Degree Adjusted Large-Scale Network Analysis Reveals Novel Putative Metabolic Disease Genes
Abstract
Simple Summary
Abstract
1. Introduction
2. Results
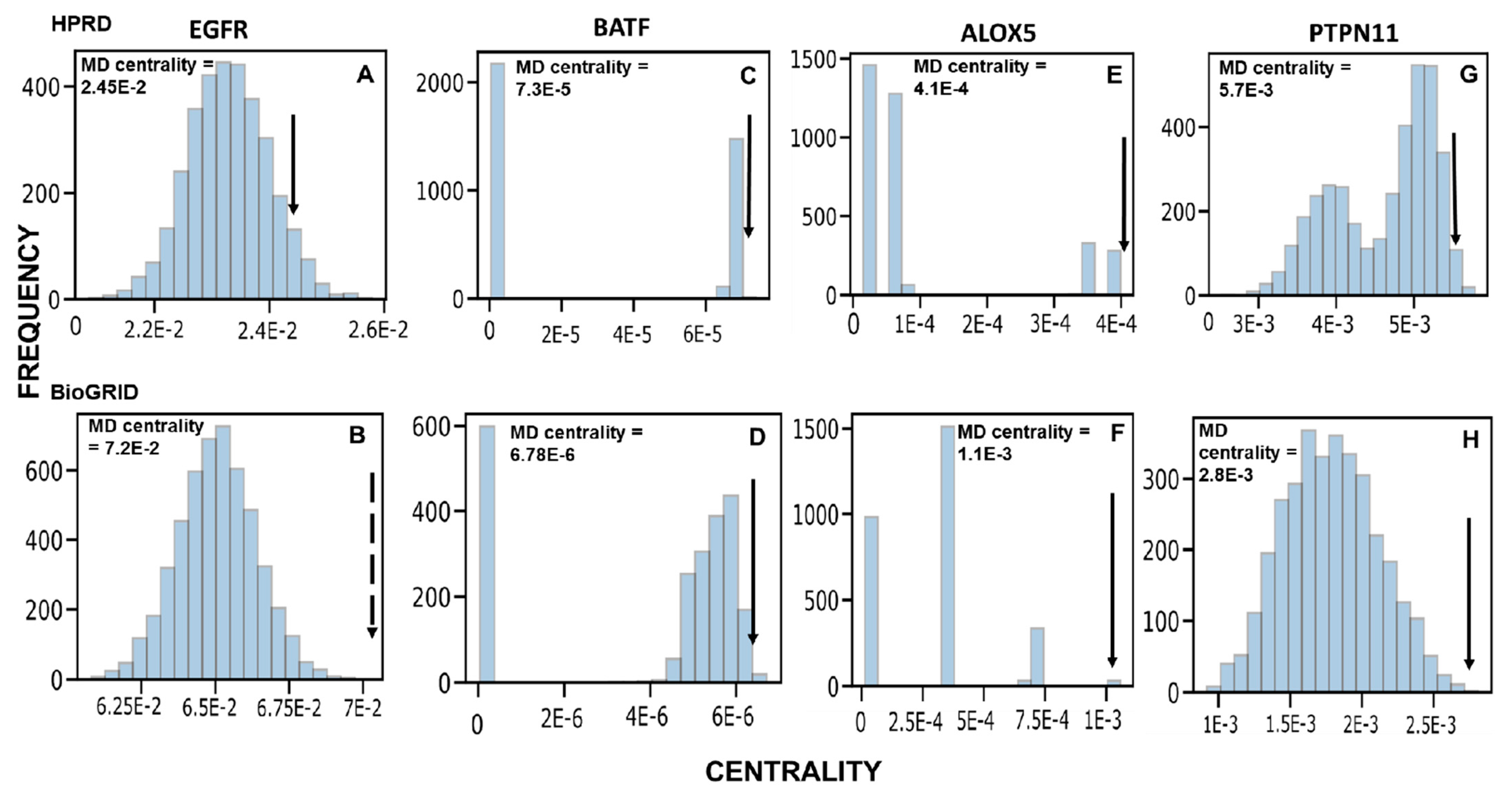
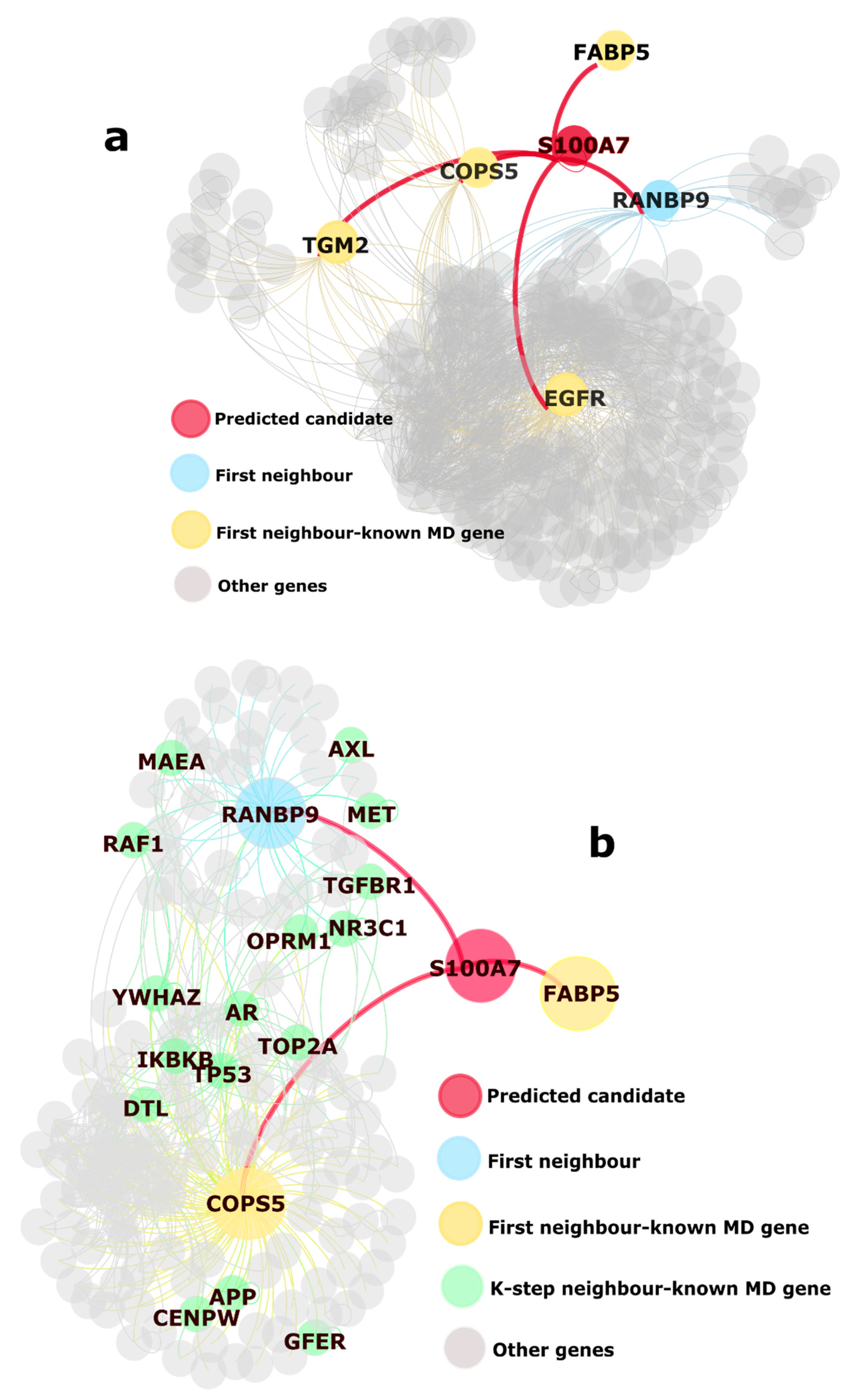
2.1. Identification of Novel Putative MD Genes
2.2. Pathway Analysis
2.3. Differential Expression Analysis
3. Discussion
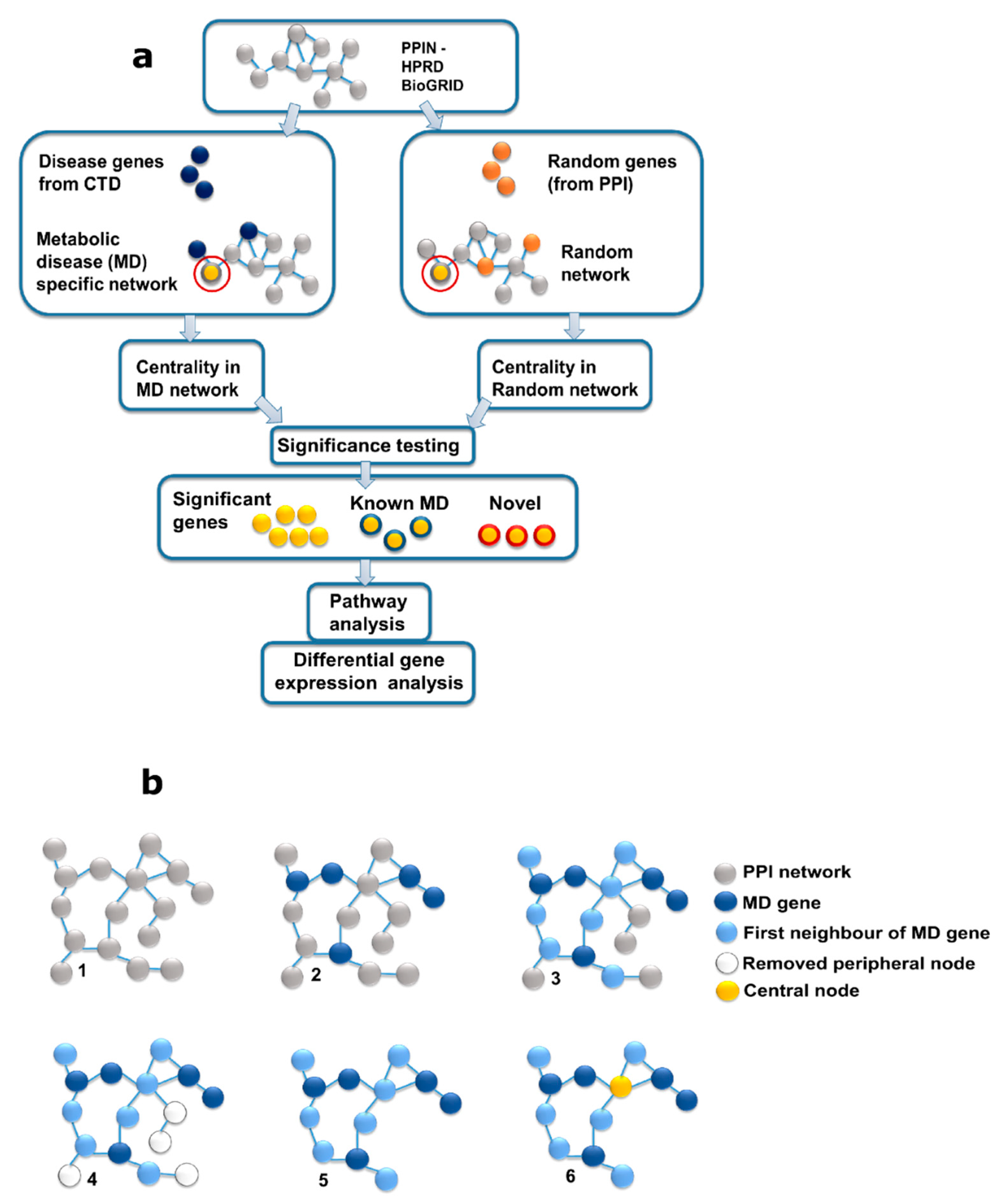
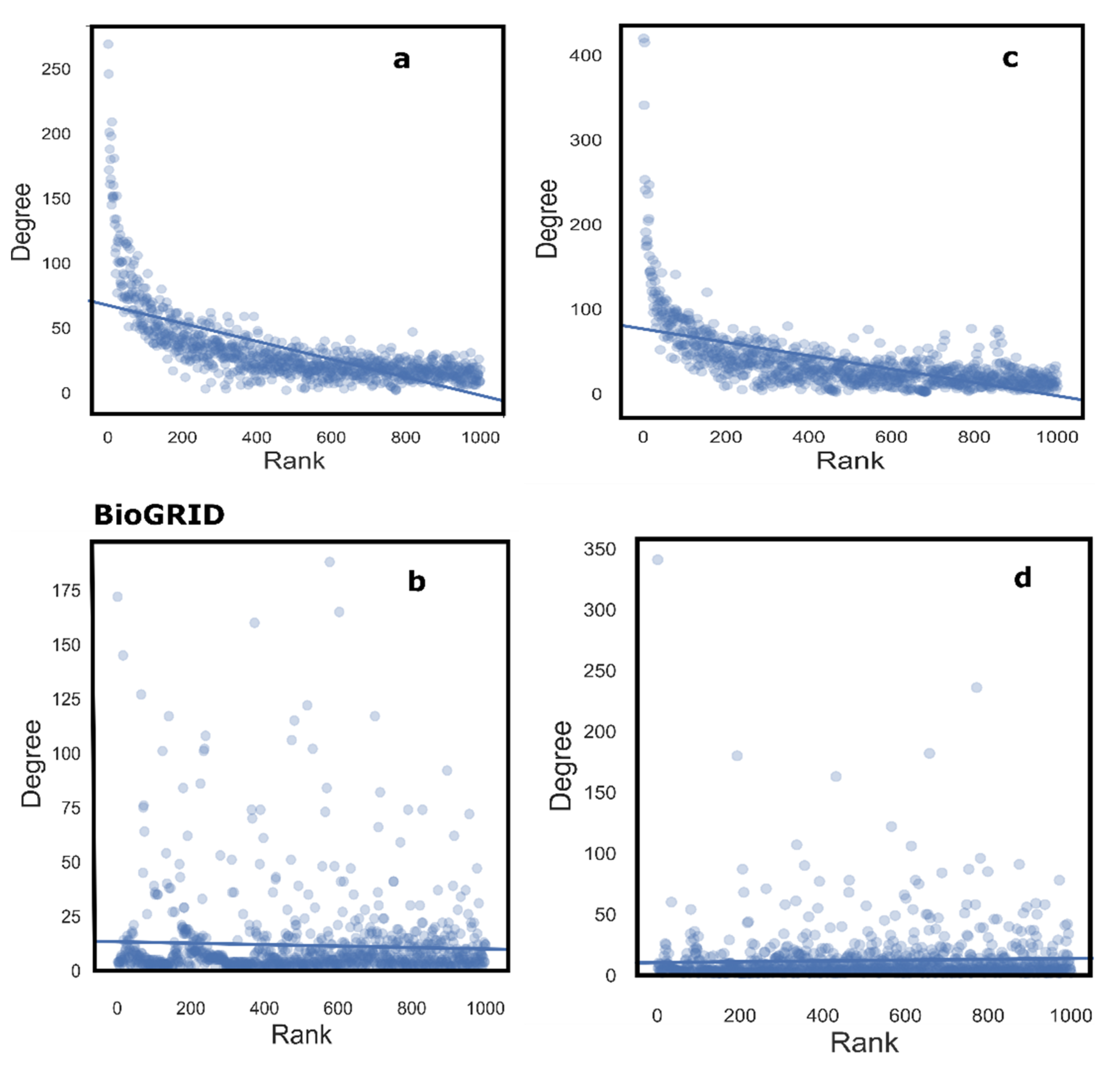
4. Methods
- (1)
- (2)
- Reconstruction and analysis of the protein interaction network for MD genes.
- (3)
- Construction and analysis of random networks for significance testing (Figure 1).
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| AD | Alzheimer’s disease |
| CTD | Comparative Toxicogenomics Database |
| BC | Betweenness centrality |
| CVD | Cardiovascular diseases |
| HGNC | HUGO Gene Nomenclature Committee |
| HPRD | Human Protein Reference Database |
| MD | Metabolic diseases |
| PPIN | Protein-protein interaction network |
| T2D | Type 2 diabetes |
References
- Eckel, R.H.; Grundy, S.M.; Zimmet, P.Z. The metabolic syndrome. Lancet 2005, 365, 1415–1428. [Google Scholar] [CrossRef]
- Dunbar, J.; Reddy, P.; Davis-Lameloise, N.; Philpot, B.; Laatikainen, T.; Kilkkinen, A.; Bunker, S.J.; Best, J.D.; Vartiainen, E.; Lo, S.K.; et al. Depression: An Important Comorbidity With Metabolic Syndrome in a General Population. Diabetes Care 2008, 31, 2368–2373. [Google Scholar] [CrossRef]
- Pradhan, A. Obesity, Metabolic Syndrome, and Type 2 Diabetes: Inflammatory Basis of Glucose Metabolic Disorders. Nutr. Rev. 2007, 65, S152–S156. [Google Scholar] [CrossRef]
- Ritchie, S.; Connell, J. The link between abdominal obesity, metabolic syndrome and cardiovascular disease. Nutr. Metab. Cardiovasc. Dis. 2007, 17, 319–326. [Google Scholar] [CrossRef]
- Pollex, R.L.; Hegele, R.A. Genetic determinants of the metabolic syndrome. Nat. Clin. Pr. Neurol. 2006, 3, 482–489. [Google Scholar] [CrossRef]
- Cornier, M.-A.; Dabelea, D.; Hernandez, T.L.; Lindstrom, R.C.; Steig, A.J.; Stob, N.R.; Van Pelt, R.E.; Wang, H.; Eckel, R.H. The Metabolic Syndrome. Endocr. Rev. 2008, 29, 777–822. [Google Scholar] [CrossRef] [PubMed]
- Seyfried, T.N.; Flores, R.E.; Poff, A.M.; D’Agostino, D.P. Cancer as a metabolic disease: Implications for novel therapeutics. Carcinogenesis 2014, 35, 515–527. [Google Scholar] [CrossRef] [PubMed]
- Abou Ziki, M.D.; Mani, A. Metabolic syndrome: Genetic insights into disease pathogenesis. Curr. Opin. Lipidol. 2016, 27, 162–171. [Google Scholar] [CrossRef]
- Lee, D.-S.; Park, J.; A Kay, K.; A Christakis, N.; Oltvai, Z.N.; Barabási, A.-L. The implications of human metabolic network topology for disease comorbidity. Proc. Natl. Acad. Sci. USA 2008, 105, 9880–9885. [Google Scholar] [CrossRef]
- Li, X.; Li, C.; Shang, D.; Li, J.; Han, J.; Miao, Y.; Wang, Y.; Wang, Q.; Li, W.; Wu, C.; et al. The Implications of Relationships between Human Diseases and Metabolic Subpathways. PLoS ONE 2011, 6, e21131. [Google Scholar] [CrossRef]
- Baumgartner, C.; Osl, M.; Netzer, M.; Baumgartner, D. Bioinformatic-driven search for metabolic biomarkers in disease. J. Clin. Bioinform. 2011, 1, 2. [Google Scholar] [CrossRef] [PubMed]
- Galhardo, M.; Sinkkonen, L.; Berninger, P.; Lin, J.; Sauter, T.; Heinäniemi, M. Integrated analysis of transcript-level regulation of metabolism reveals dis-ease-relevant nodes of the human metabolic network. Nucleic Acids Res. 2014, 42, 1474–1496. [Google Scholar] [CrossRef] [PubMed]
- Galhardo, M.; Berninger, P.; Nguyen, T.-P.; Sauter, T.; Sinkkonen, L. Cell type-selective disease-association of genes under high regulatory load. Nucleic Acids Res. 2015, 43, 8839–8855. [Google Scholar] [CrossRef] [PubMed]
- Falter-Braun, P.; Rietman, E.; Vidal, M. Networking metabolites and diseases. Proc. Natl. Acad. Sci. USA 2008, 105, 9849–9850. [Google Scholar] [CrossRef] [PubMed]
- Goh, K.-I.; Cusick, M.E.; Valle, D.; Childs, B.; Vidal, M.; Barabási, A.-L. The human disease network. Proc. Natl. Acad. Sci. USA 2007, 104, 8685–8690. [Google Scholar] [CrossRef] [PubMed]
- Amar, D.; Shamir, R. Constructing module maps for integrated analysis of heterogeneous biological networks. Nucleic Acids Res. 2014, 42, 4208–4219. [Google Scholar] [CrossRef]
- Leiserson, M.D.M.; Blokh, D.; Sharan, R.; Raphael, B.J. Simultaneous Identification of Multiple Driver Pathways in Cancer. PLoS Comput. Biol. 2013, 9, e1003054. [Google Scholar] [CrossRef]
- Lotta, L.A.; Abbasi, A.; Sharp, S.J.; Sahlqvist, A.-S.; Waterworth, D.; Brosnan, J.M.; Scott, R.A.; Langenberg, C.; Wareham, N.J. Definitions of Metabolic Health and Risk of Future Type 2 Diabetes in BMI Categories: A Systematic Review and Network Meta-analysis. Diabetes Care 2015, 38, 2177–2187. [Google Scholar] [CrossRef]
- Zolotareva, O.; Maren, K. A Survey of Gene Prioritization Tools for Mendelian and Complex Human Diseases. J. Integr. Bioinform. 2019, 16. [Google Scholar] [CrossRef]
- Silverbush, D.; Cristea, S.; Yanovich, G.; Geiger, T.; Beerenwinkel, N.; Sharan, R. Modulomics: Integrating multi-omics data to identify cancer driver modules. bioRxiv 2018. [Google Scholar] [CrossRef]
- Erten, S.; Bebek, G.; Ewing, R.M.; Koyuturk, M. DA DA: Degree-Aware Algorithms for Network-Based Disease Gene Prioritization. BioData Min. 2011, 4, 1–20. [Google Scholar] [CrossRef] [PubMed]
- Kacprowski, T.; Doncheva, N.T.; Albrecht, M. NetworkPrioritizer: A versatile tool for network-based prioritization of candidate disease genes or other molecules. Bioinformatics 2013, 29, 1471–1473. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Koschützki, D.; Schreiber, F. Centrality Analysis Methods for Biological Networks and Their Application to Gene Regulatory Networks. Gene Regul. Syst. Biol. 2008, 2, GRSB-S702. [Google Scholar] [CrossRef] [PubMed]
- Joy, M.P.; Brock, A.; Ingber, D.E.; Huang, S. High-Betweenness Proteins in the Yeast Protein Interaction Network. J. Biomed. Biotechnol. 2005, 2005, 96–103. [Google Scholar] [CrossRef]
- Badkas, A.; De Landtsheer, S.; Sauter, T. Topological network measures for drug repositioning. Briefings Bioinform. 2020. [Google Scholar] [CrossRef]
- Barabási, A.-L.; Oltvai, Z.N. Network biology: Understanding the cell’s functional organization. Nat. Rev. Genet. 2004, 5, 101–113. [Google Scholar] [CrossRef]
- Yook, S.-H.; Oltvai, Z.N.; Barabási, A.-L. Functional and topological characterization of protein interaction networks. Proteomics 2004, 4, 928–942. [Google Scholar] [CrossRef]
- Chen, J.; Bardes, E.E.; Aronow, B.J.; Jegga, A.G. ToppGene Suite for gene list enrichment analysis and candidate gene prioritization. Nucleic Acids Res. 2009, 37, W305–W311. [Google Scholar] [CrossRef]
- Biran, H.; Kupiec, M.; Sharan, R. Comparative Analysis of Normalization Methods for Network Propagation. Front. Genet. 2019, 10, 4. [Google Scholar] [CrossRef]
- Davis, A.P.; Murphy, C.G.; Saraceni-Richards, C.A.; Rosenstein, M.C.; Wiegers, T.C.; Mattingly, C.J. Comparative Toxicogenomics Database: A knowledgebase and discovery tool for chemical-gene-disease networks. Nucleic Acids Res. 2008, 37, D786–D792. [Google Scholar] [CrossRef]
- Prasad, T.S.K.; Goel, R.; Kandasamy, K.; Keerthikumar, S.; Kumar, S.; Mathivanan, S.; Telikicherla, D.; Raju, R.; Shafreen, B.; Venugopal, A.; et al. Human Protein Reference Database--2009 update. Nucleic Acids Res. 2008, 37, D767–D772. [Google Scholar] [CrossRef] [PubMed]
- Oughtred, R.; Stark, C.; Breitkreutz, B.J.; Rust, J.; Boucher, L.; Chang, C.; Kolas, N.; O’Donnell, L.; Leung, G.; McAdam, R.; et al. The BioGRID interaction database: 2019 update. Nucleic Acids Res. 2019, 47, D529–D541. [Google Scholar] [CrossRef] [PubMed]
- Kuleshov, M.V.; Jones, M.R.; Rouillard, A.D.; Fernandez, N.F.; Duan, Q.; Wang, Z.; Koplev, S.; Jenkins, S.L.; Jagodnik, K.M.; Lachmann, A.; et al. Enrichr: A comprehensive gene set enrichment analysis web server 2016 update. Nucleic Acids Res. 2016, 44, W90–W97. [Google Scholar] [CrossRef] [PubMed]
- Kamburov, A.; Stelzl, U.; Lehrach, H.; Herwig, R. The ConsensusPathDB interaction database: 2013 update. Nucleic Acids Res. 2012, 41, D793–D800. [Google Scholar] [CrossRef]
- Rouillard, A.D.; Gundersen, G.W.; Fernandez, N.F.; Wang, Z.; Monteiro, C.D.; McDermott, M.G.; Ma’Ayan, A. The harmonizome: A collection of processed datasets gathered to serve and mine knowledge about genes and proteins. Database 2016, 2016. [Google Scholar] [CrossRef]
- Chung, C.P.; Avalos, I.; Oeser, A.; Gebretsadik, T.; Shintani, A.; Raggi, P.; Stein, C.M. High prevalence of the metabolic syndrome in patients with systemic lupus erythemato-sus: Association with disease characteristics and cardiovascular risk factors. Ann. Rheum. Dis. 2007, 66, 208–214. [Google Scholar] [CrossRef]
- Boyer, L.; Richieri, R.; Dassa, D.; Boucekine, M.; Fernandez, J.; Vaillant, F.; Padovani, R.; Auquier, P.; Lancon, C. Association of metabolic syndrome and inflammation with neurocognition in patients with schizophrenia. Psychiatry Res. 2013, 210, 381–386. [Google Scholar] [CrossRef]
- Leonard, B.E.; Schwarz, M.J.; Myint, A.M. The metabolic syndrome in schizophrenia: Is inflammation a contributing cause? J. Psychopharmacol. 2012, 26, 33–41. [Google Scholar] [CrossRef]
- Soto-Angona, Ó.; Anmella, G.; Valdés-Florido, M.J.; De Uribe-Viloria, N.; Carvalho, A.F.; Penninx, B.W.J.H.; Berk, M. Non-alcoholic fatty liver disease (NAFLD) as a neglected metabolic companion of psychiatric disorders: Common pathways and future approaches. BMC Med. 2020, 18, 1–14. [Google Scholar] [CrossRef]
- Wang, D.; Li, Y.; Zhang, C.; Li, X.; Yu, J. MiR-216a-3p inhibits colorectal cancer cell proliferation through direct targeting COX-2 and ALOX5. J. Cell. Biochem. 2018, 119, 1755–1766. [Google Scholar] [CrossRef]
- Wculek, S.K.; Malanchi, I. Neutrophils support lung colonization of metastasis-initiating breast cancer cells. Nat. Cell Biol. 2015, 528, 413–417. [Google Scholar] [CrossRef] [PubMed]
- Gläser, R.; Meyer-Hoffert, U.; Harder, J.; Cordes, J.; Wittersheim, M.; Kobliakova, J.; Fölster-Holst, R.; Proksch, E.; Schröder, J.-M.; Schwarz, T. The Antimicrobial Protein Psoriasin (S100A7) Is Upregulated in Atopic Dermatitis and after Experimental Skin Barrier Disruption. J. Investig. Dermatol. 2009, 129, 641–649. [Google Scholar] [CrossRef] [PubMed]
- Matsuda, S.; Kobayashi, M.; Kitagishi, Y. Roles for PI3K/AKT/PTEN Pathway in Cell Signaling of Nonalcoholic Fatty Liver Dis-ease. ISRN Endocrinol. 2013, 2013, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Hotamisligil, G.S. Inflammation and metabolic disorders. Nat. Cell Biol. 2006, 444, 860–867. [Google Scholar] [CrossRef]
- Tiffin, N.; Adie, E.; Turner, F.; Brunner, H.G.; van Driel, M.A.; Oti, M.; Lopez-Bigas, N.; Ouzounis, C.; Perez-Iratxeta, C.; Andrade-Navarro, M.A.; et al. Computational disease gene identification: A concert of methods prioritizes type 2 diabetes and obesity candidate genes. Nucleic Acids Res. 2006, 34, 3067–3081. [Google Scholar] [CrossRef]
- de la Monte, S.M.; Longato, L.; Tong, M.; Wands, J.R. Insulin resistance and neurodegeneration: Roles of obesity, type 2 diabetes mellitus, and non-alcoholic steatohepatitis. Curr. Opin. Investig. Drugs 2009, 10, 1049–1060. [Google Scholar]
- Esser, N.; Legrand-Poels, S.; Piette, J.; Scheen, A.J.; Paquot, N. Inflammation as a link between obesity, metabolic syndrome and type 2 diabetes. Diabetes Res. Clin. Pr. 2014, 105, 141–150. [Google Scholar] [CrossRef]
- Shi, H.; Kokoeva, M.V.; Inouye, K.; Tzameli, I.; Yin, H.; Flier, J.S. TLR4 links innate immunity and fatty acid–induced insulin resistance. J. Clin. Investig. 2006, 116, 3015–3025. [Google Scholar] [CrossRef]
- Gregory, C.D.; Devitt, A. The macrophage and the apoptotic cell: An innate immune interaction viewed simplistically? Immunology 2004, 113, 1–14. [Google Scholar] [CrossRef]
- Webb, A.E.; Brunet, A. FOXO transcription factors: Key regulators of cellular quality control. Trends Biochem. Sci. 2014, 39, 159–169. [Google Scholar] [CrossRef]
- Holscher, C. Diabetes as a risk factor for Alzheimer’s disease: Insulin signalling impairment in the brain as an alternative model of Alzheimer’s disease. Biochem. Soc. Trans. 2011, 39, 891–897. [Google Scholar] [CrossRef] [PubMed]
- Posse de Chaves, E.; Sipione, S. Sphingolipids and gangliosides of the nervous system in membrane function and dysfunction. FEBS Lett. 2010, 584, 1748–1759. [Google Scholar] [CrossRef] [PubMed]
- Spielman, L.J.; Little, J.P.; Klegeris, A. Inflammation and insulin/IGF-1 resistance as the possible link between obesity and neuro-degeneration. J. Neuroimmunol. 2014, 273, 8–21. [Google Scholar] [CrossRef] [PubMed]
- Yarchoan, M.; Arnold, S.E. Repurposing diabetes drugs for brain insulin resistance in Alzheimer’s disease. Diabetes 2014, 63, 2253–2261. [Google Scholar] [CrossRef]
- Aguirre-Plans, J.; Pinero, J.; Menche, J.; Sanz, F.; I Furlong, L.; Schmidt, H.H.H.W.; Oliva, B.; Guney, E. Proximal Pathway Enrichment Analysis for Targeting Comorbid Diseases via Network Endopharmacology. Pharmaceuticals 2018, 11, 61. [Google Scholar] [CrossRef]
- Skov, V.; Glintborg, D.; Knudsen, S.; Jensen, T.; Kruse, T.A.; Tan, Q.; Brusgaard, K.; Beck-Nielsen, H.; Højlund, K.; Skov, V. Reduced Expression of Nuclear-Encoded Genes Involved in Mitochondrial Oxidative Metabolism in Skeletal Muscle of Insulin-Resistant Women With Polycystic Ovary Syndrome. Diabetes 2007, 56, 2349–2355. [Google Scholar] [CrossRef]
- Wain, H.M.; Lovering, R.; Bruford, E.; Wright, M.; Lush, M.; Wain, H. The HUGO Gene Nomenclature Committee (HGNC). Qual. Life Res. 2001, 109, 678–680. [Google Scholar] [CrossRef]
- Benjamini, Y.; Hochberg, Y. Controlling the False Discovery Rate—A Practical and Powerful Approach to Multiple Testing. J. R. Stat. Soc. Ser. B Methodol. 1995, 57, 289–300. [Google Scholar] [CrossRef]
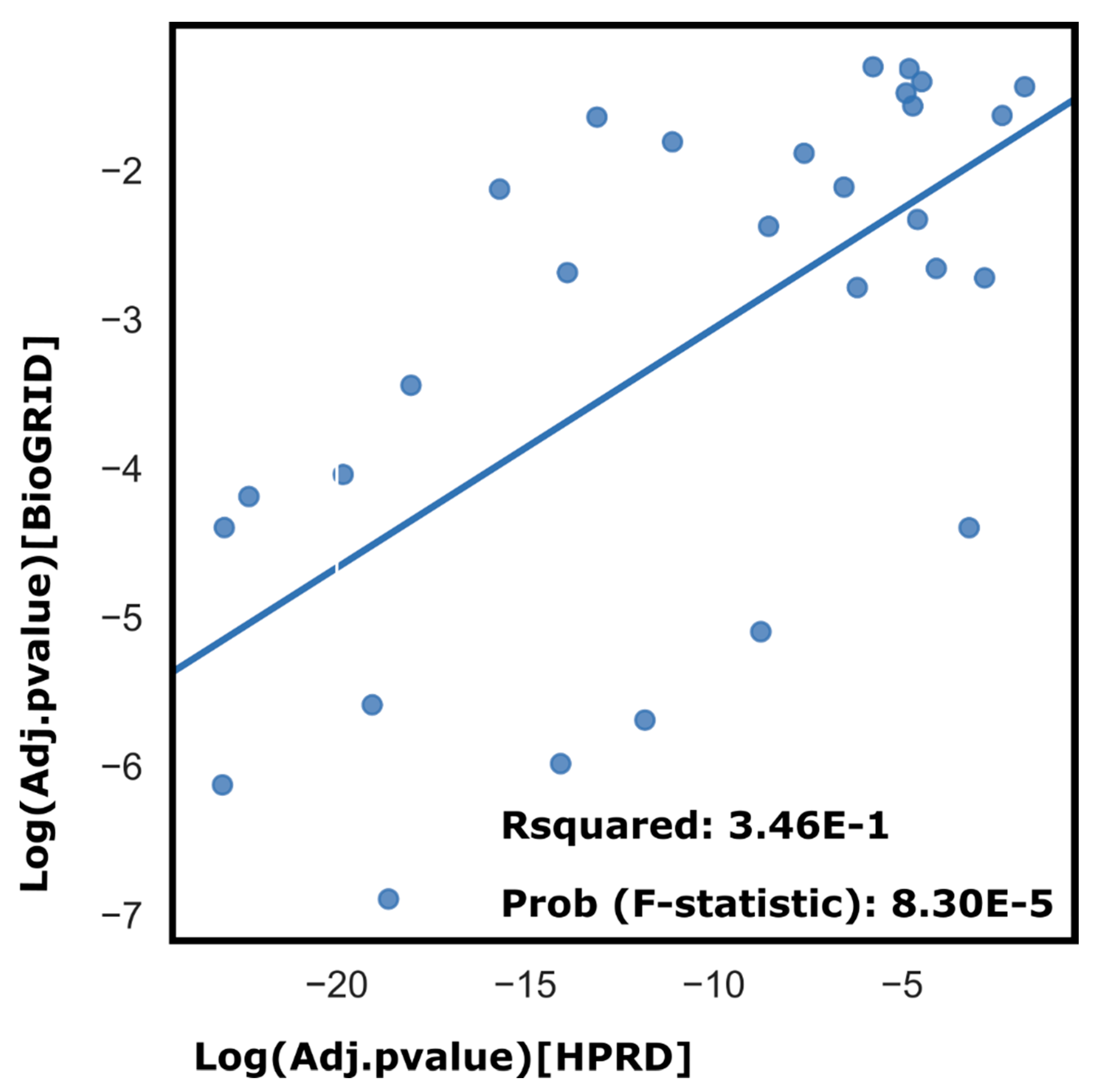
| Symbol | Name |
|---|---|
| ALOX5 | arachidonate 5-lipoxygenase |
| BATF | basic leucine zipper transcription factor, ATF-like |
| BNIPL | BCL2/adenovirus E1B 19kD interacting protein-like |
| DUSP22 | dual specificity phosphatase 22 |
| FBLN5 | fibulin 5 |
| GPC1 | glypican 1 |
| IL5RA | interleukin 5 receptor, alpha |
| OPRK1 | opioid receptor, kappa 1 |
| PLSCR3 | phospholipid scramblase 3 |
| PMPCB | peptidase (mitochondrial processing) beta |
| PTPN11 | protein tyrosine phosphatase, non-receptor type 11 |
| RNF128 | ring finger protein 128, E3 ubiquitin protein ligase |
| S100A7 | S100 calcium-binding protein A7 |
| SNCG | synuclein, gamma (breast cancer-specific protein 1) |
| STIM2 | stromal interaction molecule 2 |
| TFE3 | transcription factor binding to IGHM enhancer 3 |
| Gene | Upregulation | Downregulation |
|---|---|---|
| ALOX5 | Senescence, Rotavirus infection of children, Down Syndrome, Neurological pain disorder, Severe combined immunodeficiency (SCID) | Macular degeneration, Human immunodeficiency virus infection (HIV), Atherosclerosis |
| BATF | Glaucoma, Human immunodeficiency virus infection (HIV), Appendicitis, Oligodendroglioma, Multiple Sclerosis (MS), Severe acute respiratory syndrome (SARS), Diabetic Nephropathy | Chronic obstructive pulmonary disease (COPD), Cardiac Hypertrophy, Scleroderma, Retinoschisis |
| IL5RA | Cardiac Failure, Pauciarticular juvenile arthritis | Down Syndrome, Lung Injury, Familial combined hyperlipidaemia |
| PLSCR3 | Erectile dysfunction, Breast Cancer, Bipolar Disorder, Appendicitis, Papillary Carcinoma of the Thyroid, Bipolar Disorder | Atherosclerosis, Cardiomyopathy, Myocardial Infarction |
| S100A7 | Type 2 diabetes mellitus, Post-traumatic stress disorder (PTSD), Eczema | - |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Badkas, A.; Nguyen, T.-P.; Caberlotto, L.; Schneider, J.G.; De Landtsheer, S.; Sauter, T. Degree Adjusted Large-Scale Network Analysis Reveals Novel Putative Metabolic Disease Genes. Biology 2021, 10, 107. https://doi.org/10.3390/biology10020107
Badkas A, Nguyen T-P, Caberlotto L, Schneider JG, De Landtsheer S, Sauter T. Degree Adjusted Large-Scale Network Analysis Reveals Novel Putative Metabolic Disease Genes. Biology. 2021; 10(2):107. https://doi.org/10.3390/biology10020107
Chicago/Turabian StyleBadkas, Apurva, Thanh-Phuong Nguyen, Laura Caberlotto, Jochen G. Schneider, Sébastien De Landtsheer, and Thomas Sauter. 2021. "Degree Adjusted Large-Scale Network Analysis Reveals Novel Putative Metabolic Disease Genes" Biology 10, no. 2: 107. https://doi.org/10.3390/biology10020107
APA StyleBadkas, A., Nguyen, T.-P., Caberlotto, L., Schneider, J. G., De Landtsheer, S., & Sauter, T. (2021). Degree Adjusted Large-Scale Network Analysis Reveals Novel Putative Metabolic Disease Genes. Biology, 10(2), 107. https://doi.org/10.3390/biology10020107






