Changes in Transcriptomic Profiles in Different Reproductive Periods in Yaks
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Sample Collection
2.2. H&E Staining
2.3. RNA Sequencing
2.4. Sequencing Analysis
2.5. Real-Time Quantitative PCR (RT-qPCR)
3. Results
3.1. Ovarian Surface Observation
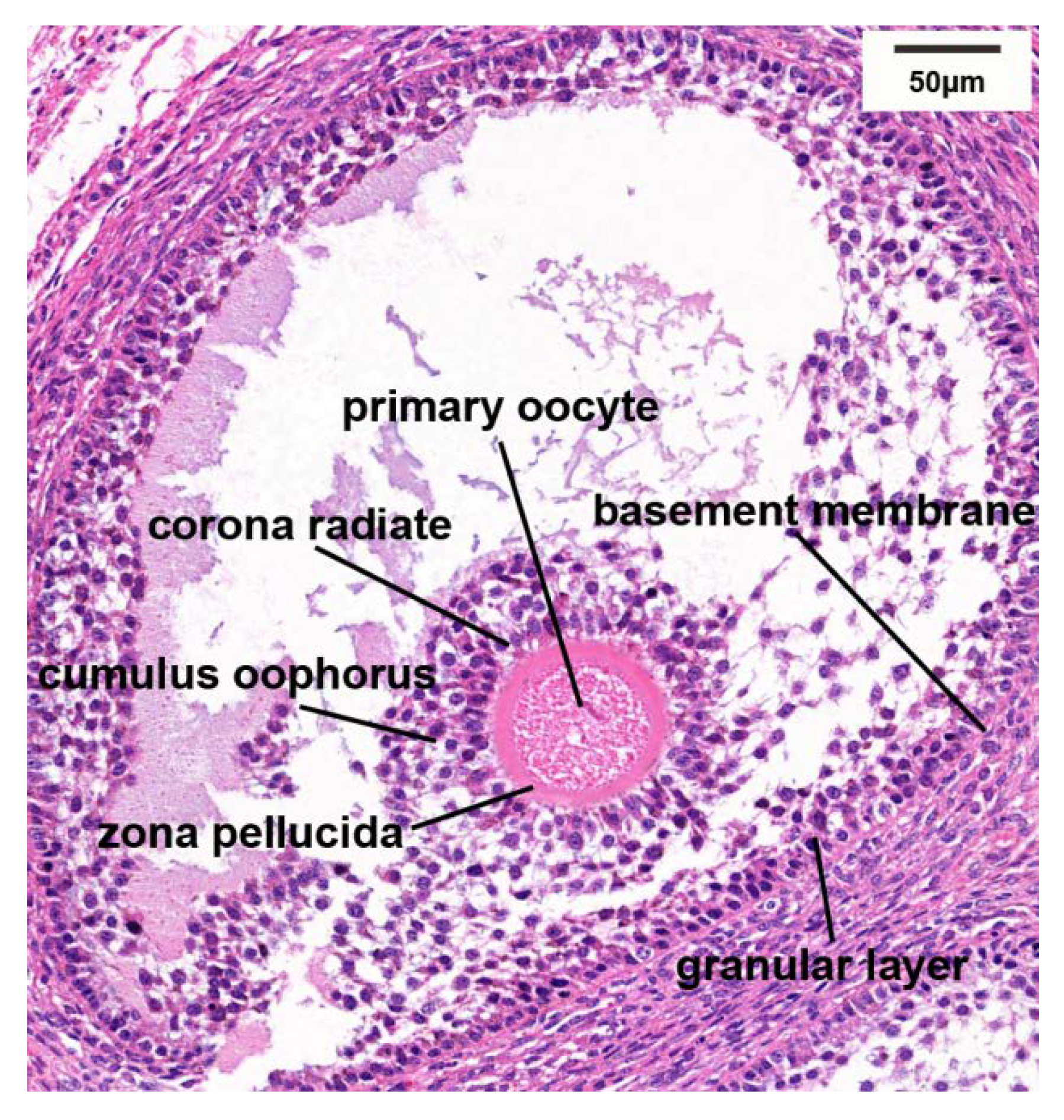
3.2. Histological Characteristics of Follicles
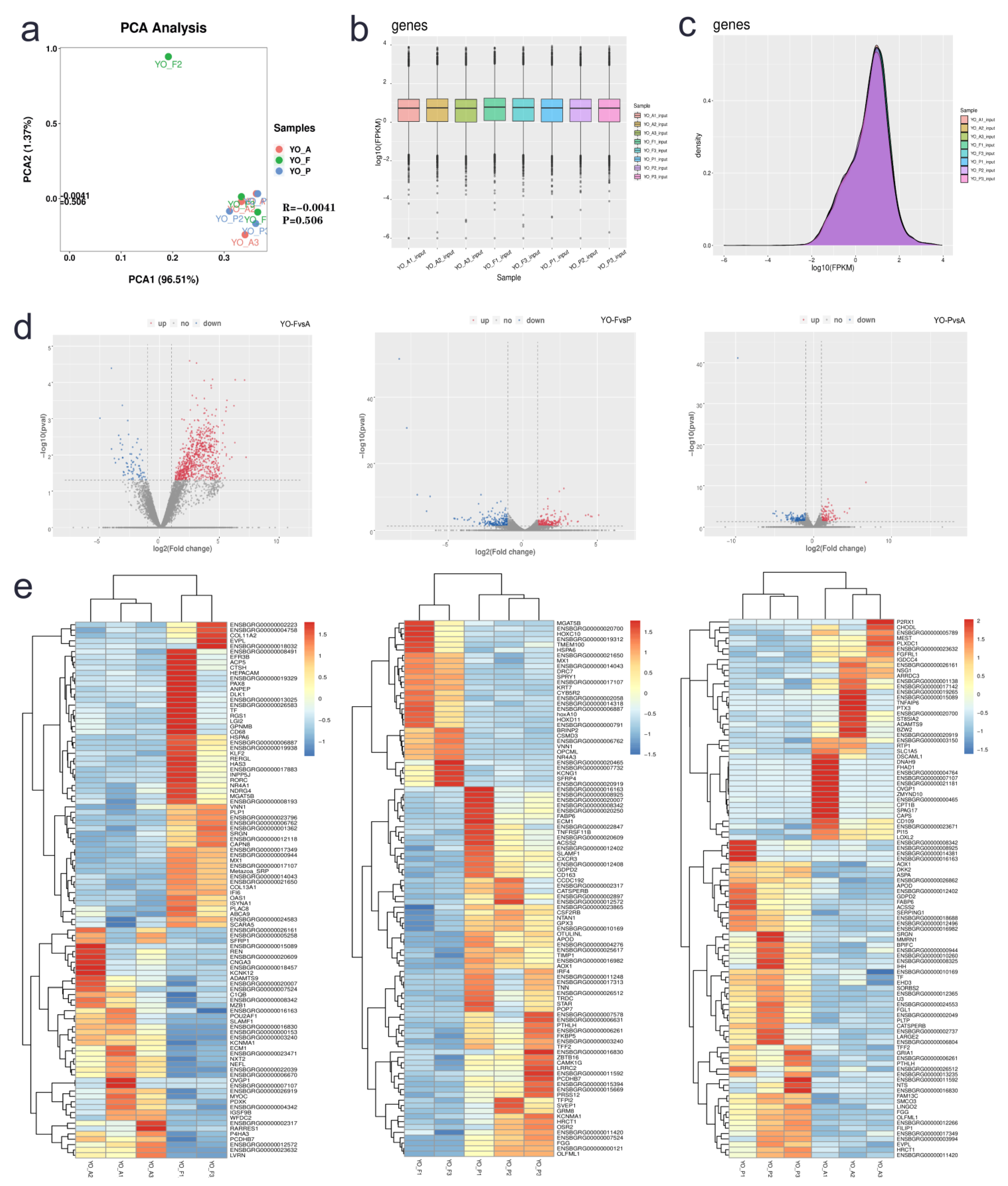
3.3. Raw Sequencing Data and Descriptive Statistics
3.4. Differentially Expressed Gene Identification
3.5. Gene Ontology Analysis of Differentially Expressed Genes
3.6. KEGG Pathway Analysis of Differentially Expressed Genes
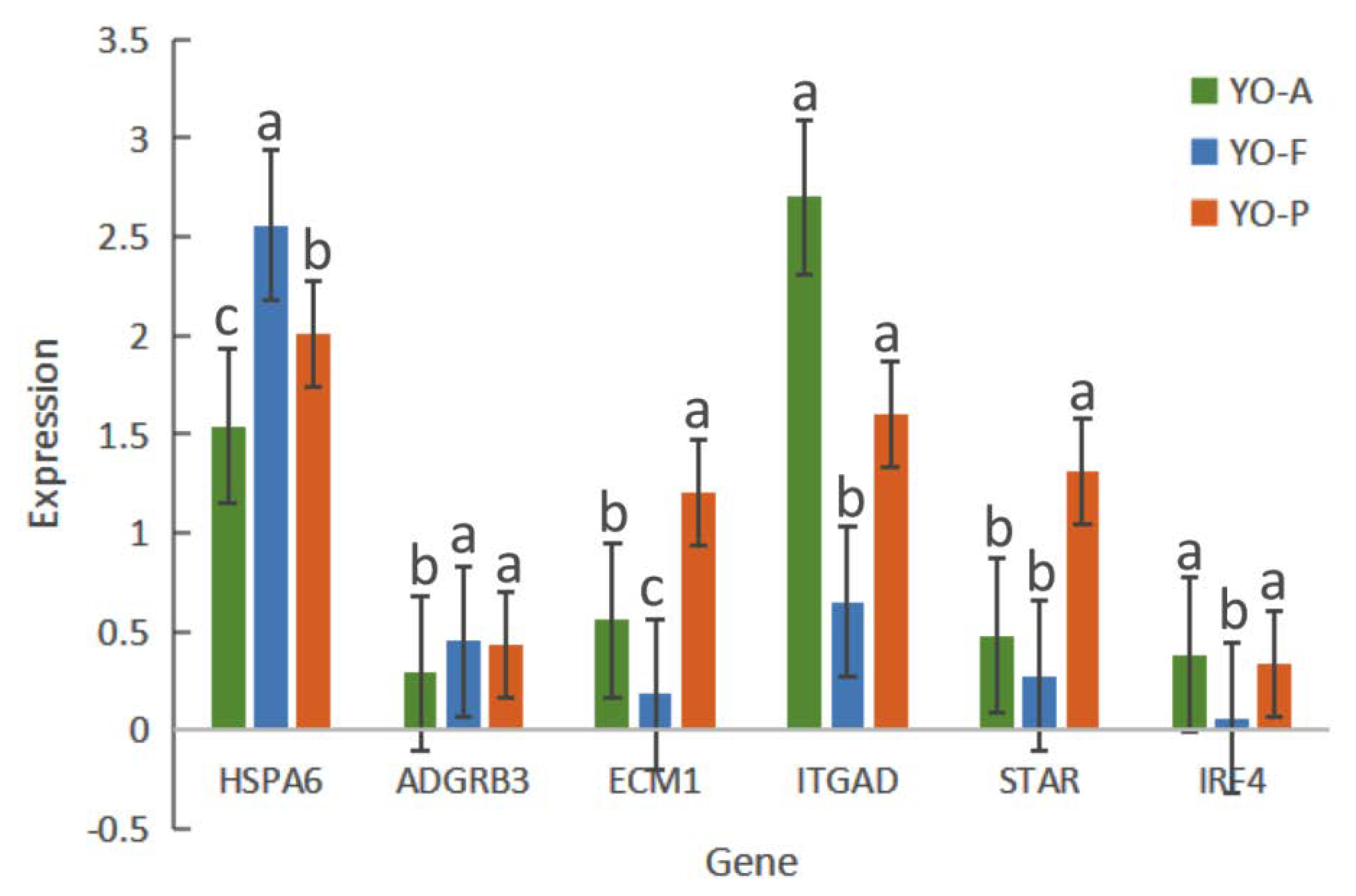
3.7. RT-qPCR Validation
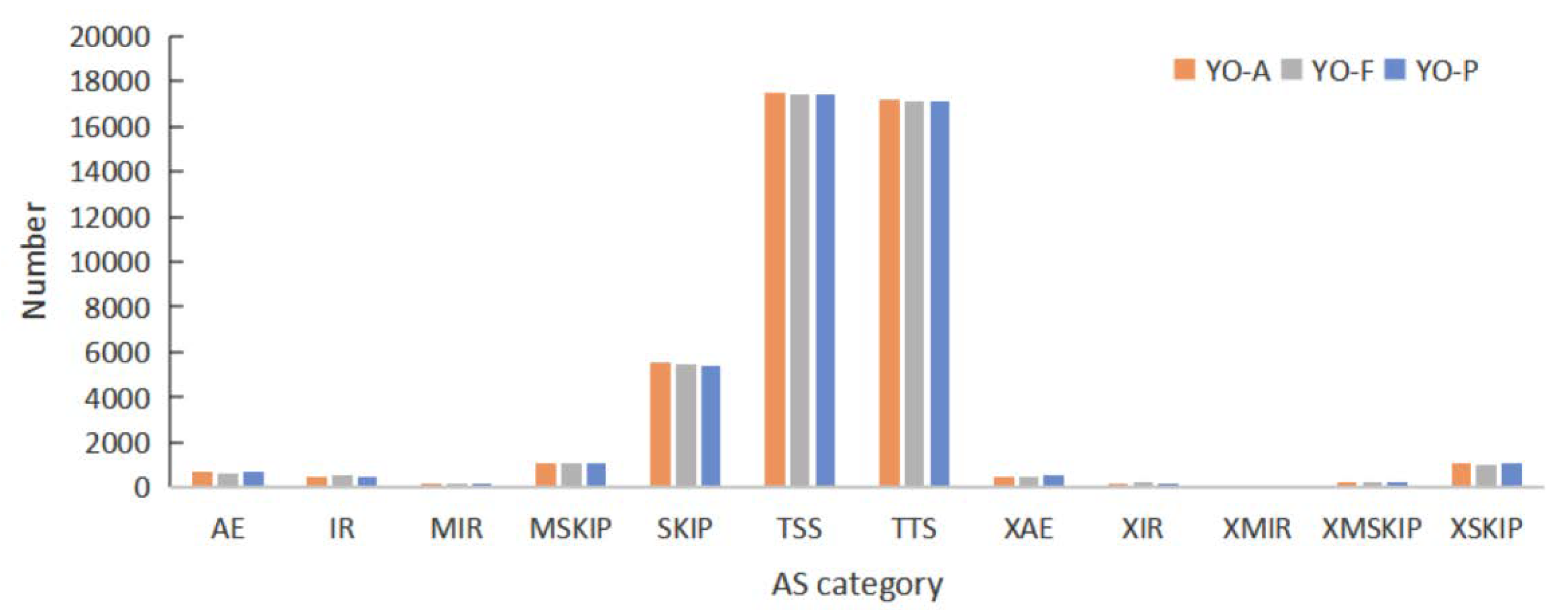
3.8. Alternative Splicing Analysis
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Qiu, Q.; Zhang, G.; Ma, T.; Qian, W.; Wang, J.; Ye, Z.; Cao, C.; Hu, Q.; Kim, J.; Larkin, D.M.; et al. The yak genome and adaptation to life at high altitude. Nat. Genet. 2012, 44, 946–949. [Google Scholar] [CrossRef]
- Xiong, X.; Li, J.; Wang, L.; Zhong, J.; Zi, X.; Wang, Y. Low Oxygen Tension and Relative Defined Culture Medium with 3,4-Dihydroxyflavone are Beneficial for Yak–Bovine Interspecies Somatic Cell Nuclear Transfer Embryo. Reprod. Domest. Anim. 2013, 49, 126–133. [Google Scholar] [CrossRef]
- Zi, X.-D. Reproduction in female yaks (Bos grunniens) and opportunities for improvement. Theriogenology 2003, 59, 1303–1312. [Google Scholar] [CrossRef]
- Prakash, B.S.; Sarkar, M.; Mondal, M. An Update on Reproduction in Yak and Mithun. Reprod. Domest. Anim. 2008, 43, 217–223. [Google Scholar] [CrossRef] [PubMed]
- Xu, F.; Qi, J.; Zhou, Y.; Zhang, Y. The production, breeding and utilization of Chinese yak. J. Grassl. Forage Sci. 2002, 03, 53–55. [Google Scholar]
- Zi, X.; Zhong, J. Progress in yak reproduction science. J. Southwest Minzu Univ. 2007, 01, 79–83. [Google Scholar]
- Cui, Y.; Yu, S. Ovarian Morphology and Follicular Systems in Yaks of Different Ages. Vet. J. 1999, 157, 197–205. [Google Scholar] [CrossRef]
- Cui, Y.; Yu, S. An Anatomical Study of the Internal Genital Organs of the Yak at Different Ages. Vet. J. 1999, 157, 192–196. [Google Scholar] [CrossRef] [PubMed]
- Yu, S.; Yong, Y.; Cui, Y. Oocyte Morphology from Primordial to Early Tertiary Follicles of Yak. Reprod. Domest. Anim. 2009, 45, 779–785. [Google Scholar] [CrossRef]
- Meng, X.L.; Cui, Y.; Yu, S.J.; Liu, X.R. Histological observations of follicular development in ovaries of yaks during estrous cycle. Vet. Sci. China 2006, 36, 57–61. [Google Scholar] [CrossRef]
- Lan, D.; Xiong, X.; Wei, Y.; Xu, T.; Zhong, J.; Zhi, X.; Wang, Y.; Li, J. RNA-Seq analysis of yak ovary: Improving yak gene structure information and mining reproduction-related genes. Sci. China Life Sci. 2014, 57, 925–935. [Google Scholar] [CrossRef] [PubMed]
- Lan, D.; Xiong, X.; Huang, C.; Mipam, T.D.; Li, J. Toward Understanding the Genetic Basis of Yak Ovary Reproduction: A Characterization and Comparative Analyses of Estrus Ovary Transcriptiome in Yak and Cattle. PLoS ONE 2016, 11, e0152675. [Google Scholar] [CrossRef]
- Xu, S.-R.; Wei, P.; Yang, Q.-L.; Jia, G.-X.; Ma, S.-K.; Jun, Z.; Zhang, R.-N. Transcriptome analysis revealed key signaling networks regulating ovarian activities in the domestic yak. Theriogenology 2020, 147, 50–56. [Google Scholar] [CrossRef]
- Chen, S.; Zhou, Y.; Chen, Y.; Gu, J. fastp: An ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 2018, 34, i884–i890. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.; Langmead, B.; Salzberg, S.L. HISAT: A fast spliced aligner with low memory requirements. Nat. Methods 2015, 12, 357–360. [Google Scholar] [CrossRef] [PubMed]
- Pertea, M.; Pertea, G.M.; Antonescu, C.M.; Chang, T.-C.; Mendell, J.T.; Salzberg, S.L. StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nat. Biotechnol. 2015, 33, 290–295. [Google Scholar] [CrossRef]
- Robinson, M.D.; McCarthy, D.J.; Smyth, G.K. edgeR: A Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 2010, 26, 139–140. [Google Scholar] [CrossRef]
- Baralle, F.E.; Giudice, J. Alternative splicing as a regulator of development and tissue identity. Nat. Rev. Mol. Cell Biol. 2017, 18, 437–451. [Google Scholar] [CrossRef]
- Lanes, C.F.C.; Bizuayehu, T.T.; Fernandes, J.M.D.O.; Kiron, V.; Babiak, I. Transcriptome of Atlantic Cod (Gadus morhua L.) Early Embryos from Farmed and Wild Broodstocks. Mar. Biotechnol. 2013, 15, 677–694. [Google Scholar] [CrossRef]
- Bent, Z.W.; Brazel, D.; Tran-Gyamfi, M.B.; Hamblin, R.Y.; VanderNoot, V.A.; Branda, S.S. Use of a Capture-Based Pathogen Transcript Enrichment Strategy for RNA-Seq Analysis of the Francisella Tularensis LVS Transcriptome during Infection of Murine Macrophages. PLoS ONE 2013, 8, e77834. [Google Scholar] [CrossRef]
- Lass, A.; Brinsden, P. The role of ovarian volume in reproductive medicine. Hum. Reprod. Updat. 1999, 5, 256–266. [Google Scholar] [CrossRef]
- McNatty, K.P.; Heath, D.A.; Lundy, T.; Fidler, A.E.; Quirke, L.; O’Connell, A.; Smith, P.; Groome, N.; Tisdall, D.J. Control of early ovarian follicular development. J. Reprod. Fertil. Suppl. 2019, 54, 3–16. [Google Scholar] [CrossRef]
- Webb, R.; Nicholas, B.; Gong, J.G.; Campbell, B.K.; Gutierrez, C.G.; Garverick, H.A.; Armstrong, D.G. Mechanisms regulating follicular development and selection of the dominant follicle. Reprod. Suppl. 2019, 61, 71–90. [Google Scholar] [CrossRef][Green Version]
- Webb, R.; Campbell, B.K. Development of the dominant follicle: Mechanisms of selection and maintenance of oocyte quality. Soc. Reprod. Fertil. Suppl. 2007, 6, 141–163. [Google Scholar] [CrossRef] [PubMed]
- Hatzirodos, N.; Hummitzsch, K.; Irving-Rodgers, H.F.; Harland, M.L.; Morris, S.E.; Rodgers, R.J. Transcriptome profiling of granulosa cells from bovine ovarian follicles during atresia. BMC Genom. 2014, 15, 40. [Google Scholar] [CrossRef]
- Qi, L.; Guo, N.; Wei, Q.; Jin, P.; Wang, W.; Mao, D. The involvement of NR4A1 and NR4A2 in the regulation of the luteal function in rats. Acta Histochem. 2018, 120, 713–719. [Google Scholar] [CrossRef] [PubMed]
- Berisha, B.; Schams, D.; Rodler, D.; Sinowatz, F.; Pfaffl, M.W. Expression and localization of members of the thrombospondin family during final follicle maturation and corpus luteum formation and function in the bovine ovary. J. Reprod. Dev. 2016, 62, 501–510. [Google Scholar] [CrossRef]
- De Cesaro, M.P.; Macedo, M.P.; Santos, J.T.; Rosa, P.R.; Ludke, C.A.; Rissi, V.B.; Gasperin, B.G.; Gonçalves, P.B. Natriuretic peptides stimulate oocyte meiotic resumption in bovine. Anim. Reprod. Sci. 2015, 159, 52–59. [Google Scholar] [CrossRef]
- De Cesaro, M.; dos Santos, J.; Ferst, J.; Nóbrega, J.; Rosa, P.; Rovani, M.; Ilha, G.; Bohrer, R.; Ferreira, R.; Gasperin, B.; et al. Natriuretic peptide system regulation in granulosa cells during follicle deviation and ovulation in cattle. Reprod. Domest. Anim. 2018, 53, 710–717. [Google Scholar] [CrossRef]
- Liu, P.-P.; Chang, H.-M.; Cheng, J.-C.; Leung, P.C.K. Activin A upregulates PTGS2 expression and increases PGE2 production in human granulosa-lutein cells. Reproduction 2016, 152, 655–664. [Google Scholar] [CrossRef]
- Jiménez, L.M.; Binelli, M.; Bertolin, K.; Pelletier, R.M.; Murphy, B.D. Scavenger receptor-B1 and luteal function in mice. J. Lipid Res. 2010, 51, 2362–2371. [Google Scholar] [CrossRef] [PubMed]
- Przygrodzka, E.; Witek, K.; Kaczmarek, M.; Andronowska, A.; Ziecik, A. Expression of factors associated with apoptosis in the porcine corpus luteum throughout the luteal phase of the estrous cycle and early pregnancy: Their possible involvement in acquisition of luteolytic sensitivity. Theriogenology 2015, 83, 535–545. [Google Scholar] [CrossRef] [PubMed]
- Urs, D.B.S.; Wu, W.-H.; Komrskova, K.; Postlerova, P.; Lin, Y.-F.; Tzeng, C.-R.; Kao, S.-H. Mitochondrial Function in Modulating Human Granulosa Cell Steroidogenesis and Female Fertility. Int. J. Mol. Sci. 2020, 21, 3592. [Google Scholar] [CrossRef]
- Fang, L.; Li, Y.; Wang, S.; Yu, Y.; Li, Y.; Guo, Y.; Yan, Y.; Sun, Y.-P. Melatonin induces progesterone production in human granulosa-lutein cells through upregulation of StAR expression. Aging 2019, 11, 9013–9024. [Google Scholar] [CrossRef] [PubMed]
- Mattar, D.; Samir, M.; Laird, M.; Knight, P.G. Modulatory effects of TGF-β1 and BMP6 on thecal angiogenesis and steroidogenesis in the bovine ovary. Reproduction 2020, 159, 397–408. [Google Scholar] [CrossRef] [PubMed]
- Emori, C.; Sugiura, K. Role of oocyte-derived paracrine factors in follicular development. Anim. Sci. J. 2014, 85, 627–633. [Google Scholar] [CrossRef]
- Abir, R.; Fisch, B.; Johnson, M.H. BMP15, fertility and the ovary. Reprod. Biomed. Online 2014, 29, 525–526. [Google Scholar] [CrossRef]
- Mamsen, L.S.; Ernst, E.H.; Borup, R.; Larsen, A.; Olesen, R.H.; Ernst, E.; Anderson, R.A.; Kristensen, S.G.; Andersen, C.Y. Temporal expression pattern of genes during the period of sex differentiation in human embryonic gonads. Sci. Rep. 2017, 7, 15961. [Google Scholar] [CrossRef]
- Hummitzsch, K.; Hatzirodos, N.; Irving-Rodgers, H.F.; Hartanti, M.D.; Perry, V.; Anderson, R.A.; Rodgers, R.J. Morphometric analyses and gene expression related to germ cells, gonadal ridge epithelial-like cells and granulosa cells during development of the bovine fetal ovary. PLoS ONE 2019, 14, e0214130. [Google Scholar] [CrossRef]
- Davoodi, S.; Cooke, R.; Fernandes, A.; Cappellozza, B.; Vasconcelos, J.L.M.; Cerri, R. Expression of estrus modifies the gene expression profile in reproductive tissues on Day 19 of gestation in beef cows. Theriogenology 2016, 85, 645–655. [Google Scholar] [CrossRef]
- Hayashi, K.; Erikson, D.W.; Tilford, S.A.; Bany, B.M.; Maclean, J.A.; Rucker, E.B.; Johnson, G.A.; Spencer, T.E. Wnt Genes in the Mouse Uterus: Potential Regulation of Implantation1. Biol. Reprod. 2009, 80, 989–1000. [Google Scholar] [CrossRef]
- Wang, L.J.; Sun, X.W.; Guo, F.Y.; Zhao, Y.J.; Zhang, J.H.; Zhao, Z.Q. Transcriptome analysis of the uniparous and multiparous goats ovaries. Reprod. Domest. Anim. 2016, 51, 877–885. [Google Scholar] [CrossRef] [PubMed]
- Silvestre, F.; Boni, R.; Fissore, R.; Tosti, E. Ca2+ signaling during maturation of cumulus-oocyte complex in mammals. Mol. Reprod. Dev. 2011, 78, 744–756. [Google Scholar] [CrossRef] [PubMed]
- Chu, Q.; Zhou, B.; Xu, F.; Chen, R.; Shen, C.; Liang, T.; Li, Y.; Schinckel, A.P. Genome-wide differential mRNA expression profiles in follicles of two breeds and at two stages of estrus cycle of gilts. Sci. Rep. 2017, 7, 1–13. [Google Scholar] [CrossRef]
- Walter, J.; Monthoux, C.; Fortes, C.; Grossmann, J.; Roschitzki, B.; Meili, T.; Riond, B.; Hofmann-Lehmann, R.; Naegeli, H.; Bleul, U. The bovine cumulus proteome is influenced by maturation condition and maturational competence of the oocyte. Sci. Rep. 2020, 10, 1–15. [Google Scholar] [CrossRef] [PubMed]
- He, C.; Ma, T.; Shi, J.; Zhang, Z.; Wang, J.; Zhu, K.; Li, Y.; Yang, M.; Song, Y.; Liu, G. Melatonin and its receptor MT1 are involved in the downstream reaction to luteinizing hormone and participate in the regulation of luteinization in different species. J. Pineal Res. 2016, 61, 279–290. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Otsu, K.; Saito, H.; Hiroi, M.; Ishikawa, K. Sandwich Configuration of Type I Collagen Suppresses Progesterone Production in Primary Cultured Porcine Granulosa Cells by Reducing Gene Expression of Cytochrome P450 Cholesterol Side-Chain Cleavage Enzyme. Arch. Biochem. Biophys. 2000, 376, 117–123. [Google Scholar] [CrossRef]
- Cole, T.J.; Short, K.L.; Hooper, S.B. The science of steroids. Semin. Fetal Neonatal Med. 2019, 24, 170–175. [Google Scholar] [CrossRef]
- Adashi, E.Y. Endocrinology of the ovary*. Hum. Reprod. 1994, 9, 815–827. [Google Scholar] [CrossRef]
- Kranc, W.; Brązert, M.; Ożegowska, K.; Nawrocki, M.J.; Budna, J.; Celichowski, P.; Dyszkiewicz-Konwińska, M.; Jankowski, M.; Jeseta, M.; Pawelczyk, L.; et al. Expression Profile of Genes Regulating Steroid Biosynthesis and Metabolism in Human Ovarian Granulosa Cells—A Primary Culture Approach. Int. J. Mol. Sci. 2017, 18, 2673. [Google Scholar] [CrossRef]
- Zamberlam, G.; Lapointe, E.; Abedini, A.; Rico, C.; Godin, P.; Paquet, M.; DeMayo, F.J.; Boerboom, D. SFRP4 Is a Negative Regulator of Ovarian Follicle Development and Female Fertility. Endocrinology 2019, 160, 1561–1572. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.Y.; Shen, H.; Jia, B.; Zhang, Y.S.; Wang, X.H.; Zeng, X.C. Differential Gene Expression in Ovaries of Qira Black Sheep and Hetian Sheep Using RNA-Seq Technique. PLoS ONE 2015, 10, e0120170. [Google Scholar] [CrossRef]
- Wiegel, R.E.; Danser, A.H.J.; Steegers-Theunissen, R.P.M.; Laven, J.S.E.; Willemsen, S.P.; Baker, V.L.; Steegers, E.A.P.; von Versen-Höynck, F. Determinants of Maternal Renin-Angiotensin-Aldosterone-System Activation in Early Pregnancy: Insights From 2 Cohorts. J. Clin. Endocrinol. Metab. 2020, 105, 3505–3517. [Google Scholar] [CrossRef]
- Wiegel, R.E.; von Versen-Höynck, F.; Steegers-Theunissen, R.P.; Steegers, E.A.; Danser, A.J. Prorenin periconceptionally and in pregnancy: Does it have a physiological role? Mol. Cell. Endocrinol. 2020, 522, 111118. [Google Scholar] [CrossRef] [PubMed]
- Fan, H.-Y.; Richards, J.S. Minireview: Physiological and Pathological Actions of RAS in the Ovary. Mol. Endocrinol. 2010, 24, 286–298. [Google Scholar] [CrossRef]
- Robayna, I.G.; Falender, A.E.; Ochsner, S.; Firestone, G.L.; Richards, J.S. Follicle-Stimulating Hormone (FSH) Stimulates Phosphorylation and Activation of Protein Kinase B (PKB/Akt) and Serum and Glucocorticoid-Induced Kinase (Sgk): Evidence for A Kinase-Independent Signaling by FSH in Granulosa Cells. Mol. Endocrinol. 2000, 14, 1283–1300. [Google Scholar] [CrossRef]
- Wang, L.-Q.; Liu, J.-C.; Chen, C.-L.; Cheng, S.-F.; Sun, X.-F.; Zhao, Y.; Yin, S.; Hou, Z.-M.; Pan, B.; Ding, C.; et al. Regulation of primordial follicle recruitment by cross-talk between the Notch and phosphatase and tensin homologue (PTEN)/AKT pathways. Reprod. Fertil. Dev. 2016, 28, 700–712. [Google Scholar] [CrossRef]
- Terenina, E.; Fabre, S.; Bonnet, A.; Monniaux, D.; Robert-Granie, C.; SanCristobal, M.; Sarry, J.; Vignoles, F.; Gondret, F.; Monget, P.; et al. Differentially expressed genes and gene networks involved in pig ovarian follicular atresia. Physiol. Genom. 2017, 49, 67–80. [Google Scholar] [CrossRef]
- Bacich, D.; Earl, C.; O’Keefe, D.; Norman, R.; Rodgers, R. Characterization of the translated products of the alternatively spliced luteinizing hormone receptor in the ovine ovary throughout the oestrous cycle. Mol. Cell. Endocrinol. 1999, 147, 113–124. [Google Scholar] [CrossRef]
- Sullivan, R.R.; Faris, B.R.; Eborn, D.; Grieger, D.M.; Cino-Ozuna, A.G.; Rozell, T.G. Follicular expression of follicle stimulating hormone receptor variants in the ewe. Reprod. Biol. Endocrinol. 2013, 11, 113. [Google Scholar] [CrossRef]
- Qiu, Y.; Seager, M.; Osman, A.; Castle-Miller, J.; Bevan, H.; Tortonese, D.J.; Murphy, D.; Harper, S.J.; Fraser, H.M.; Donaldson, L.F.; et al. Ovarian VEGF165b expression regulates follicular development, corpus luteum function and fertility. Reproduction 2012, 143, 501–511. [Google Scholar] [CrossRef] [PubMed]
- Tang, L.-T.; Ran, X.-Q.; Mao, N.; Zhang, F.-P.; Niu, X.; Ruan, Y.-Q.; Yi, F.-L.; Li, S.; Wang, J.-F. Analysis of alternative splicing events by RNA sequencing in the ovaries of Xiang pig at estrous and diestrous. Theriogenology 2018, 119, 60–68. [Google Scholar] [CrossRef] [PubMed]
- Fayad, T.; Lefebvre, R.; Nimpf, J.; Silversides, D.W.; Lussier, J.G. Low-Density Lipoprotein Receptor-Related Protein 8 (LRP8) Is Upregulated in Granulosa Cells of Bovine Dominant Follicle: Molecular Characterization and Spatio-Temporal Expression Studies. Biol. Reprod. 2007, 76, 466–475. [Google Scholar] [CrossRef] [PubMed][Green Version]
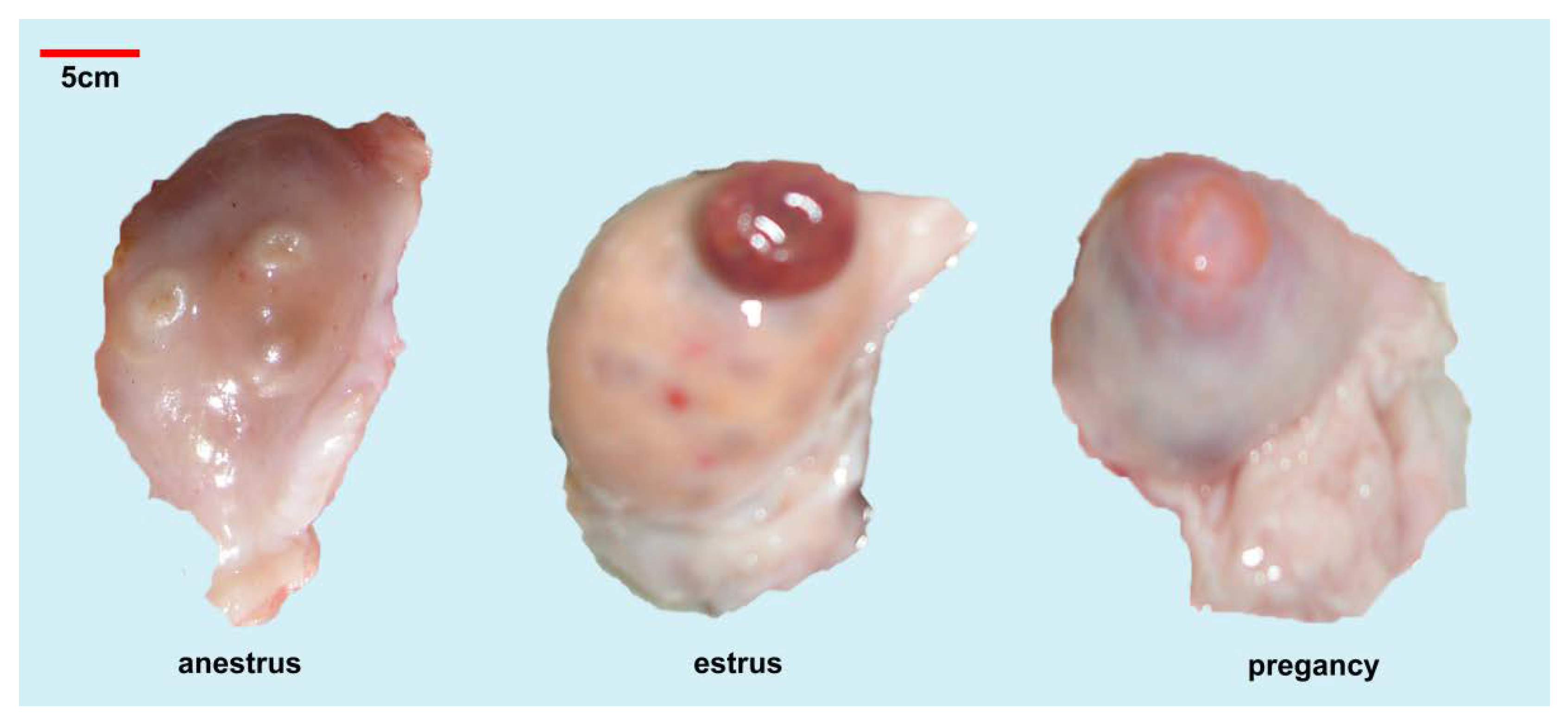


| Period | Size | Corpus Luteum or Corpus Rubrum | Number of Differing Follicles | ||
|---|---|---|---|---|---|
| 2–3 mm | 4–5 mm | >5 mm | |||
| Anestrus | 22.5 mm | none | 9 | 1.5 | 1 |
| Estrus | 23.5 mm | corpus rubrum/corpus luteum | 5 | 2 | 1 |
| Pregnancy | 24.7 mm | corpus luteum | 4.7 | 3.5 | 0.7 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Guo, S.; Cao, M.; Wang, X.; Xiong, L.; Wu, X.; Bao, P.; Chu, M.; Liang, C.; Yan, P.; Pei, J.; et al. Changes in Transcriptomic Profiles in Different Reproductive Periods in Yaks. Biology 2021, 10, 1229. https://doi.org/10.3390/biology10121229
Guo S, Cao M, Wang X, Xiong L, Wu X, Bao P, Chu M, Liang C, Yan P, Pei J, et al. Changes in Transcriptomic Profiles in Different Reproductive Periods in Yaks. Biology. 2021; 10(12):1229. https://doi.org/10.3390/biology10121229
Chicago/Turabian StyleGuo, Shaoke, Mengli Cao, Xingdong Wang, Lin Xiong, Xiaoyun Wu, Pengjia Bao, Min Chu, Chunnian Liang, Ping Yan, Jie Pei, and et al. 2021. "Changes in Transcriptomic Profiles in Different Reproductive Periods in Yaks" Biology 10, no. 12: 1229. https://doi.org/10.3390/biology10121229
APA StyleGuo, S., Cao, M., Wang, X., Xiong, L., Wu, X., Bao, P., Chu, M., Liang, C., Yan, P., Pei, J., & Guo, X. (2021). Changes in Transcriptomic Profiles in Different Reproductive Periods in Yaks. Biology, 10(12), 1229. https://doi.org/10.3390/biology10121229






