De Novo Reconstruction of Transcriptome Identified Long Non-Coding RNA Regulator of Aging-Related Brown Adipose Tissue Whitening in Rabbits
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Ethics Approval
2.2. Tissue Sample Preparation, Histological Analysis and Immunohistochemistry (IHC)
2.3. SsRNA-seq and lncRNA Identification
2.4. Transcriptomic Quantification and Differential Expression Analysis
2.5. Cell Culture and Plasmid-Mediated Overexpression
2.6. Quantitative Real-Time PCR (qRT-PCR)
2.7. Statistical Analysis
3. Results
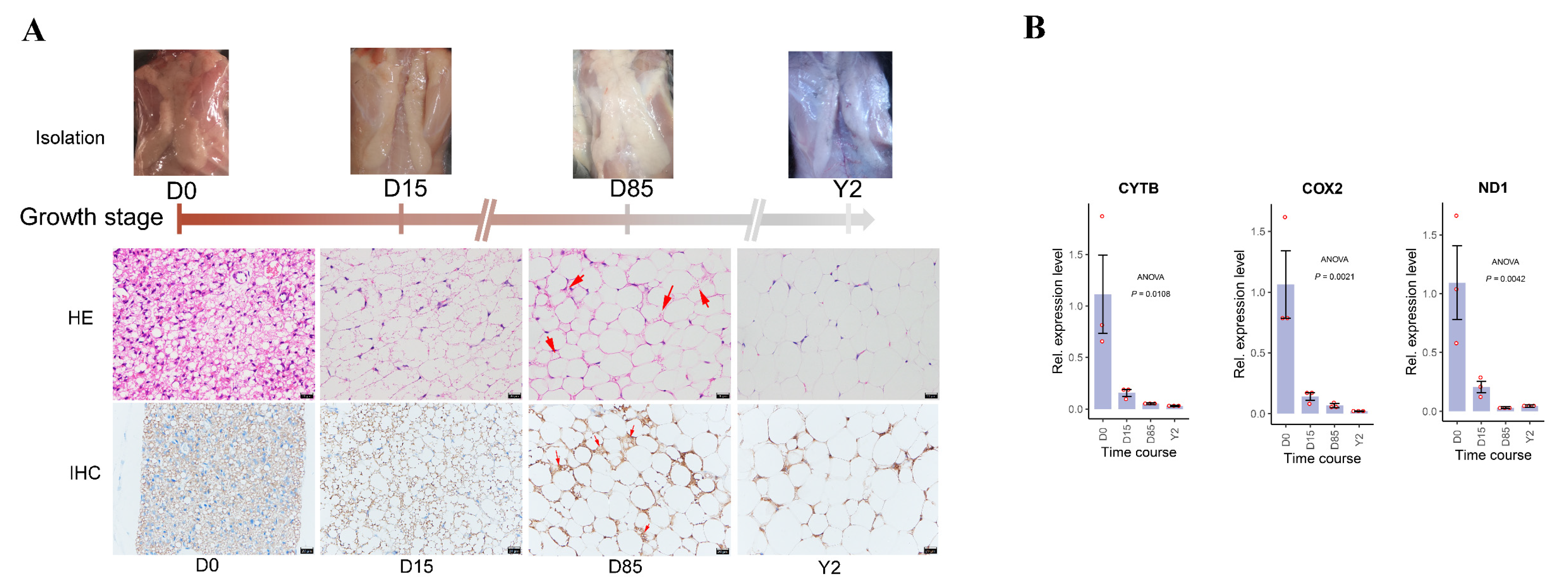
3.1. Histological Dynamics of ARBW in Rabbits
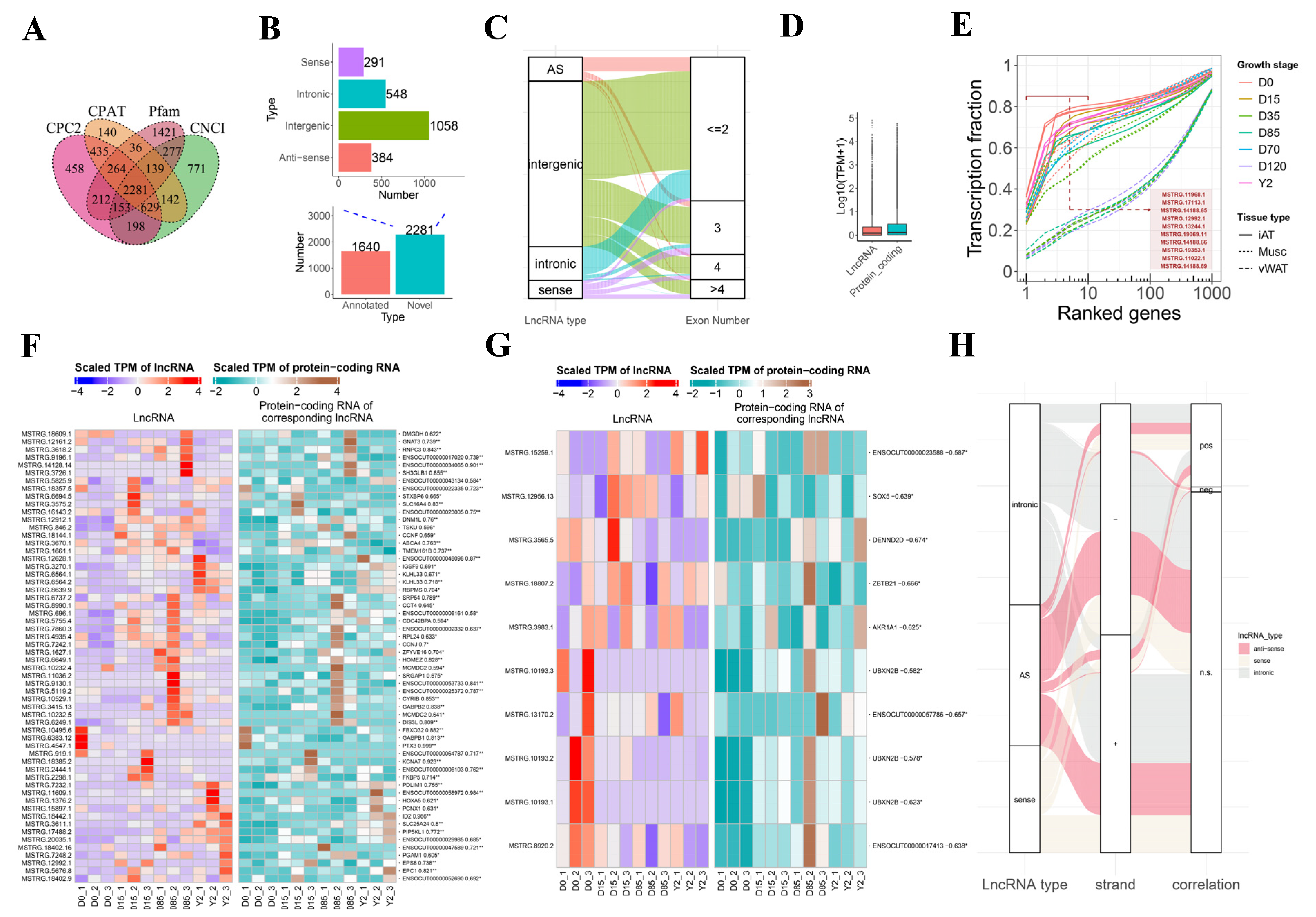
3.2. Identification and Characterization of lncRNAs in Rabbits iATs
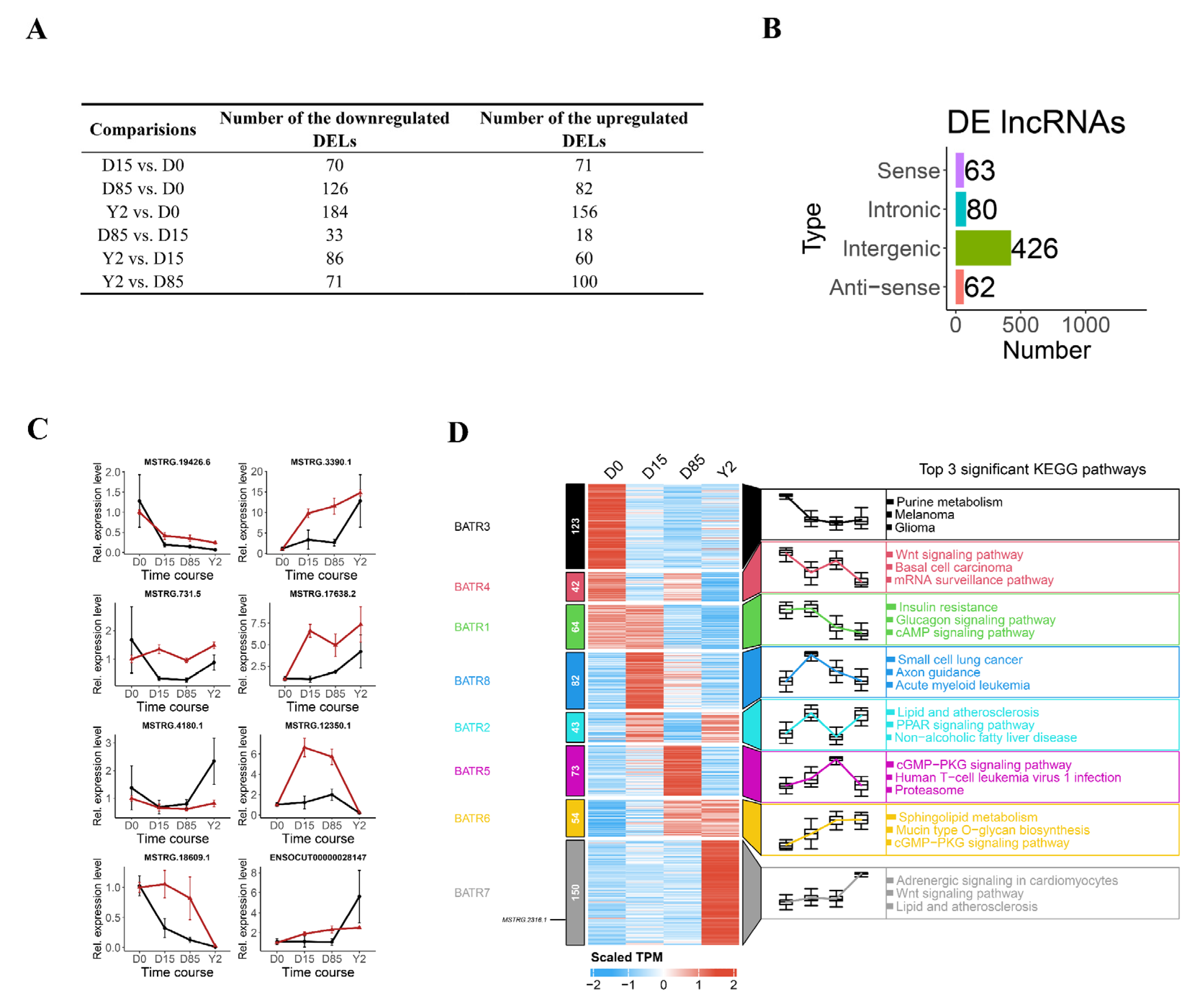
3.3. Dynamics of lncRNA Expression during ARBW of Rabbit iATs
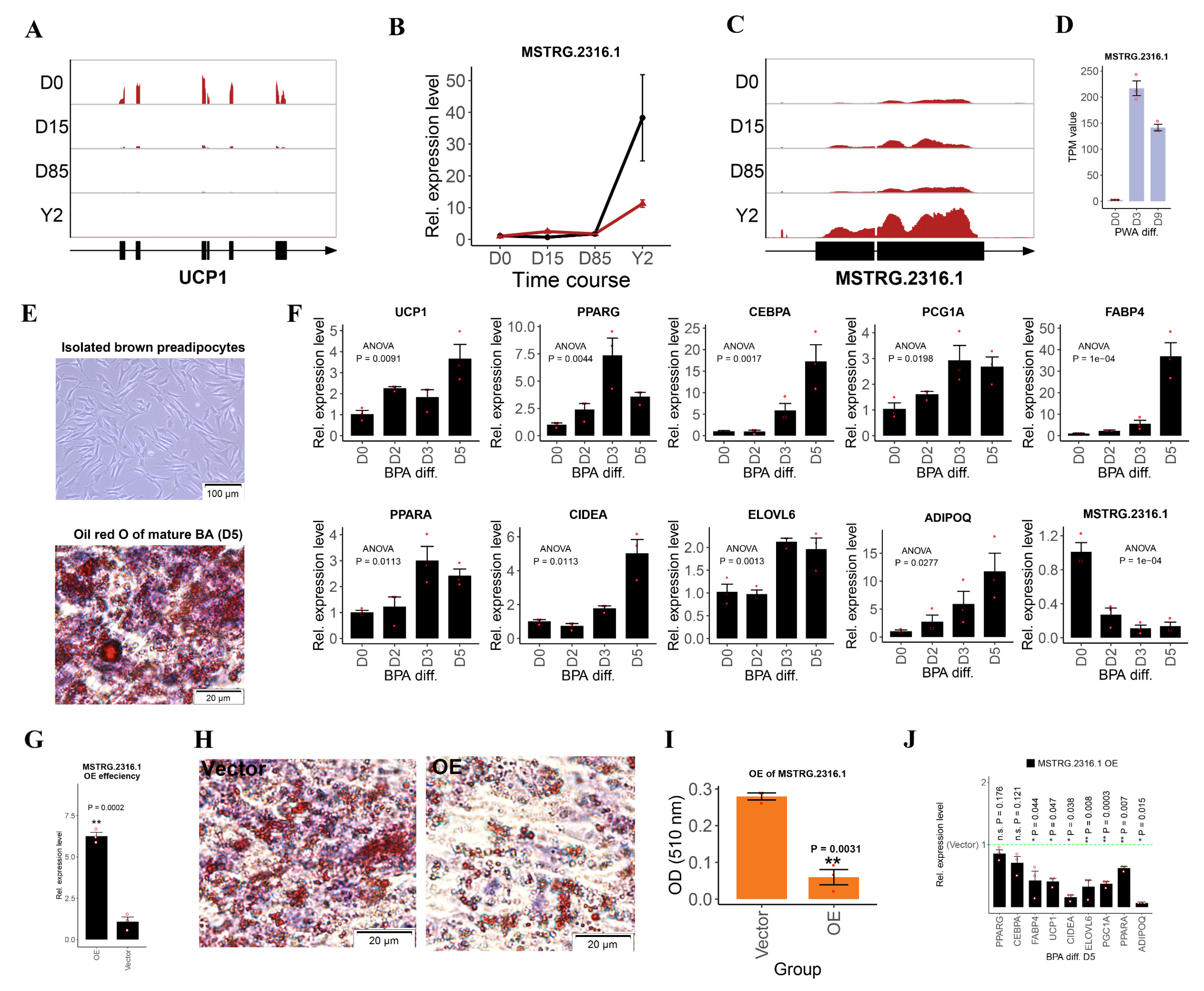
3.4. Selection of lncRNA Candidates and Functional Validation of lncRNA MSTRG.2316.1
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Peirce, V.; Carobbio, S.; Vidal-Puig, A. The different shades of fat. Nature 2014, 510, 76–83. [Google Scholar] [CrossRef]
- Alvarez-Dominguez, J.R.; Bai, Z.; Xu, D.; Yuan, B.; Lo, K.A.; Yoon, M.J.; Lim, Y.C.; Knoll, M.; Slavov, N.; Chen, S.; et al. De Novo Reconstruction of Adipose Tissue Transcriptomes Reveals Long Non-coding RNA Regulators of Brown Adipocyte Development. Cell Metab. 2015, 21, 764–776. [Google Scholar] [CrossRef] [Green Version]
- van Marken Lichtenbelt, W.D.; Vanhommerig, J.W.; Smulders, N.M.; Drossaerts, J.M.; Kemerink, G.J.; Bouvy, N.D.; Schrauwen, P.; Teule, G.J. Cold-activated brown adipose tissue in healthy men. N. Engl. J. Med. 2009, 360, 1500–1508. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Saito, M.; Okamatsu-Ogura, Y.; Matsushita, M.; Watanabe, K.; Yoneshiro, T.; Nio-Kobayashi, J.; Iwanaga, T.; Miyagawa, M.; Kameya, T.; Nakada, K.; et al. High incidence of metabolically active brown adipose tissue in healthy adult humans: Effects of cold exposure and adiposity. Diabetes 2009, 58, 1526–1531. [Google Scholar] [CrossRef] [Green Version]
- Bartelt, A.; Widenmaier, S.B.; Schlein, C.; Johann, K.; Goncalves, R.L.S.; Eguchi, K.; Fischer, A.W.; Parlakgül, G.; Snyder, N.A.; Nguyen, T.B.; et al. Brown adipose tissue thermogenic adaptation requires Nrf1-mediated proteasomal activity. Nat. Med. 2018, 24, 292–303. [Google Scholar] [CrossRef] [PubMed]
- Cypess, A.M.; Lehman, S.; Williams, G.; Tal, I.; Rodman, D.; Goldfine, A.B.; Kuo, F.C.; Palmer, E.L.; Tseng, Y.H.; Doria, A.; et al. Identification and importance of brown adipose tissue in adult humans. N. Engl. J. Med. 2009, 360, 1509–1517. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Derry, D.M.; Morrow, E.; Sadre, N.; Flattery, K.V. Brown and white fat during the life of the rabbit. Dev. Biol. 1972, 27, 204–216. [Google Scholar] [CrossRef]
- Lim, S.; Honek, J.; Xue, Y.; Seki, T.; Cao, Z.; Andersson, P.; Yang, X.; Hosaka, K.; Cao, Y. Cold-induced activation of brown adipose tissue and adipose angiogenesis in mice. Nat. Protoc. 2012, 7, 606–615. [Google Scholar] [CrossRef]
- Richard, M.A.; Pallubinsky, H.; Blondin, D.P. Functional characterization of human brown adipose tissue metabolism. Biochem. J. 2020, 477, 1261–1286. [Google Scholar] [CrossRef] [Green Version]
- Dawkins, M.J.; Hull, D. Brown Adipose Tissue and the Response of New-Born Rabbits to Cold. J. Physiol. 1964, 172, 216–238. [Google Scholar] [CrossRef] [Green Version]
- Schlein, C.; Fischer, A.W.; Sass, F.; Worthmann, A.; Tödter, K.; Jaeckstein, M.Y.; Behrens, J.; Lynes, M.D.; Kiebish, M.A.; Narain, N.R.; et al. Endogenous Fatty Acid Synthesis Drives Brown Adipose Tissue Involution. Cell Rep. 2021, 34, 108624. [Google Scholar] [CrossRef]
- Batista, P.J.; Chang, H.Y. Long noncoding RNAs: Cellular address codes in development and disease. Cell 2013, 152, 1298–1307. [Google Scholar] [CrossRef] [Green Version]
- St Laurent, G.; Wahlestedt, C.; Kapranov, P. The Landscape of long noncoding RNA classification. Trends Genet. 2015, 31, 239–251. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kuang, L.; Lei, M.; Li, C.; Zhang, X.; Ren, Y.; Zheng, J.; Guo, Z.; Zhang, C.; Yang, C.; Mei, X.; et al. Identification of Long Non-Coding RNAs Related to Skeletal Muscle Development in Two Rabbit Breeds with Different Growth Rate. Int. J. Mol. Sci. 2018, 19, 2046. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Robinson, E.K.; Covarrubias, S.; Carpenter, S. The how and why of lncRNA function: An innate immune perspective. Biochim. Biophys. Acta Gene Regul. Mech. 2020, 1863, 194419. [Google Scholar] [CrossRef] [PubMed]
- Peng, W.X.; Koirala, P.; Mo, Y.Y. LncRNA-mediated regulation of cell signaling in cancer. Oncogene 2017, 36, 5661–5667. [Google Scholar] [CrossRef] [PubMed]
- Bai, Z.; Chai, X.R.; Yoon, M.J.; Kim, H.J.; Lo, K.A.; Zhang, Z.C.; Xu, D.; Siang, D.T.C.; Walet, A.C.E.; Xu, S.H.; et al. Dynamic transcriptome changes during adipose tissue energy expenditure reveal critical roles for long noncoding RNA regulators. PLoS Biol. 2017, 15, e2002176. [Google Scholar] [CrossRef] [PubMed]
- Mi, L.; Zhao, X.Y.; Li, S.; Yang, G.; Lin, J.D. Conserved function of the long noncoding RNA Blnc1 in brown adipocyte differentiation. Mol. Metab. 2017, 6, 101–110. [Google Scholar] [CrossRef]
- Ding, C.; Lim, Y.C.; Chia, S.Y.; Walet, A.C.E.; Xu, S.; Lo, K.A.; Zhao, Y.; Zhu, D.; Shan, Z.; Chen, Q.; et al. De novo reconstruction of human adipose transcriptome reveals conserved lncRNAs as regulators of brown adipogenesis. Nat. Commun. 2018, 9, 1329. [Google Scholar] [CrossRef] [Green Version]
- Wang, G.Z.; Du, K.; Hu, S.Q.; Chen, S.Y.; Jia, X.B.; Cai, M.C.; Shi, Y.; Wang, J.; Lai, S.J. Genome-wide identification and characterization of long non-coding RNAs during postnatal development of rabbit adipose tissue. Lipids Health Dis. 2018, 17, 271. [Google Scholar] [CrossRef] [Green Version]
- Du, K.; Wang, G.Z.; Ren, A.Y.; Cai, M.C.; Luo, G.; Jia, X.B.; Hu, S.Q.; Wang, J.; Chen, S.Y.; Lai, S.J. Genome-wide identification and characterization of long non-coding RNAs during differentiation of visceral preadipocytes in rabbit. Funct. Integr. Genom. 2020, 20, 409–419. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; He, Z.; Zhang, D.; Li, H.; Wang, Z. Using oxidation kinetic models to predict the quality indices of rabbit meat under different storage temperatures. Meat Sci. 2020, 162, 108042. [Google Scholar] [CrossRef] [PubMed]
- Cullere, M.; Dalle Zotte, A. Rabbit meat production and consumption: State of knowledge and future perspectives. Meat Sci. 2018, 143, 137–146. [Google Scholar] [CrossRef] [PubMed]
- Dalle Zotte, A.; Szendro, Z. The role of rabbit meat as functional food. Meat Sci. 2011, 88, 319–331. [Google Scholar] [CrossRef] [PubMed]
- Wang, G.; Du, K.; Xie, Z.; Tang, R.; Jia, X.; Chen, S.; Lai, S. Screening and Identification of Differentially Expressed and Adipose Growth-Related Protein-Coding Genes During the Deposition of Perirenal Adipose Tissue in Rabbits. Diabetes Metab. Syndr. Obes. 2020, 13, 4669–4680. [Google Scholar] [CrossRef] [PubMed]
- Brown, J.; Pirrung, M.; McCue, L.A. FQC Dashboard: Integrates FastQC results into a web-based, interactive, and extensible FASTQ quality control tool. Bioinformatics 2017, 33, 3137–3139. [Google Scholar] [CrossRef] [Green Version]
- Martin, M. Cutadapt removes adapter sequences from high-throughput sequencing reads. Embnet J. 2011, 17, 10–12. [Google Scholar] [CrossRef]
- Kim, D.; Langmead, B.; Salzberg, S.L. HISAT: A fast spliced aligner with low memory requirements. Nat. Methods 2015, 12, 357–360. [Google Scholar] [CrossRef] [Green Version]
- Pertea, M.; Pertea, G.M.; Antonescu, C.M.; Chang, T.C.; Mendell, J.T.; Salzberg, S.L. StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nat. Biotechnol. 2015, 33, 290–295. [Google Scholar] [CrossRef] [Green Version]
- Trapnell, C.; Roberts, A.; Goff, L.; Pertea, G.; Kim, D.; Kelley, D.R.; Pimentel, H.; Salzberg, S.L.; Rinn, J.L.; Pachter, L. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat. Protoc. 2012, 7, 562–578. [Google Scholar] [CrossRef] [Green Version]
- Kang, Y.J.; Yang, D.C.; Kong, L.; Hou, M.; Meng, Y.Q.; Wei, L.; Gao, G. CPC2: A fast and accurate coding potential calculator based on sequence intrinsic features. Nucleic Acids Res. 2017, 45, W12–W16. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, L.; Park, H.J.; Dasari, S.; Wang, S.; Kocher, J.P.; Li, W. CPAT: Coding-Potential Assessment Tool using an alignment-free logistic regression model. Nucleic Acids Res. 2013, 41, e74. [Google Scholar] [CrossRef] [PubMed]
- Sun, L.; Luo, H.; Bu, D.; Zhao, G.; Yu, K.; Zhang, C.; Liu, Y.; Chen, R.; Zhao, Y. Utilizing sequence intrinsic composition to classify protein-coding and long non-coding transcripts. Nucleic Acids Res. 2013, 41, e166. [Google Scholar] [CrossRef] [PubMed]
- Finn, R.D.; Bateman, A.; Clements, J.; Coggill, P.; Eberhardt, R.Y.; Eddy, S.R.; Heger, A.; Hetherington, K.; Holm, L.; Mistry, J. Pfam: The protein families database. Nucleic Acids Res. 2014, 42, 222–230. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kutter, C.; Watt, S.; Stefflova, K.; Wilson, M.D.; Goncalves, A.; Ponting, C.P.; Odom, D.T.; Marques, A.C.; Bartel, D.P. Rapid Turnover of Long Noncoding RNAs and the Evolution of Gene Expression. PLoS Genet. 2012, 8, e1002841. [Google Scholar] [CrossRef]
- Yan, P.; Luo, S.; Lu, J.Y.; Shen, X. Cis- and trans-acting lncRNAs in pluripotency and reprogramming. Curr. Opin. Genet. Dev. 2017, 46, 170–178. [Google Scholar] [CrossRef] [PubMed]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef] [Green Version]
- Gu, Z.; Eils, R.; Schlesner, M. Complex heatmaps reveal patterns and correlations in multidimensional genomic data. Bioinformatics 2016, 32, 2847–2849. [Google Scholar] [CrossRef] [Green Version]
- Yu, G.; Wang, L.G.; Han, Y.; He, Q.Y. clusterProfiler: An R package for comparing biological themes among gene clusters. Omics 2012, 16, 284–287. [Google Scholar] [CrossRef]
- Wang, L.; Yang, X.; Zhu, Y.; Zhan, S.; Chao, Z.; Zhong, T.; Guo, J.; Wang, Y.; Li, L.; Zhang, H. Genome-Wide Identification and Characterization of Long Noncoding RNAs of Brown to White Adipose Tissue Transformation in Goats. Cells 2019, 8, 904. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Casteilla, L.; Champigny, O.; Bouillaud, F.; Robelin, J.; Ricquier, D. Sequential changes in the expression of mitochondrial protein mRNA during the development of brown adipose tissue in bovine and ovine species. Sudden occurrence of uncoupling protein mRNA during embryogenesis and its disappearance after birth. Biochem. J. 1989, 257, 665–671. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mure, L.S.; Le, H.D.; Benegiamo, G.; Chang, M.W.; Rios, L.; Jillani, N.; Ngotho, M.; Kariuki, T.; Dkhissi-Benyahya, O.; Cooper, H.M.; et al. Diurnal transcriptome atlas of a primate across major neural and peripheral tissues. Science 2018, 359, eaao0318. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cai, R.; Sun, Y.; Qimuge, N.; Wang, G.; Wang, Y.; Chu, G.; Yu, T.; Yang, G.; Pang, W. Adiponectin AS lncRNA inhibits adipogenesis by transferring from nucleus to cytoplasm and attenuating Adiponectin mRNA translation. Biochim. Biophys. Acta Mol. Cell Biol. Lipids 2018, 1863, 420–432. [Google Scholar] [CrossRef]
- You, L.; Zhou, Y.; Cui, X.; Wang, X.; Sun, Y.; Gao, Y.; Wang, X.; Wen, J.; Xie, K.; Tang, R.; et al. GM13133 is a negative regulator in mouse white adipocytes differentiation and drives the characteristics of brown adipocytes. J. Cell. Physiol. 2018, 233, 313–324. [Google Scholar] [CrossRef] [PubMed]
- Lai, S.J.; Du, K.; Shi, Y.; Li, C.; Wnag, G.Z.; Hu, S.Q.; Jia, X.B.; Wang, J.; Chen, S.Y. Long Non-Coding RNAs in Brown Adipose Tissue. Diabetes Metab. Syndr. Obes. 2020, 13, 3193–3204. [Google Scholar] [CrossRef]
- Bast-Habersbrunner, A.; Fromme, T. Purine Nucleotides in the Regulation of Brown Adipose Tissue Activity. Front. Endocrinol. (Lausanne) 2020, 11, 118. [Google Scholar] [CrossRef]
- Kang, S.; Bajnok, L.; Longo, K.A.; Petersen, R.K.; Hansen, J.B.; Kristiansen, K.; MacDougald, O.A. Effects of Wnt signaling on brown adipocyte differentiation and metabolism mediated by PGC-1alpha. Mol. Cell. Biol. 2005, 25, 1272–1282. [Google Scholar] [CrossRef] [Green Version]
- Kuipers, E.N.; Held, N.M.; In Het Panhuis, W.; Modder, M.; Ruppert, P.M.M.; Kersten, S.; Kooijman, S.; Guigas, B.; Houtkooper, R.H.; Rensen, P.C.N.; et al. A single day of high-fat diet feeding induces lipid accumulation and insulin resistance in brown adipose tissue in mice. Am. J. Physiol. Endocrinol. Metab. 2019, 317, E820–E830. [Google Scholar] [CrossRef]
- Liu, D.; Ceddia, R.P.; Collins, S. Cardiac natriuretic peptides promote adipose ‘browning’ through mTOR complex-1. Mol. Metab. 2018, 9, 192–198. [Google Scholar] [CrossRef]
- Xiong, Y.; Yue, F.; Jia, Z.; Gao, Y.; Jin, W.; Hu, K.; Zhang, Y.; Zhu, D.; Yang, G.; Kuang, S. A novel brown adipocyte-enriched long non-coding RNA that is required for brown adipocyte differentiation and sufficient to drive thermogenic gene program in white adipocytes. Biochim. Biophys. Acta Mol. Cell Biol. Lipids 2018, 1863, 409–419. [Google Scholar] [CrossRef]
- da Silva, I.V.; Díaz-Sáez, F.; Zorzano, A.; Gumà, A.; Camps, M.; Soveral, G. Aquaglyceroporins Are Differentially Expressed in Beige and White Adipocytes. Int. J. Mol. Sci. 2020, 21, 610. [Google Scholar] [CrossRef] [PubMed] [Green Version]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Du, K.; Bai, X.; Yang, L.; Shi, Y.; Chen, L.; Wang, H.; Cai, M.; Wang, J.; Chen, S.; Jia, X.; et al. De Novo Reconstruction of Transcriptome Identified Long Non-Coding RNA Regulator of Aging-Related Brown Adipose Tissue Whitening in Rabbits. Biology 2021, 10, 1176. https://doi.org/10.3390/biology10111176
Du K, Bai X, Yang L, Shi Y, Chen L, Wang H, Cai M, Wang J, Chen S, Jia X, et al. De Novo Reconstruction of Transcriptome Identified Long Non-Coding RNA Regulator of Aging-Related Brown Adipose Tissue Whitening in Rabbits. Biology. 2021; 10(11):1176. https://doi.org/10.3390/biology10111176
Chicago/Turabian StyleDu, Kun, Xue Bai, Li Yang, Yu Shi, Li Chen, Haoding Wang, Mingchen Cai, Jie Wang, Shiyi Chen, Xianbo Jia, and et al. 2021. "De Novo Reconstruction of Transcriptome Identified Long Non-Coding RNA Regulator of Aging-Related Brown Adipose Tissue Whitening in Rabbits" Biology 10, no. 11: 1176. https://doi.org/10.3390/biology10111176
APA StyleDu, K., Bai, X., Yang, L., Shi, Y., Chen, L., Wang, H., Cai, M., Wang, J., Chen, S., Jia, X., & Lai, S. (2021). De Novo Reconstruction of Transcriptome Identified Long Non-Coding RNA Regulator of Aging-Related Brown Adipose Tissue Whitening in Rabbits. Biology, 10(11), 1176. https://doi.org/10.3390/biology10111176




