H.; Leng, Y.; Qian, K.; Huang, Q.; Zhang, C.; Lu, Z.; Chen, J.; Sun, T.; et al. MiR-873 regulates ERα transcriptional activity and tamoxifen resistance via targeting CDK3 in breast cancer cells. Oncogene 2015, 34, 4018. [Google Scholar] [CrossRef]
- Nagata, Y.; Lan, K.; Zhou, X.; Tan, M.; Esteva, F.J.; Sahin, A.A.; Klos, K.S.; Li, P.; Monia, B.P.; Nguyen, N.T.; et al. PTEN activation contributes to tumor inhibition by trastuzumab, and loss of PTEN predicts trastuzumab resistance in patients. Cancer Cell 2004, 6, 117–127. [Google Scholar] [CrossRef]
- Nicholson, R.I.; Johnston, S.R. Endocrine therapy—Current benefits and limitations. Breast Cancer Res. Treat. 2005, 93, 3–10. [Google Scholar] [CrossRef]
- Normanno, N.; Di Maio, M.; De Maio, E.; De Luca, A.; de Matteis, A.; Giordano, A.; Perrone, F. Mechanisms of endocrine resistance and novel therapeutic strategies in breast cancer. Endocr. Relat. Cancer 2005, 12, 721–747. [Google Scholar] [CrossRef]
- Nemeth, B.T.; Varga, Z.V.; Wu, W.J.; Pacher, P. Trastuzumab cardiotoxicity: From clinical trials to experimental studies. Br. J. Pharmacol. 2016, 174, 3727–3748. [Google Scholar] [CrossRef]
- Park, J.W.; Hwang, S.R.; Yoon, I.S. Advanced Growth Factor Delivery Systems in Wound Management and Skin Regeneration. Molecules 2017, 22, 1259. [Google Scholar] [CrossRef]
- Wanek, T.; Kuntner, C.; Bankstahl, J.P.; Bankstahl, M.; Stanek, J.; Sauberer, M.; Mairinger, S.; Strommer, S.; Wacheck, V.; Löscher, W.; et al. A comparative small-animal PET evaluation of [11C]tariquidar, [11C]elacridar and (R)-[11C]verapamil for detection of P-glycoprotein-expressing murine breast cancer. Eur. J. Nucl. Med. Mol. Imaging 2011, 39, 149–159. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Chen, W.X.; Hu, Q.; Qiu, M.T.; Zhong, S.L.; Xu, J.J.; Tang, J.H.; Zhao, J.H. miR-221/222: Promising biomarkers for breast cancer. Tumour Biol. 2013, 34, 1361–1370. [Google Scholar] [CrossRef] [PubMed]
- Cuk, K.; Zucknick, M.; Madhavan, D.; Schott, S.; Golatta, M.; Heil, J.; Marmé, F.; Turchinovich, A.; Sinn, P.; Sohn, C.; et al. Plasma microRNA panel for minimally invasive detection of breast cancer. PLoS ONE 2013, 8, e76729. [Google Scholar] [CrossRef]
- Gao, J.; Zhang, Q.; Xu, J.; Guo, L.; Li, X. Clinical significance of serum miR-21 in breast cancer compared with CA153 and CEA. Chin. J. Cancer Res. 2013, 25, 743–748. [Google Scholar] [CrossRef] [PubMed]
- Shaker, O.; Maher, M.; Nassar, Y.; Morcos, G.; Gad, Z. Role of microRNAs -29b-2, -155, -197 and -205 as diagnostic biomarkers in serum of breast cancer females. Gene 2015, 560, 77–82. [Google Scholar] [CrossRef]
- He, Y.; Deng, F.; Yang, S.; Wang, D.; Chen, X.; Zhong, S.; Zhao, J.; Tang, J. Exosomal microRNA: A novel biomarker for breast cancer. Biomark. Med. 2018, 12, 177–188. [Google Scholar] [CrossRef] [PubMed]
- Stevic, I.; Müller, V.; Weber, K.; Fasching, P.A.; Karn, T.; Marmé, F.; Schem, C.; Stickeler, E.; Denkert, C.; van Mackelenbergh, M.; et al. Specific microRNA signatures in exosomes of triple-negative and HER2-positive breast cancer patients undergoing neoadjuvant therapy within the GeparSixto trial. BMC Med. 2018, 16, 179. [Google Scholar] [CrossRef] [PubMed]
- Rodríguez-Martínez, A.; de Miguel-Pérez, D.; Ortega, F.G.; García-Puche, J.L.; Robles-Fernández, I.; Exposito, J.; Martorell-Marugan, J.; Carmona-Sáez, P.; Garrido-Navas, M.D.C.; Rolfo, C.; et al. Exosomal miRNA profile as complementary tool in the diagnostic and prediction of treatment response in localized breast cancer under neoadjuvant chemotherapy. Breast Cancer Res. 2019, 21, 21. [Google Scholar] [CrossRef]
- Piasecka, D.; Braun, M.; Kordek, R.; Sadej, R.; Romanska, H. MicroRNAs in regulation of triple-negative breast cancer progression. J. Cancer Res. Clin. Oncol. 2018, 144, 1401–1411. [Google Scholar] [CrossRef] [PubMed]
- O’Brien, K.P.; Khan, S.; Thompson, K.; Joyce, D.; Dwyer, R.M. Abstract P6-04-01: Engineering mesenchymal stem cells(MSCs) to secrete tumour-suppressing exosomes for breast cancer therapy. Cancer Res. 2017, 77. [Google Scholar] [CrossRef]
- Hannafon, B.N.; Carpenter, K.J.; Berry, W.L.; Janknecht, R.; Dooley, W.C.; Ding, W.Q. Exosome-mediated microRNA signaling from breast cancer cells is altered by the anti-angiogenesis agent docosahexaenoic acid (DHA). Mol. Cancer 2015, 14, 133. [Google Scholar] [CrossRef]
- Gao, G.; Liang, X.; Ma, W. Sinomenine restrains breast cancer cells proliferation, migration and invasion via modulation of miR-29/PDCD-4 axis. Artif. Cells Nanomed. Biotechnol. 2019, 47, 3839–3846. [Google Scholar] [CrossRef]
- Wei, Y.; Li, M.; Cui, S.; Wang, D.; Zhang, C.Y.; Zen, K.; Li, L. Shikonin Inhibits the Proliferation of Human Breast Cancer Cells by Reducing Tumor-Derived Exosomes. Molecules 2016, 21, 777. [Google Scholar] [CrossRef]
- Li, L.Q.; Li, X.L.; Wang, L.; Du, W.J.; Guo, R.; Liang, H.H.; Liu, X.; Liang, D.S.; Lu, Y.J.; Shan, H.L.; et al. Matrine inhibits breast cancer growth via miR-21/PTEN/Akt pathway in MCF-7 cells. Cell. Physiol. Biochem. 2012, 30, 631–641. [Google Scholar] [CrossRef]
- Zhong, W.; Chen, S.; Qin, Y.; Zhang, H.; Wang, H.; Meng, J.; Huai, L.; Zhang, Q.; Yin, T.; Lei, Y.; et al. Doxycycline inhibits breast cancer EMT and metastasis through PAR-1/NF-κB/miR-17/E-cadherin pathway. Oncotarget 2017, 8, 104855–104866. [Google Scholar] [CrossRef]
- Yardley, D.A. Visceral disease in patients with metastatic breast cancer: Efficacy and safety of treatment with ixabepilone and other chemotherapeutic agents. Clin. Breast Cancer 2010, 10, 64–73. [Google Scholar] [CrossRef]
- Liu, Q.; Peng, F.; Chen, J. The Role of Exosomal MicroRNAs in the Tumor Microenvironment of Breast Cancer. Int. J. Mol. Sci. 2019, 20, 3884. [Google Scholar] [CrossRef]
- Le, M.T.; Hamar, P.; Guo, C.; Basar, E.; Perdigão-Henriques, R.; Balaj, L.; Lieberman, J. miR-200-containing extracellular vesicles promote breast cancer cell metastasis. J. Clin. Investig. 2014, 124, 5109–5128. [Google Scholar] [CrossRef]
- Tang, Y.; Cui, Y.; Li, Z.; Jiao, Z.; Zhang, Y.; He, Y.; Chen, G.; Zhou, Q.; Wang, W.; Zhou, X.; et al. Radiation-induced miR-208a increases the proliferation and radioresistance by targeting p21 in human lung cancer cells. J. Exp. Clin. Cancer Res. 2016, 35, 7. [Google Scholar] [CrossRef] [PubMed]
- Han, F.; Huang, D.; Huang, X.; Wang, W.; Yang, S.; Chen, S. Exosomal microRNA-26b-5p down-regulates ATF2 to enhance radiosensitivity of lung adenocarcinoma cells. J. Cell. Mol. Med. 2020, 24, 7730–7742. [Google Scholar] [CrossRef]
- Mizushima, N.; Yoshimori, T.; Ohsumi, Y. The role of Atg proteins in autophagosome formation. Annu. Rev. Cell Dev. Biol. 2011, 27, 107–132. [Google Scholar] [CrossRef] [PubMed]
- Moraes, L.A.; Kar, S.; Foo, S.L.; Gu, T.; Toh, Y.Q.; Ampomah, P.B.; Sachaphibulkij, K.; Yap, G.; Zharkova, O.; Lukman, H.M.; et al. Annexin-A1 enhances breast cancer growth and migration by promoting alternative macrophage polarization in the tumour microenvironment. Sci. Rep. 2017, 7, 17925. [Google Scholar] [CrossRef] [PubMed]
- Kenific, C.M.; Thorburn, A.; Debnath, J. Autophagy and metastasis: Another double-edged sword. Curr. Opin. Cell Biol. 2010, 22, 241–245. [Google Scholar] [CrossRef]
- Liu, F.; Sang, M.; Meng, L.; Gu, L.; Liu, S.; Li, J.; Geng, C. miR-92b promotes autophagy and suppresses viability and invasion in breast cancer by targeting EZH2. Int. J. Oncol. 2018, 53, 1505–1515. [Google Scholar] [CrossRef]
- Ai, H.; Zhou, W.; Wang, Z.; Qiong, G.; Chen, Z.; Deng, S. microRNAs-107 inhibited autophagy, proliferation, and migration of breast cancer cells by targeting HMGB1. J. Cell. Biochem. 2018. [Google Scholar] [CrossRef]
- Satyavarapu, E.M.; Das, R.; Mandal, C.; Mukhopadhyay, A.; Mandal, C. Autophagy-independent induction of LC3B through oxidative stress reveals its non-canonical role in anoikis of ovarian cancer cells. Cell Death Dis. 2018, 9, 934. [Google Scholar] [CrossRef]
- Paoli, P.; Giannoni, E.; Chiarugi, P. Anoikis molecular pathways and its role in cancer progression. Biochim. Biophys. Acta Mol. Cell Res. 2013, 1833, 3481–3498. [Google Scholar] [CrossRef] [PubMed]
- Howe, E.N.; Cochrane, D.R.; Richer, J.K. Targets of miR-200c mediate suppression of cell motility and anoikis resistance. Breast Cancer Res. 2011, 13, R45. [Google Scholar] [CrossRef] [PubMed]
- Joyce, D.P.; Kerin, M.J.; Dwyer, R.M. Exosome-encapsulated microRNAs as circulating biomarkers for breast cancer. Int. J. Cancer 2016, 139, 1443–1448. [Google Scholar] [CrossRef] [PubMed]
- Hannafon, B.N.; Trigoso, Y.D.; Calloway, C.L.; Zhao, Y.D.; Lum, D.H.; Welm, A.L.; Zhao, Z.J.; Blick, K.E.; Dooley, W.C.; Ding, W.Q. Plasma exosome microRNAs are indicative of breast cancer. Breast Cancer Res. 2016, 18, 90. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).