Presence of a Mitovirus Is Associated with Alteration of the Mitochondrial Proteome, as Revealed by Protein–Protein Interaction (PPI) and Co-Expression Network Models in Chenopodium quinoa Plants
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Materials
2.2. Mitochondrial Enrichment Protocol
2.3. Sample Preparation for LC-MS
2.4. Nano LC-MS/MS Analysis
2.5. Processing of Raw Mass Spectra
2.6. Protein Profiles Preprocessing, Statistical Evaluations and Quantitative Analysis
2.7. Chenopodium quinoa Protein–Protein Interaction (PPI) and Co-Expression (Co-Exp) Network Model Reconstruction
2.8. Real-Time PCR
3. Results
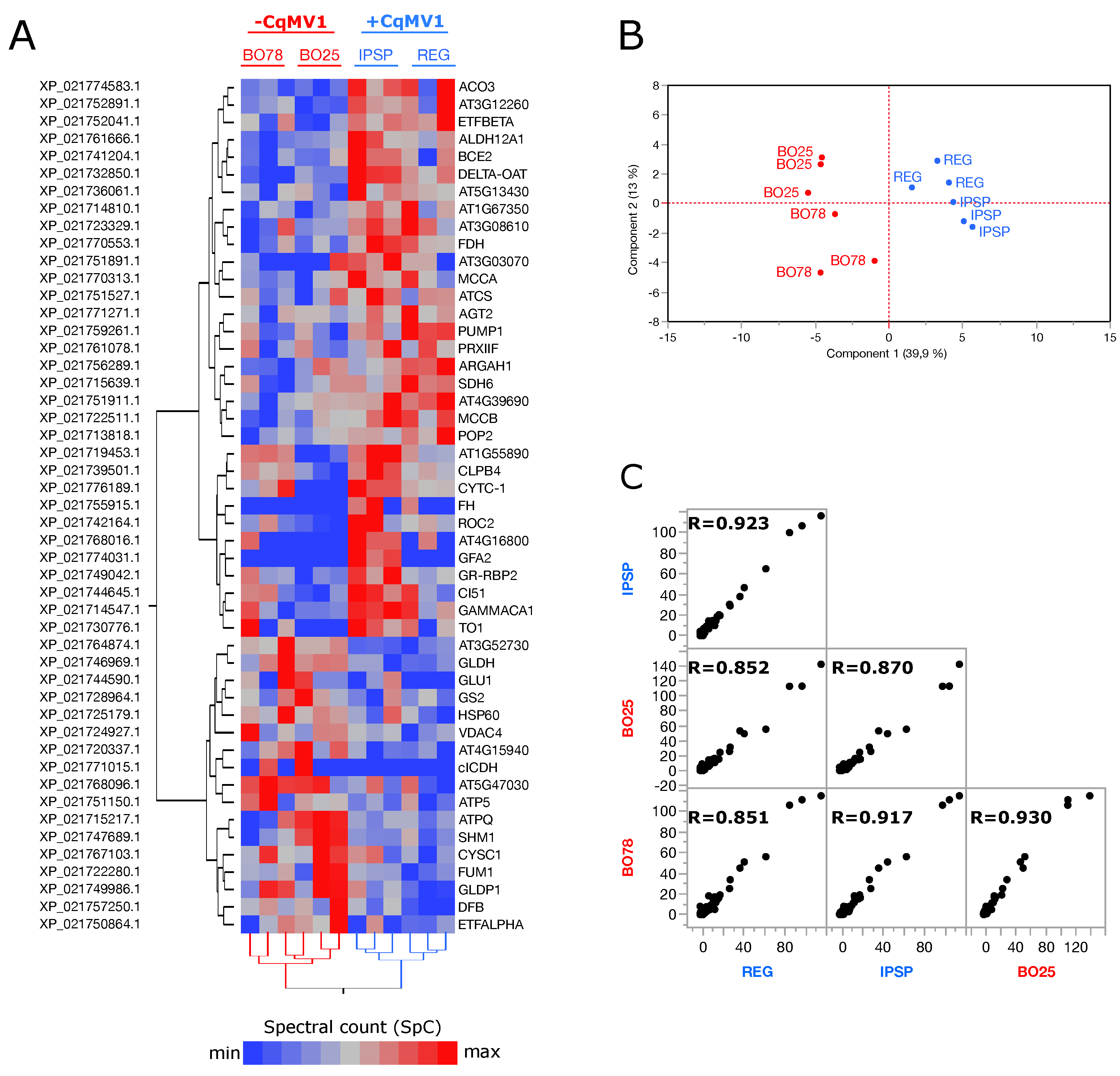
3.1. Mitochondrial Proteome from Leaves of Chenopodium quinoa, −CqMV1 and +CqMV1
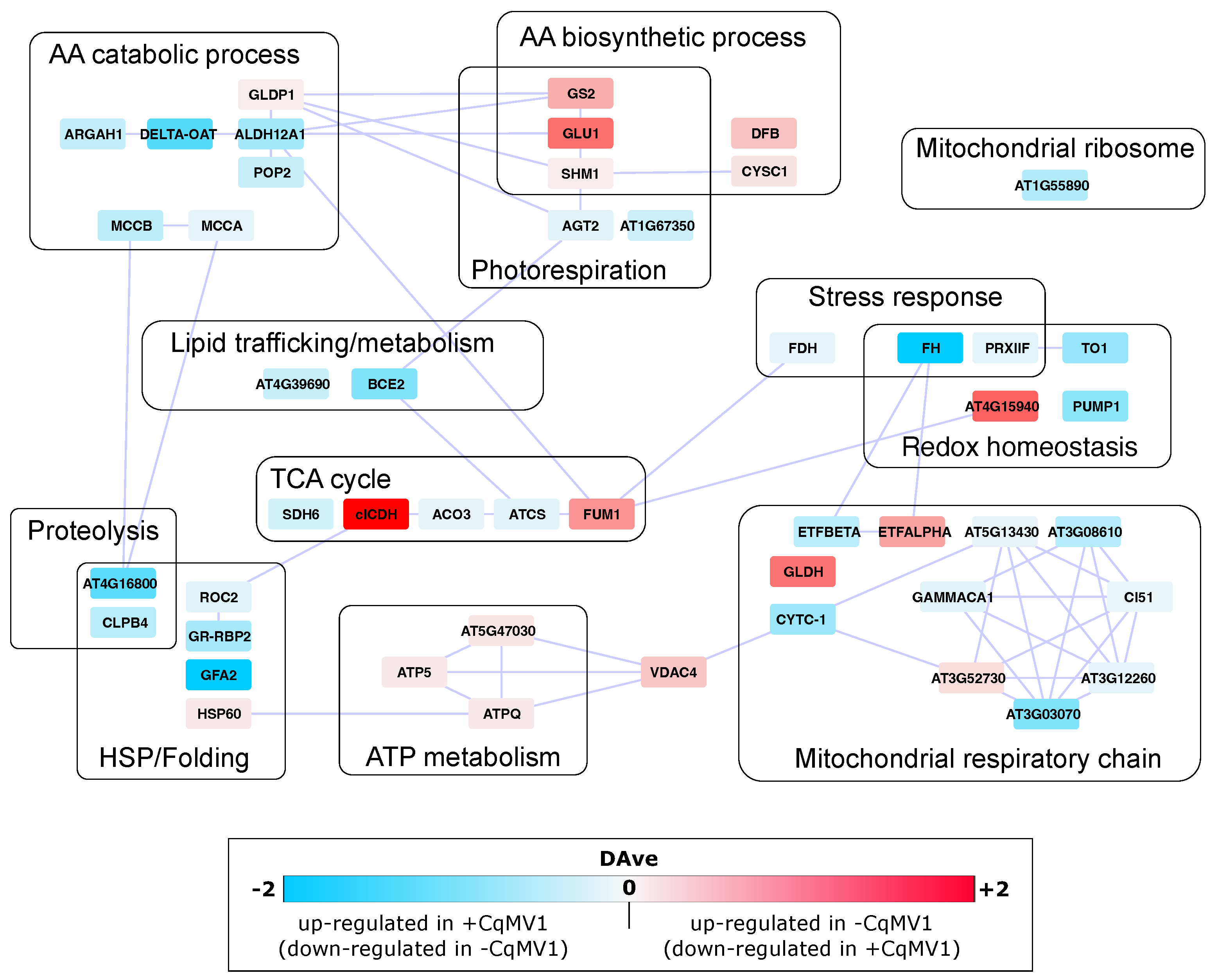
3.2. Mitochondrial Proteins Differentially Expressed in −CqMV1 vs. +CqMV1
3.3. Network Analysis
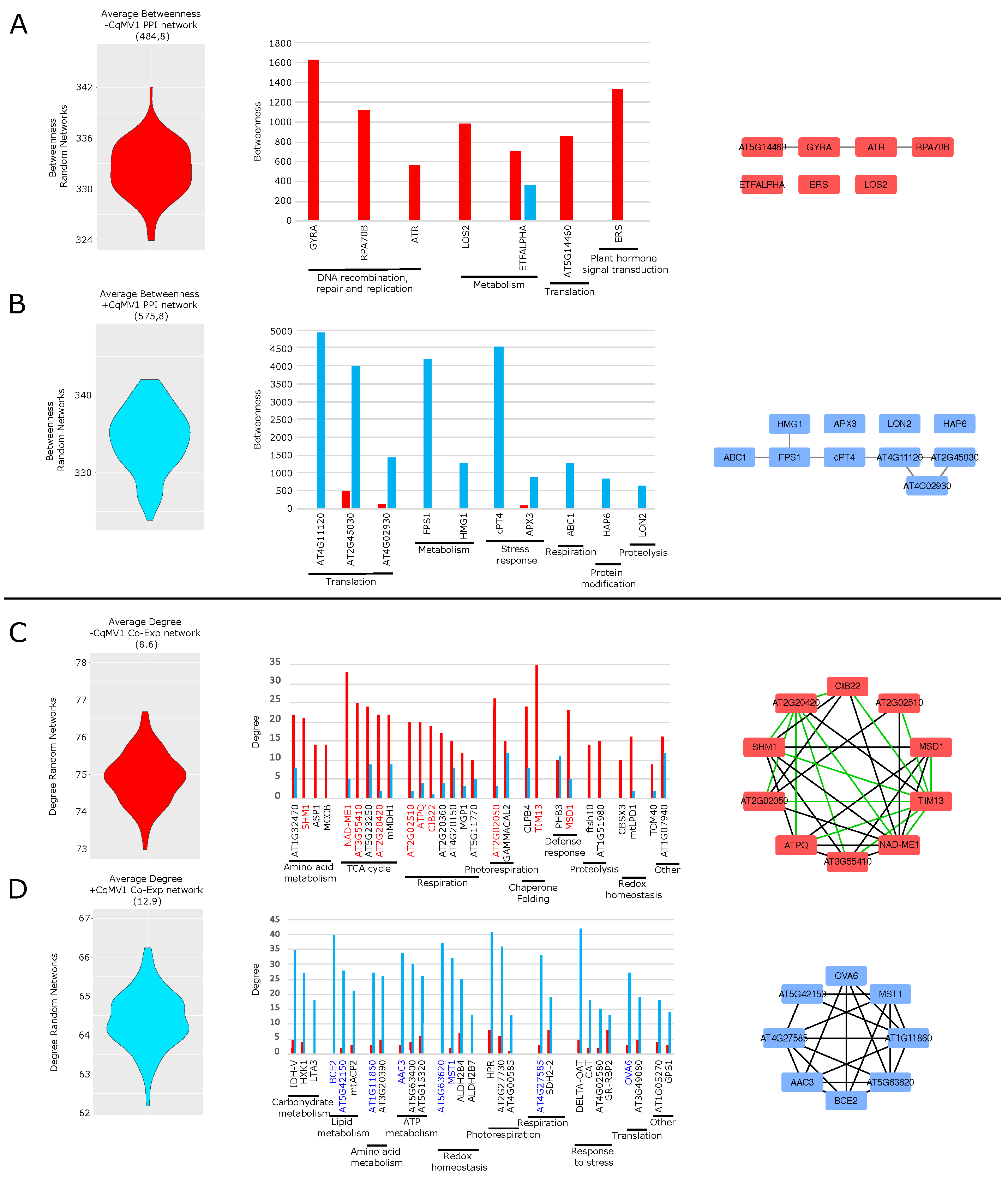
Chenopodium quinoa PPI and Co-Expression Network Hubs
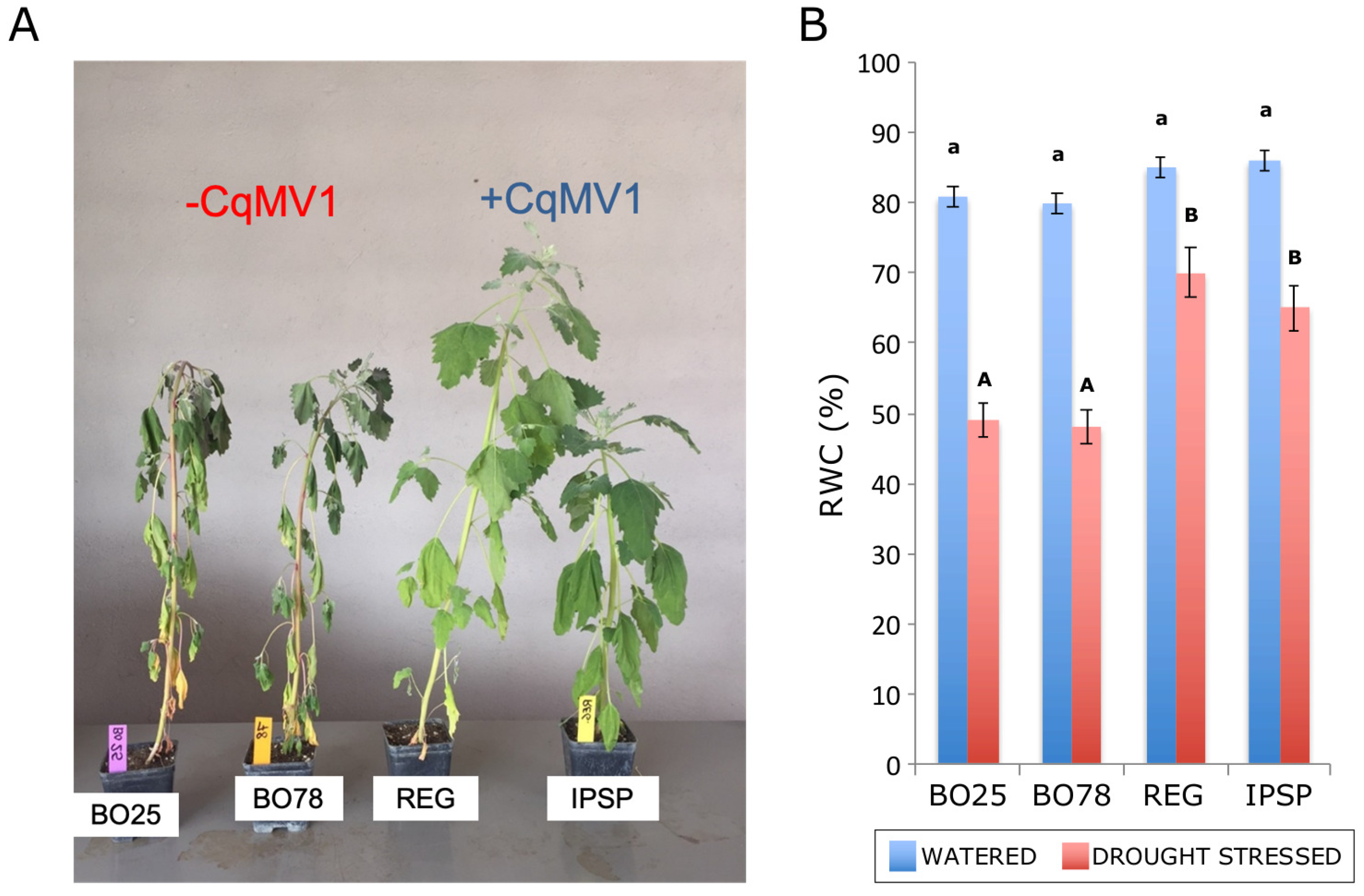
3.4. Abiotic Stress Differentially Affected Infected and Not Infected Line
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Koonin, E.V.; Dolja, V.V.; Krupovic, M. Origins and evolution of viruses of eukaryotes: The ultimate modularity. Virology 2015, 479–480, 2–25. [Google Scholar] [CrossRef] [Green Version]
- Hillman, B.I.; Cai, G. The family narnaviridae: Simplest of RNA viruses. Adv. Virus Res. 2013, 86, 149–176. [Google Scholar] [CrossRef] [PubMed]
- Horie, M.; Honda, T.; Suzuki, Y.; Kobayashi, Y.; Daito, T.; Oshida, T.; Ikuta, K.; Jern, P.; Gojobori, T.; Coffin, J.M.; et al. Endogenous non-retroviral RNA virus elements in mammalian genomes. Nature 2010, 463, 84–87. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Aris, K.; Gifford, R.J. Endogenous Eviral Eelements in Eanimal Egenomes. PLoS Genet. 2010, 6, e1001191. [Google Scholar] [CrossRef]
- Bruenn, J.A.; Benjamin, E.; Warner, P.Y. Widespread mitovirus sequences in plant genomes. PeerJ 2015, 3, e876. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nibert, M.L.; Vong, M.; Fugate, K.K.; Debat, H.J. Evidence for contemporary plant mitoviruses. Virology 2018, 518, 14–24. [Google Scholar] [CrossRef] [PubMed]
- Nerva, L.; Vigani, G.; Di Silvestre, D.; Ciuffo, M.; Forgia, M.; Chitarra, W.; Turina, M. Biological and Molecular Characterization of Chenopodium quinoa Mitovirus 1 Reveals a Distinct Small RNA Response Compared to Those of Cytoplasmic RNA Viruses. J. Virol. 2019, 93, e01998-18. [Google Scholar] [CrossRef] [Green Version]
- Fonseca, P.; Ferreira, F.; da Silva, F.; Oliveira, L.S.; Marques, J.T.; Goes-Neto, A.; Aguiar, E.; Gruber, A. Characterization of a Novel Mitovirus of the Sand Fly , javax.xml.bind.JAXBElement@507ff0b7, Using Genomic and Virus-Host Interaction Signatures. Viruses 2020, 13, 9. [Google Scholar] [CrossRef]
- Polashock, J.J.; Hillman, B.I. A small mitochondrial double-stranded (ds) RNA element associated with a hypovirulent strain of the chestnut blight fungus and ancestrally related to yeast cytoplasmic T and W dsRNAs. Proc. Natl. Acad. Sci. USA 1994, 91, 8680–8684. [Google Scholar] [CrossRef] [Green Version]
- Vella, D.; Zoppis, I.; Mauri, G.; Mauri, P.; Silvestre, D. From protein–protein interactions to protein co-expression networks: A new perspective to evaluate large-scale proteomic data. EURASIP J. Bioinform. Syst. Biol. 2017, 2017, 6. [Google Scholar] [CrossRef] [Green Version]
- Capriotti, A.L.; Cavaliere, C.; Piovesana, S.; Stampachiacchiere, S.; Ventura, S.; Zenezini Chiozzi, R.; Laganà, A. Characterization of quinoa seed proteome combining different protein precipitation techniques: Improvement of knowledge of nonmodel plant proteomics. J. Sep. Sci. 2015, 38, 1017–1025. [Google Scholar] [CrossRef]
- Burrieza, H.P.; Rizzo, A.J.; Moura Vale, E.; Silveira, V.; Maldonado, S. Shotgun proteomic analysis of quinoa seeds reveals novel lysine-rich seed storage globulins. Food Chem. 2019, 293, 299–306. [Google Scholar] [CrossRef]
- Rasouli, F.; Kiani-Pouya, A.; Shabala, L.; Li, L.; Tahir, A.; Yu, M.; Hedrich, R.; Chen, Z.; Wilson, R.; Zhang, H.; et al. Salinity Effects on Guard Cell Proteome in Chenopodium quinoa. Int. J. Mol. Sci. 2021, 22, 428. [Google Scholar] [CrossRef]
- Di Silvestre, D.; Bergamaschi, A.; Bellini, E.; Mauri, P. Large Scale Proteomic Data and Network-Based Systems Biology Approaches to Explore the Plant World. Proteomes 2018, 6, 27. [Google Scholar] [CrossRef] [Green Version]
- Ding, Z.; Kihara, D. Computational identification of protein–protein interactions in model plant proteomes. Sci. Rep. 2019, 9, 8740. [Google Scholar] [CrossRef] [PubMed]
- Zhao, J.; Lei, Y.; Hong, J.; Zheng, C.; Zhang, L. AraPPINet: An Updated Interactome for the Analysis of Hormone Signaling Crosstalk in Arabidopsis thaliana. Front. Plant Sci. 2019, 10, 870. [Google Scholar] [CrossRef] [PubMed]
- Yang, S.; Li, H.; He, H.; Zhou, Y.; Zhang, Z. Critical assessment and performance improvement of plant-pathogen protein–protein interaction prediction methods. Briefings Bioinform. 2019, 20, 274–287. [Google Scholar] [CrossRef] [PubMed]
- Di Silvestre, D.; Vigani, G.; Mauri, P.; Hammadi, S.; Morandini, P.; Murgia, I. Network Topological Analysis for the Identification of Novel Hubs in Plant Nutrition. Front. Plant Sci. 2021, 12, 629013. [Google Scholar] [CrossRef]
- Lee, T.; Kim, H.; Lee, I. Network-assisted crop systems genetics: Network inference and integrative analysis. Curr. Opin. Plant Biol. 2015, 24, 61–70. [Google Scholar] [CrossRef] [PubMed]
- Rao, X.; Dixon, R.A. Co-expression networks for plant biology: Why and how. Acta Biochim. Biophys. Sin. 2019, 51, 981–988. [Google Scholar] [CrossRef]
- Zegaoui, Z.; Planchais, S.; Cabassa, C.; Djebbar, R.; Belbachir, O.A.; Carol, P. Variation in relative water content, proline accumulation and stress gene expression in two cowpea landraces under drought. J. Plant Physiol. 2017, 218, 26–34. [Google Scholar] [CrossRef]
- Vigani, G.; Bohic, S.; Faoro, F.; Vekemans, B.; Vincze, L.; Terzano, R. Cellular Fractionation and Nanoscopic X-ray Fluorescence Imaging Analyses Reveal Changes of Zinc Distribution in Leaf Cells of Iron-Deficient Plants. Front. Plant Sci. 2018, 9, 1112. [Google Scholar] [CrossRef] [PubMed]
- Sievers, F.; Wilm, A.; Dineen, D.; Gibson, T.J.; Karplus, K.; Li, W.; Lopez, R.; McWilliam, H.; Remmert, M.; Söding, J.; et al. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol. Syst. Biol. 2011, 7, 539. [Google Scholar] [CrossRef] [PubMed]
- Szklarczyk, D.; Gable, A.L.; Lyon, D.; Junge, A.; Wyder, S.; Huerta-Cepas, J.; Simonovic, M.; Doncheva, N.T.; Morris, J.H.; Bork, P.; et al. STRING v11: Protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 2019, 47, D607–D613. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xiao, N.; Cao, D.S.; Zhu, M.F.; Xu, Q.S. protr/ProtrWeb: R package and web server for generating various numerical representation schemes of protein sequences. Bioinformatics 2015, 31, 1857–1859. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sahu, S.S.; Loaiza, C.D.; Kaundal, R. Plant-mSubP: A computational framework for the prediction of single- and multi-target protein subcellular localization using integrated machine-learning approaches. AoB Plants 2020, 12, plz068. [Google Scholar] [CrossRef] [PubMed]
- Griffin, N.M.; Yu, J.; Long, F.; Oh, P.; Shore, S.; Li, Y.; Koziol, J.A.; Schnitzer, J.E. Label-free, normalized quantification of complex mass spectrometry data for proteomic analysis. Nat. Biotechnol. 2010, 28, 83–89. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vigani, G.; Di Silvestre, D.; Agresta, A.M.; Donnini, S.; Mauri, P.; Gehl, C.; Bittner, F.; Murgia, I. Molybdenum and iron mutually impact their homeostasis in cucumber (Cucumis sativus) plants. New Phytol. 2017, 213, 1222–1241. [Google Scholar] [CrossRef]
- Di Silvestre, D.; Brambilla, F.; Mauri, P.L. Multidimensional protein identification technology for direct-tissue proteomics of heart. Methods Mol. Biol. (Clifton N. J.) 2013, 1005, 25–38. [Google Scholar] [CrossRef]
- Doncheva, N.T.; Morris, J.H.; Gorodkin, J.; Jensen, L.J. Cytoscape StringApp: Network Analysis and Visualization of Proteomics Data. J. Proteome Res. 2019, 18, 623–632. [Google Scholar] [CrossRef]
- Su, G.; Morris, J.H.; Demchak, B.; Bader, G.D. Biological network exploration with Cytoscape 3. Curr. Protoc. Bioinform. 2014, 47, 8–13. [Google Scholar] [CrossRef] [Green Version]
- Scardoni, G.; Tosadori, G.; Faizan, M.; Spoto, F.; Fabbri, F.; Laudanna, C. Biological network analysis with CentiScaPe: Centralities and experimental dataset integration. F1000Research 2014, 3, 139. [Google Scholar] [CrossRef] [Green Version]
- Scardoni, G.; Laudanna, C. New Frontiers in Graph Theory; Intech: London, UK, 2012. [Google Scholar] [CrossRef] [Green Version]
- Wang, M.B.; Masuta, C.; Smith, N.A.; Shimura, H. RNA silencing and plant viral diseases. Mol. Plant-Microbe Interact. MPMI 2012, 25, 1275–1285. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Barkan, A.; Small, I. Pentatricopeptide repeat proteins in plants. Annu. Rev. Plant Biol. 2014, 65, 415–442. [Google Scholar] [CrossRef]
- Ruwe, H.; Wang, G.; Gusewski, S.; Schmitz-Linneweber, C. Systematic analysis of plant mitochondrial and chloroplast small RNAs suggests organelle-specific mRNA stabilization mechanisms. Nucleic Acids Res. 2016, 44, 7406–7417. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Araújo, W.L.; Ishizaki, K.; Nunes-Nesi, A.; Larson, T.R.; Tohge, T.; Krahnert, I.; Witt, S.; Obata, T.; Schauer, N.; Graham, I.A.; et al. Identification of the 2-hydroxyglutarate and isovaleryl-CoA dehydrogenases as alternative electron donors linking lysine catabolism to the electron transport chain of Arabidopsis mitochondria. Plant Cell 2010, 22, 1549–1563. [Google Scholar] [CrossRef] [Green Version]
- Pires, M.V.; Pereira, A.A., Jr.; Medeiros, D.B.; Daloso, D.M.; Pham, P.A.; Barros, K.A.; Engqvist, M.K.M.; Florian, A.; Krahnert, I.; Maurino, V.G.; et al. The influence of alternative pathways of respiration that utilize branched-chain amino acids following water shortage in Arabidopsis. Plant Cell Environ. 2016, 39, 1304–1319. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- You, J.; Zhang, Y.; Liu, A.; Li, D.; Wang, X.; Dossa, K.; Zhou, R.; Yu, J.; Zhang, Y.; Wang, L.; et al. Transcriptomic and metabolomic profiling of drought-tolerant and susceptible sesame genotypes in response to drought stress. BMC Plant Biol. 2019, 19, 1–16. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, J.Y.; Park, S.J.; Jang, B.; Jung, C.H.; Ahn, S.J.; Goh, C.H.; Cho, K.; Han, O.; Kang, H. Functional characterization of a glycine-rich RNA-binding protein 2 in Arabidopsis thaliana under abiotic stress conditions. Plant J. Cell Mol. Biol. 2007, 50, 439–451. [Google Scholar] [CrossRef]
- Anwar, A.; Wang, K.; Wang, J.; Shi, L.; Du, L.; Ye, X. Expression of Arabidopsis Ornithine Aminotransferase (AtOAT) encoded gene enhances multiple abiotic stress tolerances in wheat. Plant Cell Rep. 2021, 40, 1155–1170. [Google Scholar] [CrossRef]
- Boon, L.; Geerts, W.J.; Jonker, A.; Lamers, W.H.; Van Noorden, C.J. High protein diet induces pericentral glutamate dehydrogenase and ornithine aminotransferase to provide sufficient glutamate for pericentral detoxification of ammonia in rat liver lobules. Histochem. Cell Biol. 1999, 111, 445–452. [Google Scholar] [CrossRef] [PubMed]
- Majumdar, R.; Shao, L.; Minocha, R.; Long, S.; Minocha, S.C. Ornithine: The overlooked molecule in the regulation of polyamine metabolism. Plant Cell Physiol. 2013, 54, 990–1004. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yergaliyev, T.M.; Nurbekova, Z.; Mukiyanova, G.; Akbassova, A.; Sutula, M.; Zhangazin, S.; Bari, A.; Tleukulova, Z.; Shamekova, M.; Masalimov, Z.K.; et al. The involvement of ROS producing aldehyde oxidase in plant response to Tombusvirus infection. Plant Physiol. Biochem. 2016, 109, 36–44. [Google Scholar] [CrossRef] [PubMed]
- Yu, L.; Wang, W.; Zeng, S.; Chen, Z.; Yang, A.; Shi, J.; Zhao, X.; Song, B. Label-free quantitative proteomics analysis of Cytosinpeptidemycin responses in southern rice black-streaked dwarf virus-infected rice. Pestic. Biochem. Physiol. 2018, 147, 20–26. [Google Scholar] [CrossRef] [PubMed]
- Shi, J.; Yu, L.; Song, B. Proteomics analysis of Xiangcaoliusuobingmi-treated Capsicum annuum L. infected with Cucumber mosaic. Virus 2018, 149, 113–122. [Google Scholar] [CrossRef]
- Chen, S.; Yu, N.; Yang, S.; Zhong, B.; Lan, H. Identification of Telosma mosaic virus infection in Passiflora edulis and its impact on phytochemical contents. Virol. J. 2018, 15, 168. [Google Scholar] [CrossRef] [Green Version]
- Das, P.P.; Chua, G.M.; Lin, Q.; Wong, S.M. iTRAQ-based analysis of leaf proteome identifies important proteins in secondary metabolite biosynthesis and defence pathways crucial to cross-protection against TMV. J. Proteom. 2019, 196, 42–56. [Google Scholar] [CrossRef]
- Alscher, R.G.; Donahue, J.L.; Cramer, C.L. Reactive oxygen species and antioxidants: Relationships in green cells. Physiol. Plant. 1997, 100, 224–233. [Google Scholar] [CrossRef]
- Sweetlove, L.J.; Heazlewood, J.L.; Herald, V.; Holtzapffel, R.; Day, D.A.; Leaver, C.J.; Millar, A.H. The impact of oxidative stress on Arabidopsis mitochondria. Plant J. Cell Mol. Biol. 2002, 32, 891–904. [Google Scholar] [CrossRef]
- Pastore, D.; Trono, D.; Laus, M.N.; Di Fonzo, N.; Flagella, Z. Possible plant mitochondria involvement in cell adaptation to drought stress. A case study: Durum wheat mitochondria. J. Exp. Bot. 2007, 58, 195–210. [Google Scholar] [CrossRef] [Green Version]

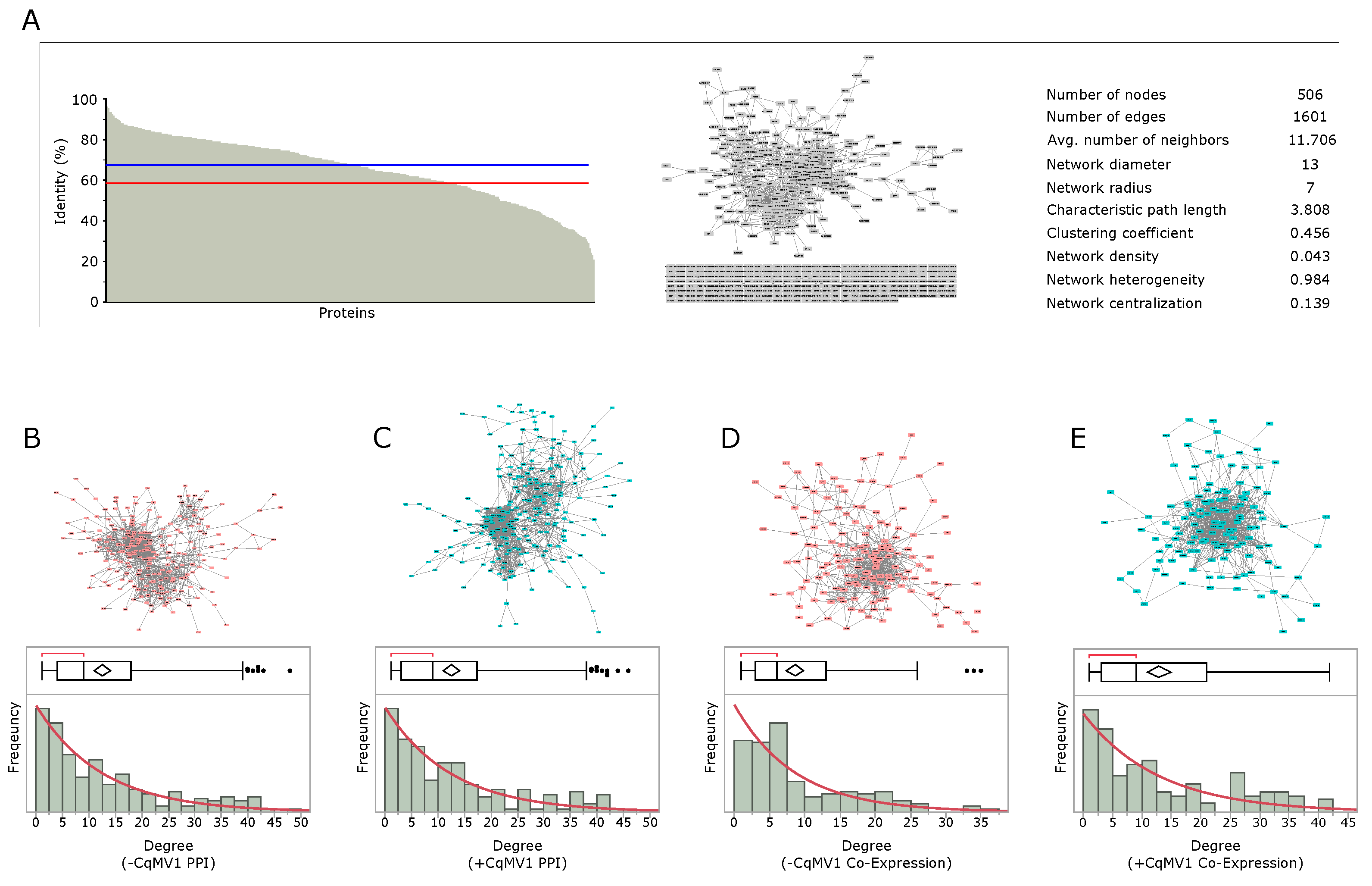
| TOP. PARAMS | PPI NETWORK | Co-Exp NETWORK | ||
|---|---|---|---|---|
| −CqMV1 | +CqMV1 | −CqMV1 | +CqMV1 | |
| Connected components | 1 | 1 | 1 | 1 |
| Numbers of nodes | 217 | 218 | 151 | 130 |
| Numbers of edges | 1350 | 1356 | 651 | 837 |
| Avg. number of neighbors | 12.442 | 12.440 | 8.623 | 12.877 |
| Network diameter | 9 | 13 | 12 | 8 |
| Network radius | 5 | 7 | 6 | 5 |
| Characteristic path length | 3.245 | 3.654 | 3.708 | 2.963 |
| Clustering coefficient | 0.494 | 0.457 | 0.354 | 0.415 |
| Network density | 0.058 | 0.057 | 0.057 | 0.100 |
| Network heterogeneity | 0.938 | 0.933 | 0.866 | 0.899 |
| Network centralization | 0.166 | 0.156 | 0.166 | 0.156 |
| Differential Expression | PPI HUBS | Co-Exp HUBS | |||
|---|---|---|---|---|---|
| DEPS | UP-reg in | −CqMV1 | +CqMV1 | −CqMV1 | +CqMV1 |
| BCE2 | +CqMV1 | X | |||
| DELTA-OAT | +CqMV1 | X | |||
| MCCB | +CqMV1 | X | |||
| CLPB4 | +CqMV1 | X | |||
| GR-RBP2 | +CqMV1 | X | |||
| ATPQ | −CqMV1 | X | |||
| SHM1 | −CqMV1 | X | |||
| ETFALPHA | −CqMV1 | X | |||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Di Silvestre, D.; Passignani, G.; Rossi, R.; Ciuffo, M.; Turina, M.; Vigani, G.; Mauri, P.L. Presence of a Mitovirus Is Associated with Alteration of the Mitochondrial Proteome, as Revealed by Protein–Protein Interaction (PPI) and Co-Expression Network Models in Chenopodium quinoa Plants. Biology 2022, 11, 95. https://doi.org/10.3390/biology11010095
Di Silvestre D, Passignani G, Rossi R, Ciuffo M, Turina M, Vigani G, Mauri PL. Presence of a Mitovirus Is Associated with Alteration of the Mitochondrial Proteome, as Revealed by Protein–Protein Interaction (PPI) and Co-Expression Network Models in Chenopodium quinoa Plants. Biology. 2022; 11(1):95. https://doi.org/10.3390/biology11010095
Chicago/Turabian StyleDi Silvestre, Dario, Giulia Passignani, Rossana Rossi, Marina Ciuffo, Massimo Turina, Gianpiero Vigani, and Pier Luigi Mauri. 2022. "Presence of a Mitovirus Is Associated with Alteration of the Mitochondrial Proteome, as Revealed by Protein–Protein Interaction (PPI) and Co-Expression Network Models in Chenopodium quinoa Plants" Biology 11, no. 1: 95. https://doi.org/10.3390/biology11010095
APA StyleDi Silvestre, D., Passignani, G., Rossi, R., Ciuffo, M., Turina, M., Vigani, G., & Mauri, P. L. (2022). Presence of a Mitovirus Is Associated with Alteration of the Mitochondrial Proteome, as Revealed by Protein–Protein Interaction (PPI) and Co-Expression Network Models in Chenopodium quinoa Plants. Biology, 11(1), 95. https://doi.org/10.3390/biology11010095









