Responses of the Soil Microbial Community to Salinity Stress in Maize Fields
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Sampling and Soil Physicochemical Characteristics
2.2. Soil DNA Extraction and Sequencing
2.3. Statistical Analyses
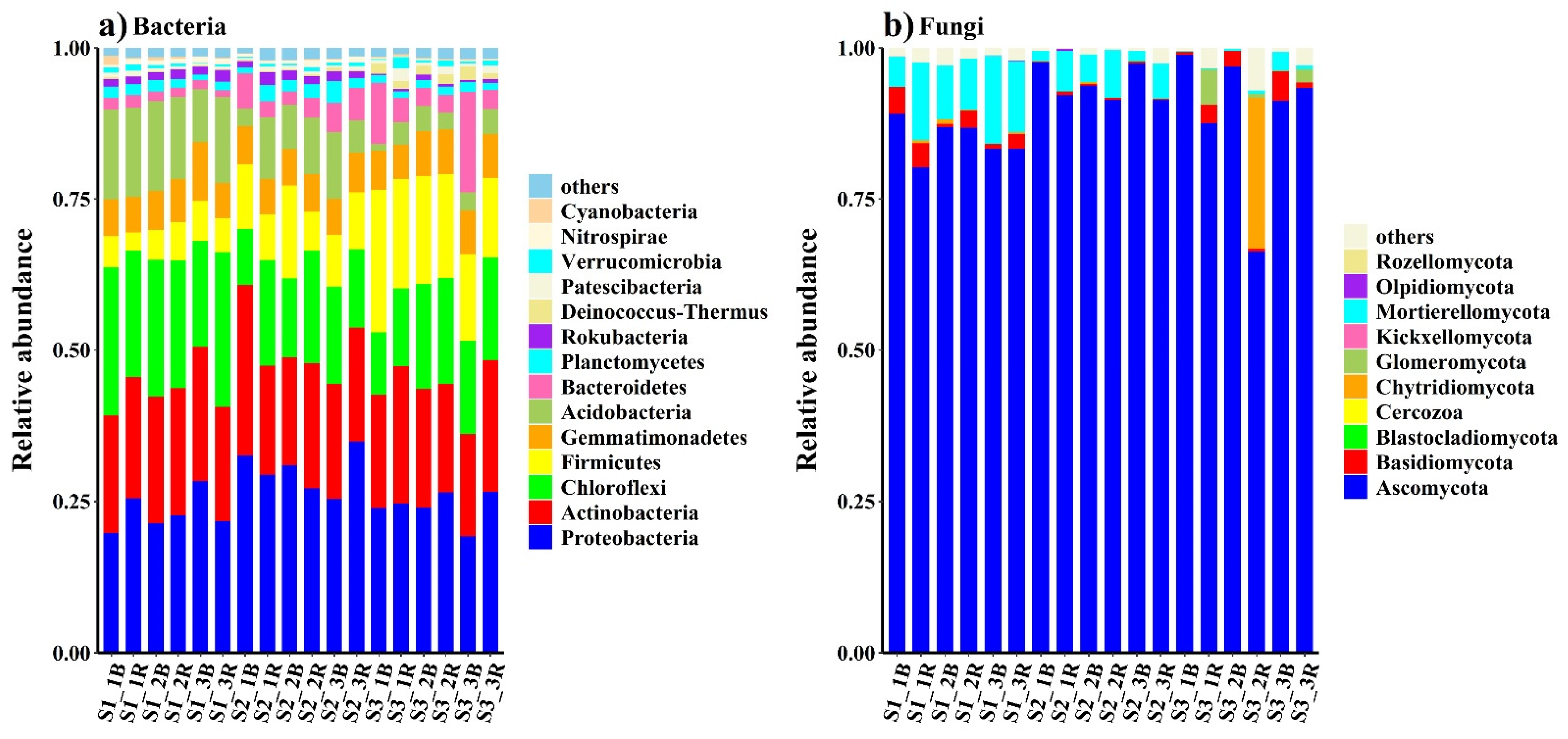
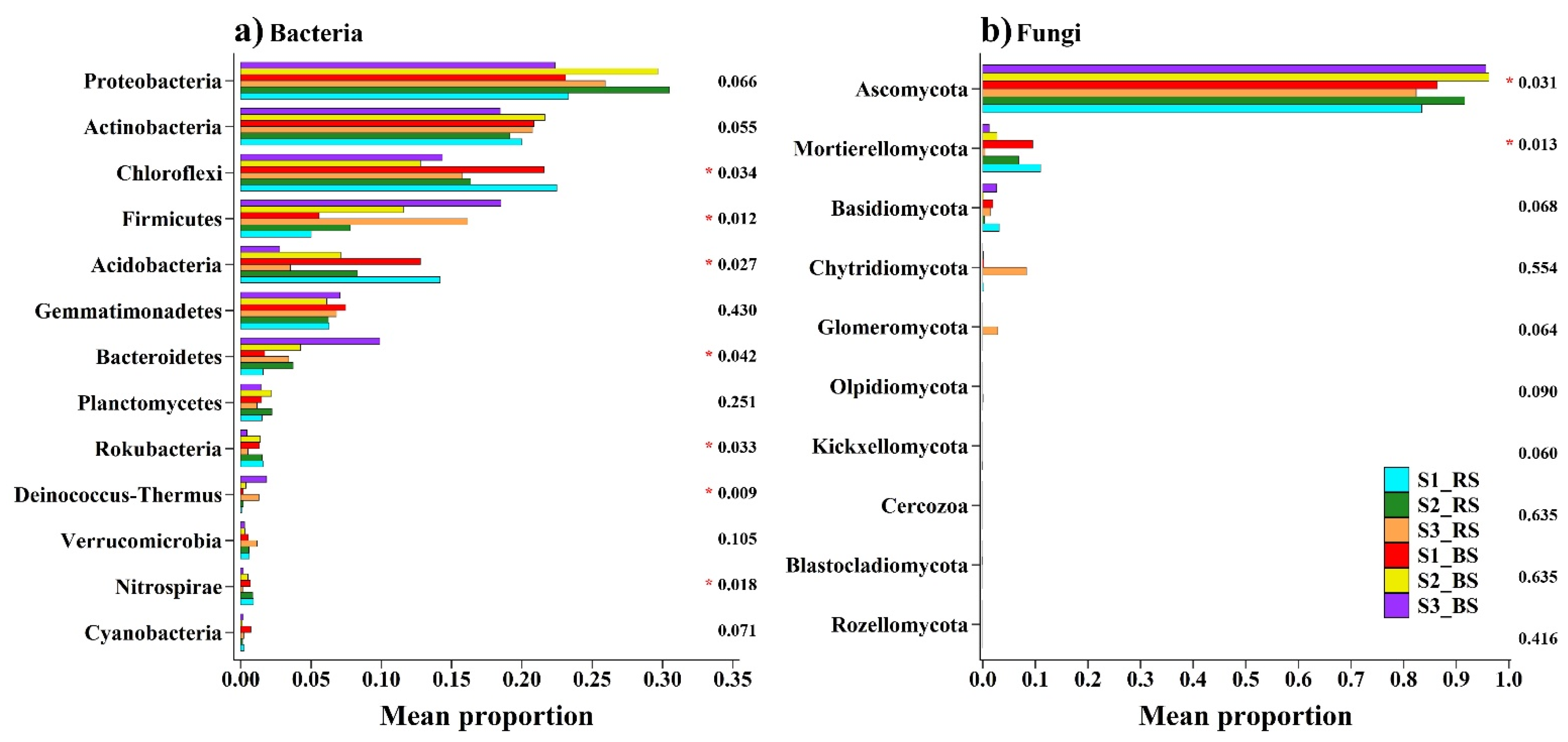
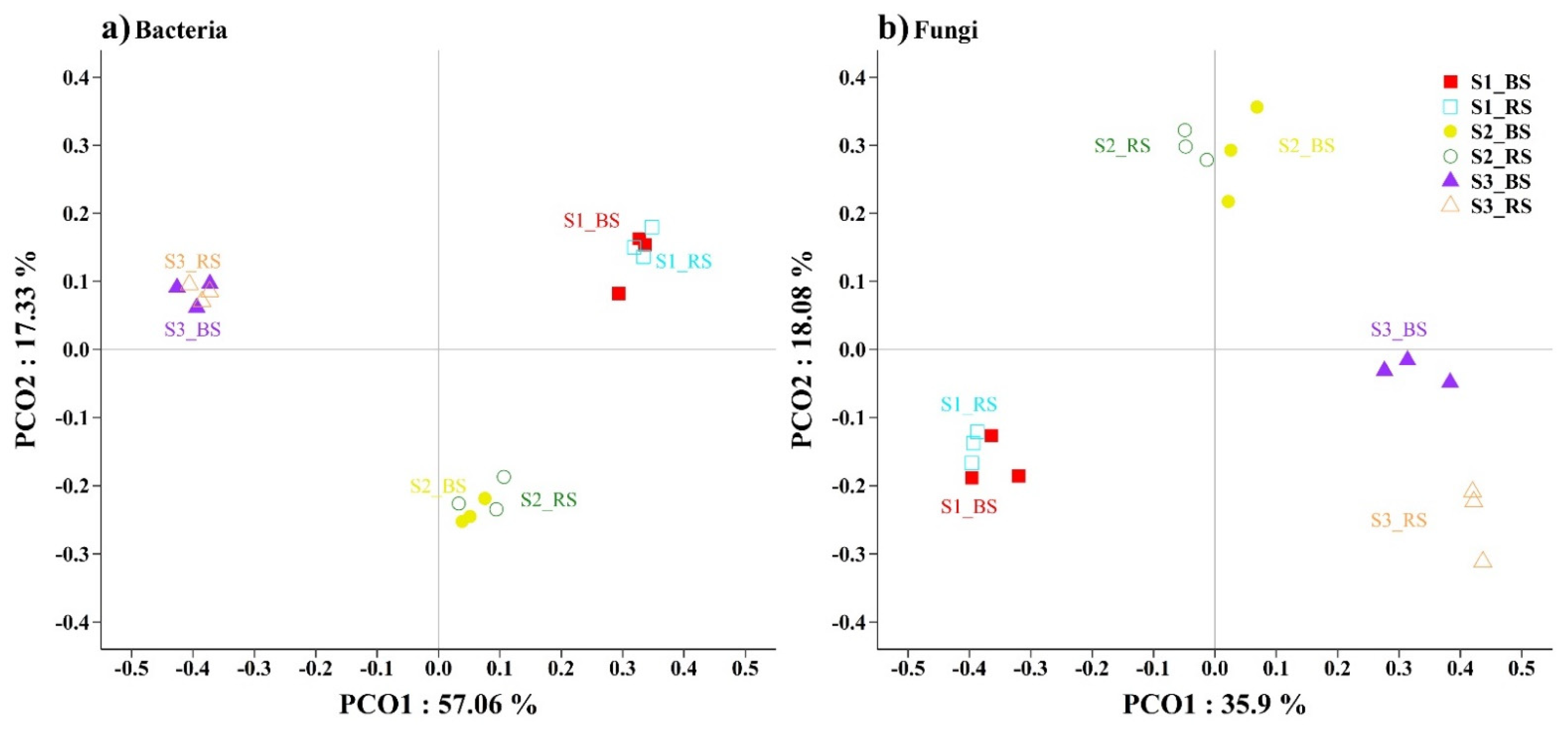
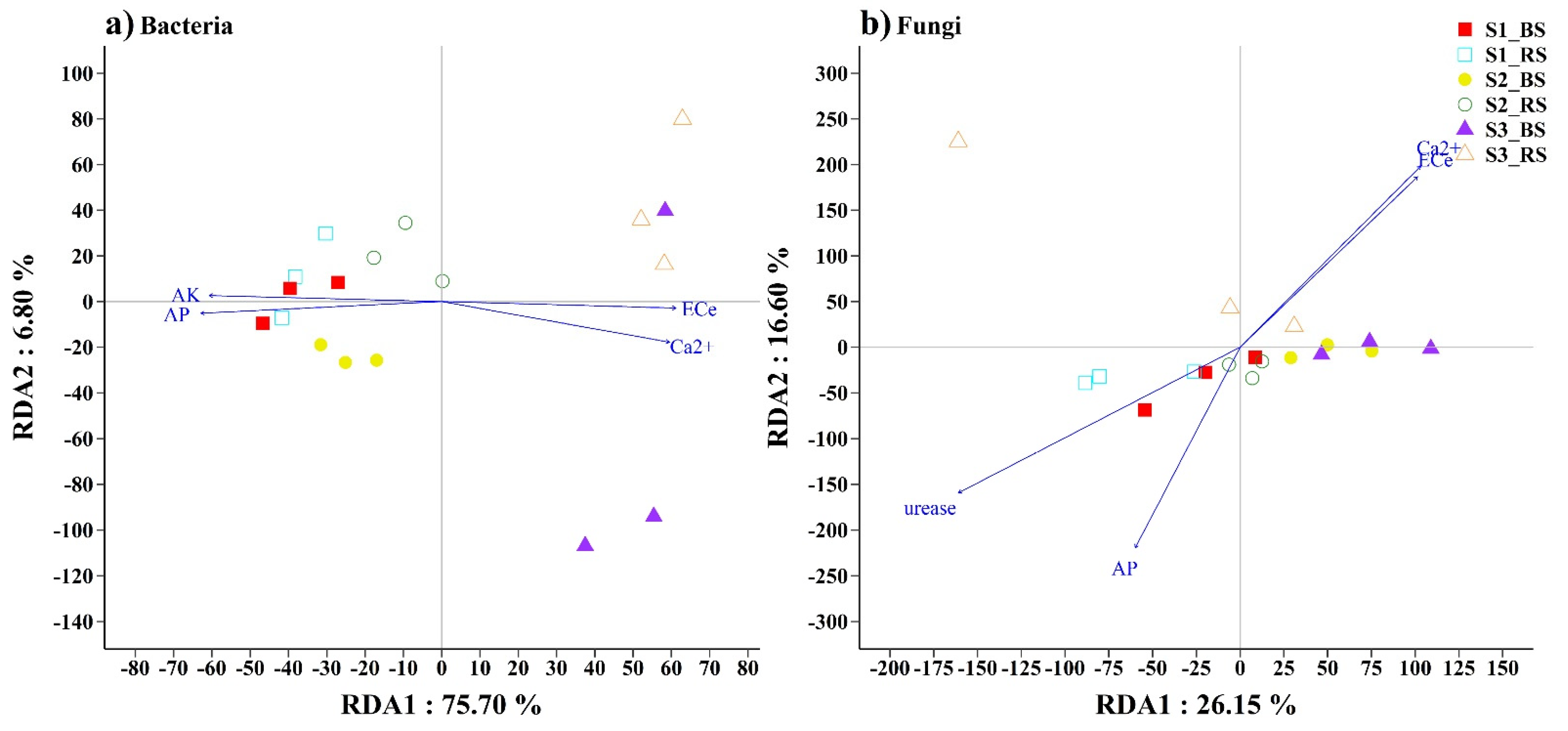
3. Results
3.1. Soil Physicochemical Properties
3.2. Soil Microbial Distribution and Diversity
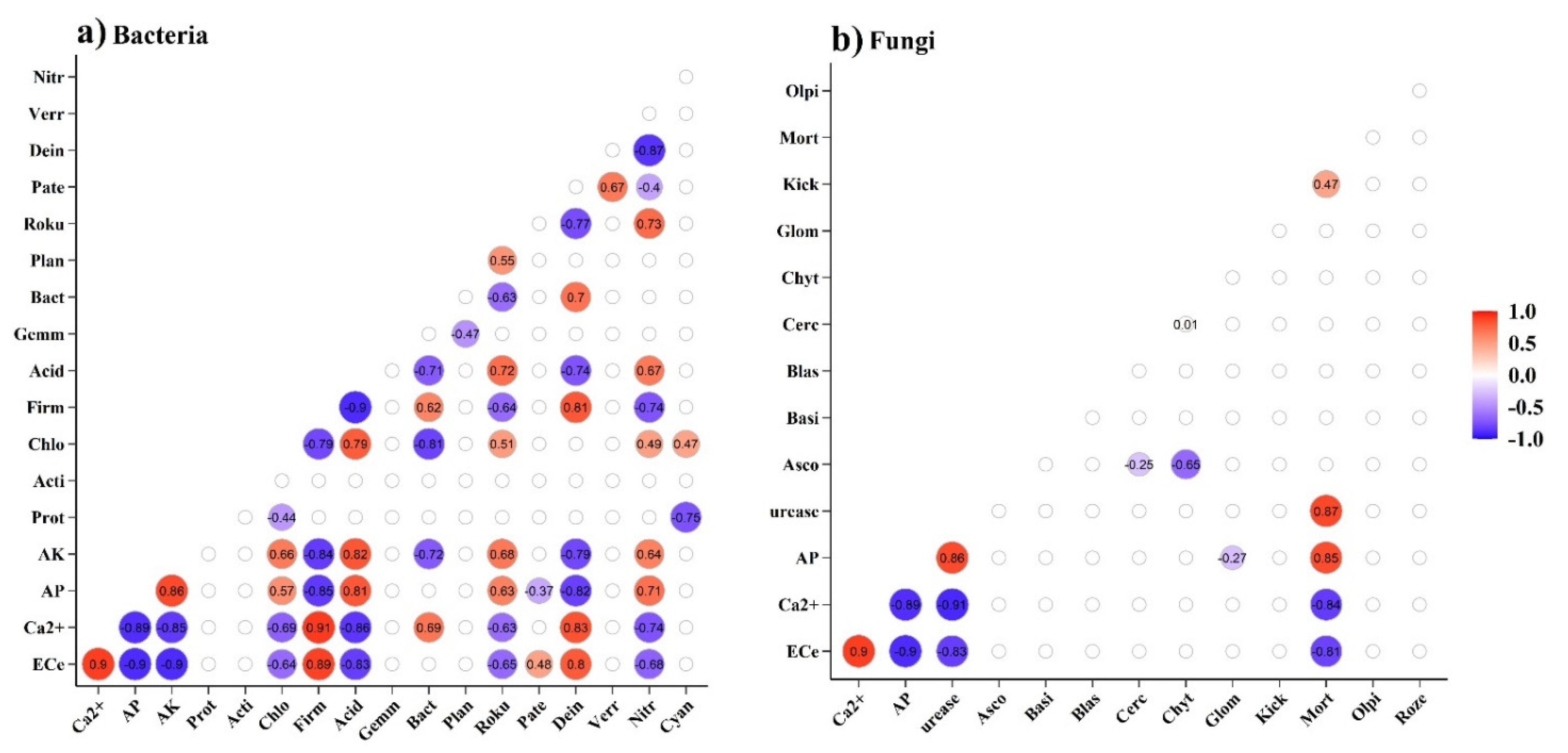
3.3. Soil Microbial Communities Associated with Environmental Factors
4. Discussion
4.1. Responses of Soil Properties to Salinity
4.2. Responses of the Bacterial and Fungal Communities to Salinity
4.3. Responses of Microbial Distribution to Physicochemical Properties in Saline Soil
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Wang, J.Z.; Ding, J.L.; Yu, D.L.; Ma, X.K.; Zhang, Z.P.; Ge, X.Y.; Teng, D.X.; Li, X.H.; Liang, J.; Lizag, A.; et al. Capability of Sentinel-2 MSI data for monitoring and mapping of soil salinity in dry and wet seasons in the Ebinur Lake region, Xinjiang, China. Geoderma 2019, 353, 172–187. [Google Scholar] [CrossRef]
- Zhao, S.; Liu, J.J.; Banerjee, S.; Zhou, N.; Zhao, Z.Y.; Zhang, K.; Hu, M.F.; Tian, C.Y. Biogeographical distribution of bacterial communities in saline agricultural soil. Geoderma 2020, 361, 114095. [Google Scholar] [CrossRef]
- Wang, W.; Wu, Z.; He, Y.; Huang, Y.; Li, X.; Ye, B.C. Plant growth promotion and alleviation of salinity stress in Capsicum annuum L. by Bacillus isolated from saline soil in Xinjiang. Ecotox. Environ. Saf. 2018, 164, 520–529. [Google Scholar] [CrossRef]
- Zhao, S.; Liu, J.J.; Banerjee, S.; Zhou, N.; Zhao, Z.Y.; Zhang, K.; Tian, C.Y. Soil pH is equally important as salinity in shaping bacterial communities in saline soils under halophytic vegetation. Sci. Rep. 2018, 8, 4550. [Google Scholar] [CrossRef]
- Yang, B.; Qi, K.B.; Bhusal, D.R.; Huang, J.S.; Chen, W.J.; Wu, Q.S.; Hussain, A.; Pang, X.Y. Soil microbial community and enzymatic activity in soil particle-size fractions of spruce plantation and secondary birch forest. Eur. J. Soil Biol. 2020, 99, 103196. [Google Scholar] [CrossRef]
- Deng, S.; Ke, T.; Li, L.; Cai, S.; Zhou, Y.; Liu, Y.; Guo, L.; Chen, L.; Zhang, D. Impacts of environmental factors on the whole microbial communities in the rhizosphere of a metal-tolerant plant: Elsholtzia haichowensis Sun. Environ. Pollut. 2018, 237, 1088–1097. [Google Scholar] [CrossRef]
- Fu, D.G.; Wu, X.N.; Qiu, Q.T.; Duan, C.Q.; Jones, D.L. Seasonal variations in soil microbial communities under different land restoration types in a subtropical mountains region, Southwest China. Appl. Soil Ecol. 2020, 153, 103634. [Google Scholar] [CrossRef]
- Chowdhury, N.; Marschner, P.; Burns, R. Response of microbial activity and community structure to decreasing soil osmotic and matric potential. Plant. Soil 2011, 344, 241–254. [Google Scholar] [CrossRef]
- Wichern, J.; Wichern, F.; Joergensen, R.G. Impact of salinity on soil microbial communities and the decomposition of maize in acidic soils. Geoderma 2006, 137, 100–108. [Google Scholar] [CrossRef]
- Kamble, P.N.; Gaikwad, V.B.; Kuchekar, S.R.; Baath, E. Microbial growth, biomass, community structure and nutrient limitation in high pH and salinity soils from Pravaranagar (India). Eur. J. Soil Biol. 2014, 65, 87–95. [Google Scholar] [CrossRef]
- Rath, K.M.; Maheshwari, A.; Bengtson, P.; Rousk, J. Comparative toxicities of salts on microbial processes in soil. Appl. Microbiol. Biotechnol. 2016, 82, 2012–2020. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rath, K.M.; Murphy, D.N.; Rousk, J. The microbial community size, structure, and process rates along natural gradients of soil salinity. Soil Biol. Biochem. 2019, 138, 107607. [Google Scholar] [CrossRef]
- Hollister, E.B.; Engledow, A.S.; Hammett, A.J.; Provin, T.L.; Wilkinson, H.H.; Gentry, T.J. Shifts in microbial community structure along an ecological gradient of hypersaline soils and sediments. ISME J. 2010, 4, 829–838. [Google Scholar] [CrossRef] [Green Version]
- Lozupone, C.A.; Knight, R. Global patterns in bacterial diversity. Proc. Natl. Acad. Sci. USA 2007, 104, 11436–11440. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, G.; Bai, J.; Tebbe, C.C.; Huang, L.; Jia, J.; Wang, W.; Wang, X.; Yu, L.; Zhao, Q. Spartina alterniflora invasions reduce soil fungal diversity and simplify co-occurrence networks in a salt marsh ecosystem. Sci. Total Environ. 2021, 758, 143667. [Google Scholar] [CrossRef] [PubMed]
- Vanegas, J.; Munoz-Garcia, A.; Perez-Parra, K.A.; Figueroa-Galvis, I.; Mestanza, O.; Polania, J. Effect of salinity on fungal diversity in the rhizosphere of the halophyte Avicennia germinans from a semi-arid mangrove. Fungal Ecol. 2019, 42, 100855. [Google Scholar] [CrossRef]
- Egamberdieva, D.; Renella, G.; Wirth, S.; Islam, R. Secondary salinity effects on soil microbial biomass. Biol. Fertil. Soils 2010, 46, 445–449. [Google Scholar] [CrossRef]
- Yan, N.; Marschner, P. Response of soil respiration and microbial biomass to changing EC in saline soils. Soil Biol. Biochem. 2013, 65, 322–328. [Google Scholar] [CrossRef]
- Casamayor, E.O.; Massana, R.; Benlloch, S.; Ovreas, L.; Diez, B.; Goddard, V.J.; Gasol, J.M.; Joint, I.; Rodriguez-Valera, F.; Pedros-Alio, C. Changes in archaeal, bacterial and eukaryal assemblages along a salinity gradient by comparison of genetic fingerprinting methods in a multipond solar saltern. Environ. Microbiol. 2002, 4, 338–348. [Google Scholar] [CrossRef]
- Pascual, J.; Blanco, S.; Ramos, J.L.; van Dillewijn, P. Responses of bulk and rhizosphere soil microbial communities to thermoclimatic changes in a Mediterranean ecosystem. Soil Biol. Biochem. 2018, 118, 130–144. [Google Scholar] [CrossRef]
- Deng, S.P.; Popova, I.E.; Dick, L.; Dick, R. Bench scale and microplate format assay of soil enzyme activities using spectroscopic and fluorometric approaches. Appl. Soil Ecol. 2013, 64, 84–90. [Google Scholar] [CrossRef]
- Diaz, F.J.; Sanchez-Hernandez, J.C.; Notario, J.S. Effects of irrigation management on arid soils enzyme activities. J. Arid. Environ. 2021, 185, 104330. [Google Scholar] [CrossRef]
- Xu, H.W.; Qu, Q.; Chen, Y.H.; Liu, G.B.; Xue, S. Responses of soil enzyme activity and soil organic carbon stability over time after cropland abandonment in different vegetation zones of the Loess Plateau of China. Catena 2021, 196, 104812. [Google Scholar] [CrossRef]
- Kozich, J.J.; Westcott, S.L.; Baxter, N.T.; Highlander, S.K.; Schloss, P.D. Development of a dual-index sequencing strategy and curation pipeline for analyzing amplicon sequence data on the MiSeq Illumina sequencing platform. Appl. Microbiol. Biotechnol. 2013, 79, 5112–5120. [Google Scholar] [CrossRef] [Green Version]
- Yang, C.; Wang, X.Z.; Miao, F.H.; Li, Z.Y.; Tang, W.; Sun, J. Assessing the effect of soil salinization on soil microbial respiration and diversities under incubation conditions. Appl. Soil Ecol. 2020, 155, 103671. [Google Scholar] [CrossRef]
- Usyk, M.; Zolnik, C.P.; Patel, H.; Levi, M.H.; Burk, R.D. Novel ITS1 Fungal Primers for Characterization of the Mycobiome. mSphere 2017, 2, e00488-17. [Google Scholar] [CrossRef] [Green Version]
- Kang, X.; Wang, Y.; Li, S.; Sun, X.; Lu, X.; Rajaofera, M.J.N.; Lu, Y.; Kang, L.; Zheng, A.; Zou, Z.; et al. Comparative Analysis of the Gut Microbiota of Adult Mosquitoes From Eight Locations in Hainan, China. Front. Cell. Infect. Microbiol. 2020, 10, 596750. [Google Scholar] [CrossRef] [PubMed]
- Shahrestani, M.M.; Marcussen, T.; Mehryarza, S.S.; Yousefi, N. Morphological and phylogenetic relationships within Viola sect. Sclerosium (Violaceae) in Iran. Flora 2015, 215, 67–74. [Google Scholar] [CrossRef]
- Anslan, S.; Mikryukov, V.; Armolaitis, K.; Ankuda, J.; Lazdina, D.; Makovskis, K.; Vesterdal, L.; Schmidt, I.K.; Tedersoo, L. Highly comparable metabarcoding results from MGI-Tech and Illumina sequencing platforms. PeerJ 2021, 9, e12254. [Google Scholar] [CrossRef]
- Rahman, M.S.; Hoque, M.N.; Puspo, J.A.; Islam, M.R.; Das, N.; Siddique, M.A.; Hossain, M.A.; Sultana, M. Microbiome signature and diversity regulates the level of energy production under anaerobic condition. Sci. Rep. 2021, 11, 19777. [Google Scholar] [CrossRef] [PubMed]
- Wang, R.S.; Wan, S.Q.; Sun, J.X.; Xiao, H.J. Soil salinity, sodicity and cotton yield parameters under different drip irrigation regimes during saline wasteland reclamation. Agric. Water Manag. 2018, 209, 20–31. [Google Scholar] [CrossRef]
- Xiong, Y.W.; Li, X.W.; Wang, T.T.; Gong, Y.; Zhang, C.M.; Xing, K.; Qin, S. Root exudates-driven rhizosphere recruitment of the plant growth-promoting rhizobacterium Bacillus flexus KLBMP 4941 and its growth-promoting effect on the coastal halophyte Limonium sinense under salt stress. Ecotox. Environ. Saf. 2020, 194, 110374. [Google Scholar] [CrossRef]
- Inal, A.; Gunes, A. Interspecific root interactions and rhizosphere effects on salt ions and nutrient uptake between mixed grown peanut/maize and peanut/barley in original saline-sodic-boron toxic soil. J. Plant Physiol. 2008, 165, 490–503. [Google Scholar] [CrossRef]
- Shi, S.H.; Tian, L.; Nasir, F.; Bahadur, A.; Batool, A.; Luo, S.S.; Yang, F.; Wang, Z.C.; Tian, C.J. Response of microbial communities and enzyme activities to amendments in saline-alkaline soils. Appl. Soil Ecol. 2019, 135, 16–24. [Google Scholar] [CrossRef]
- Marinari, S.; Carbone, S.; Vittori Antisari, L.; Grego, S.; Vianello, G. Microbial activity and functional diversity in Psamment soils in a forested coastal dune-swale system. Geoderma 2012, 173–174, 249–257. [Google Scholar] [CrossRef]
- Zhang, C.; Liu, G.B.; Xue, S.; Song, Z.L. Rhizosphere soil microbial activity under different vegetation types on the Loess Plateau, China. Geoderma 2011, 161, 115–125. [Google Scholar] [CrossRef]
- Herlemann, D.P.; Labrenz, M.; Jurgens, K.; Bertilsson, S.; Waniek, J.J.; Andersson, A.F. Transitions in bacterial communities along the 2000 km salinity gradient of the Baltic Sea. ISME J. 2011, 5, 1571–1579. [Google Scholar] [CrossRef] [Green Version]
- Rath, K.M.; Rousk, J. Salt effects on the soil microbial decomposer community and their role in organic carbon cycling: A review. Soil Biol. Biochem. 2015, 81, 108–123. [Google Scholar] [CrossRef]
- Thompson, L.R.; Sanders, J.G.; McDonald, D.; Amir, A.; Ladau, J.; Locey, K.J.; Prill, R.J.; Tripathi, A.; Gibbons, S.M.; Ackermann, G.; et al. A communal catalogue reveals Earth’s multiscale microbial diversity. Nature 2017, 551, 457–463. [Google Scholar] [CrossRef] [Green Version]
- Gregory, G.J.; Boyd, E.F. Stressed out: Bacterial response to high salinity using compatible solute biosynthesis and uptake systems, lessons from Vibrionaceae. Comp. Struct. Biotechnol. J. 2021, 19, 1014–1027. [Google Scholar] [CrossRef]
- McMillen, B.G.; Juniper, S.; Abbott, L.K. Inhibition of hyphal growth of a vesicular-arbuscular mycorrhizal fungus in soil containing sodium chloride limits the spread of infection from spores. Soil Biol. Biochem. 1998, 30, 1639–1646. [Google Scholar] [CrossRef]
- Gu, Y.; Bai, Y.; Xiang, Q.; Yu, X.; Zhao, K.; Zhang, X.; Li, C.; Liu, S.; Chen, Q. Degradation shaped bacterial and archaeal communities with predictable taxa and their association patterns in Zoige wetland at Tibet plateau. Sci. Rep. 2018, 8, 3884. [Google Scholar] [CrossRef] [Green Version]
- Zahran, H.H. Diversity, adaptation and activity of the bacterial flora in saline environments. Biol. Fertil. Soils 1997, 25, 211–223. [Google Scholar] [CrossRef]
- Sardinha, M.; Muller, T.; Schmeisky, H.; Joergensen, R.G. Microbial performance in soils along a salinity gradient under acidic conditions. Appl. Soil Ecol. 2003, 23, 237–244. [Google Scholar] [CrossRef]
- Batra, L.; Manna, M.C. Dehydrogenase activity and microbial biomass carbon in salt-affected soils of semiarid and arid regions. Arid Soil Res. Rehabil. 1997, 11, 295–303. [Google Scholar] [CrossRef]
- Pei, P.B.; Aslam, M.; Du, H.; Liang, H.H.; Wang, H.; Liu, X.J.; Chen, W.Z. Environmental factors shape the epiphytic bacterial communities of Gracilariopsis lemaneiformis. Sci. Rep. 2021, 11, 8671. [Google Scholar] [CrossRef]
- Norris, V.; Grant, S.; Freestone, P.; Canvin, J.; Sheikh, F.N.; Toth, I.; Trinei, M.; Modha, K.; Norman, R.I. Calcium signalling in bacteria. J. Bacteriol. 1996, 178, 3677–3682. [Google Scholar] [CrossRef] [Green Version]
- Xia, Z.; Bai, E.; Wang, Q.; Gao, D.; Zhou, J.; Jiang, P.; Wu, J. Biogeographic Distribution Patterns of Bacteria in Typical Chinese Forest Soils. Front. Microbiol. 2016, 7, 1106. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xue, L.; Ren, H.; Li, S.; Leng, X.; Yao, X. Soil Bacterial Community Structure and Co-occurrence Pattern during Vegetation Restoration in Karst Rocky Desertification Area. Front. Microbiol. 2017, 8, 2377. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rietz, D.N.; Haynes, R.J. Effects of irrigation-induced salinity and sodicity on soil microbial activity. Soil Biol. Biochem. 2003, 35, 845–854. [Google Scholar] [CrossRef]
- Jing, X.; Chen, X.; Fang, J.Y.; Ji, C.J.; Shen, H.H.; Zheng, C.Y.; Zhu, B. Soil microbial carbon and nutrient constraints are driven more by climate and soil physicochemical properties than by nutrient addition in forest ecosystems. Soil Biol. Biochem. 2020, 141, 107657. [Google Scholar] [CrossRef]
- Boone, D.R.; Castenholz, R.W.; Garrity, G.M. Bergey’s Manual of Systematic Bacteriology; Springer: New York, NY, USA, 2001; pp. 451–464. [Google Scholar] [CrossRef]
- Wang, C.; Liu, S.; Zhang, Y.; Liu, B.; He, F.; Xu, D.; Zhou, Q.; Wu, Z. Bacterial communities and their predicted functions explain the sediment nitrogen changes along with submerged macrophyte restoration. Microb. Ecol. 2018, 76, 625–636. [Google Scholar] [CrossRef] [PubMed]
- Hashmi, I.; Bindschedler, S.; Junier, P. Firmicutes. In Beneficial Microbes in Agro-Ecology; Academic Press: Cambridge, MA, USA, 2020; pp. 363–396. [Google Scholar] [CrossRef]
- Mukhtar, S.; Mehnaz, S.; Mirza, M.S.; Mirza, B.S.; Malik, K.A. Diversity of Bacillus-like bacterial community in the rhizospheric and non-rhizospheric soil of halophytes (Salsola stocksii and Atriplex amnicola), and characterization of osmoregulatory genes in halophilic Bacilli. Can. J. Microbiol. 2018, 64, 567–579. [Google Scholar] [CrossRef] [Green Version]
- Novello, G.; Gamalero, E.; Bona, E.; Boatti, L.; Mignone, F.; Massa, N.; Cesaro, P.; Lingua, G.; Berta, G. The Rhizosphere Bacterial Microbiota of Vitis vinifera cv. Pinot Noir in an Integrated Pest Management Vineyard. Front. Microbiol. 2017, 8, 1528. [Google Scholar] [CrossRef] [PubMed]
- Martens, E.C.; Koropatkin, N.M.; Smith, T.J.; Gordon, J.I. Complex glycan catabolism by the human gut microbiota: The Bacteroidetes Sus-like paradigm. J. Biol. Chem. 2009, 284, 24673–24677. [Google Scholar] [CrossRef] [Green Version]
- Church, M.J. Resource Control of Bacterial Dynamics in the Sea. In Microbial Ecology of the Oceans, 2nd ed.; John Wiley & Sons, Inc.: Hoboken, NJ, USA, 2008; pp. 335–382. [Google Scholar] [CrossRef]
- De Figueiredo, D.R.; Ferreira, R.V.; Cerqueira, M.; de Melo, T.C.; Pereira, M.J.; Castro, B.B.; Correia, A. Impact of water quality on bacterioplankton assemblage along Certima River Basin (central western Portugal) assessed by PCR-DGGE and multivariate analysis. Environ. Monit. Assess. 2012, 184, 471–485. [Google Scholar] [CrossRef]
- Muhammad, T.; Zhou, B.; Liu, Z.Y.; Chen, X.Z.; Li, Y.K. Effects of phosphorus-fertigation on emitter clogging in drip irrigation system with saline water. Agric. Water Manag. 2021, 243, 106392. [Google Scholar] [CrossRef]
| Physicochemical Factor | S1 | S2 | S3 | |||
|---|---|---|---|---|---|---|
| S1_RS | S1_BS | S2_RS | S2_BS | S3_RS | S3_BS | |
| ECe (ds/m) | 0.34 ± 0.01 c | 0.36 ± 0.03 c | 3.24 ± 0.39 b | 3.69 ± 0.11 b | 6.13 ± 0.72 a | 6.72 ± 0.49 a |
| pH | 7.88 ± 0.07 c | 7.99 ± 0.07 bc | 8.16 ± 0.04 bc | 8.23 ± 0.04 b | 8.32 ± 0.23 ab | 8.58 ± 0.29 a |
| clay content (%) | 0.55 ± 0.23 a | 0.57 ± 0.12 a | 0.53 ± 0.19 a | 0.67 ± 0.10 a | 0.90 ± 0.67 a | 0.61 ± 0.03 a |
| silt content (%) | 26.72 ± 2.88 a | 25.69 ± 1.57 a | 30.91 ± 2.15 a | 33.09 ± 1.37 a | 32.47 ± 9.25 a | 30.95 ± 2.70 a |
| sand content (%) | 72.73 ± 3.10 a | 73.74 ± 1.67 a | 68.56 ± 2.30 a | 66.24 ± 1.47 a | 66.63 ± 9.92 a | 68.44 ± 2.73 a |
| K+ (mg/kg) | 24.00 ± 2.29 e | 41.5 ± 2.18 d | 63.67 ± 12.75 c | 76.50 ± 6.38 b | 67.33 ± 5.84 bc | 123.83 ± 1.04 a |
| Na+ (mg/kg) | 27.00 ± 2.29 e | 97.33 ± 3.75 e | 271.00 ± 36.17 d | 2890.00 ± 193.08 b | 652.67 ± 37.44 c | 3334.67 ± 63.49 a |
| Ca2+ (mg/kg) | 158.20 ± 5.78 d | 151.64 ± 2.73 d | 495.75 ± 18.43 c | 1132.00 ± 131.76 b | 2101.50 ± 361.50 a | 2193.50 ± 161.47 a |
| Mg2+ (mg/kg) | 4.19 ± 0.92 d | 5.71 ± 1.82 d | 261.00 ± 24.98 c | 715.42 ± 65.67 b | 271.08 ± 94.11 c | 1582.50 ± 68.27 a |
| SOC (g/kg) | 12.61 ± 0.20 a | 11.23 ± 0.77 b | 6.29 ± 0.46 c | 6.20 ± 1.11 c | 5.49 ± 0.20 c | 5.67 ± 0.11 c |
| AN (mg/kg) | 28.24 ± 2.46 a | 30.57 ± 4.20 a | 20.54 ± 2.14 bc | 22.40 ± 4.28 b | 16.34 ± 1.07 c | 18.90 ± 2.14 bc |
| AP (mg/kg) | 60.33 ± 3.13 a | 62.07 ± 1.46 a | 52.00 ± 3.51 b | 50.17 ± 2.37 b | 25.33 ± 1.92 d | 34.70 ± 2.01 c |
| AK (mg/kg) | 242.67 ± 9.71 a | 242.00 ± 21.52 a | 142.33 ± 7.77 c | 205.33 ± 13.50 b | 109.67 ± 5.86 d | 98.33 ± 2.89 d |
| ALP (mg/g/d) | 3.19 ± 0.20 a | 3.49 ± 0.37 a | 1.43 ± 0.06 b | 0.88 ± 0.18 c | 0.69 ± 0.06 cd | 0.42 ± 0.02 d |
| Invertase (mg/g/d) | 12.36 ± 1.25 a | 10.97 ± 1.01 b | 4.31 ± 0.32 c | 3.24 ± 0.29 c | 1.46 ± 0.04 d | 1.07 ± 0.14 d |
| Urease (mg/g/d) | 1.57 ± 0.09 a | 1.63 ± 0.02 a | 0.87 ± 0.06 b | 0.75 ± 0.04 c | 0.49 ± 0.02 b | 0.42 ± 0.01 d |
| CAT (mL/g) | 1.47 ± 0.12 a | 1.10 ± 0.06 b | 1.00 ± 0.03 bc | 0.94 ± 0.06 c | 0.85 ± 0.02 cd | 0.80 ± 0.02 d |
| Variable | Shannon | Chao1 | OTU richness | |||
|---|---|---|---|---|---|---|
| Bacteria | Fungi | Bacteria | Fungi | Bacteria | Fungi | |
| ECe | −0.543 * | −0.652 ** | 0.609 ** | −0.922 ** | 0.627 ** | −0.937 ** |
| pH | −0.616 ** | −0.025 | −0.293 | −0.387 | −0.077 | −0.372 |
| clay | −0.090 | −0.014 | 0.131 | 0.029 | 0.019 | 0.072 |
| silt | −0.086 | −0.441 | 0.538 * | −0.350 | 0.408 | −0.317 |
| sand | 0.049 | 0.408 | −0.499 * | 0.300 | −0.381 | 0.268 |
| K+ | −0.576 * | −0.571 * | 0.501 * | −0.765 ** | 0.515 * | −0.764 ** |
| Na+ | −0.579 * | −0.591 ** | 0.606 ** | −0.779 ** | 0.528 * | −0.772 ** |
| Ca2+ | −0.586 * | −0.668 ** | 0.568 * | −0.913 ** | 0.535 * | −0.916 ** |
| Mg2+ | −0.536 * | −0.513 * | 0.662 ** | −0.724 ** | 0.535 * | −0.735 ** |
| SOC | 0.687 ** | 0.709 ** | −0.488 * | 0.920 ** | −0.400 | 0.926 ** |
| AN | 0.537 * | 0.694 ** | −0.461 | 0.783 ** | −0.464 | 0.779 ** |
| AP | 0.543 * | 0.744 ** | −0.562 * | 0.915 ** | −0.527 * | 0.936 ** |
| AK | 0.597 ** | 0.539* | −0.647 ** | 0.890 ** | −0.737 ** | 0.896 ** |
| ALP | 0.670 ** | 0.596 ** | −0.558 * | 0.891 ** | −0.526 * | 0.895 ** |
| Invertase | 0.719 ** | 0.659 ** | −0.514 * | 0.905 ** | −0.508 * | 0.908 ** |
| Urease | 0.660 ** | 0.652 ** | −0.515 * | 0.901 ** | −0.535 * | 0.890 ** |
| CAT | 0.643 ** | 0.676 ** | −0.568 * | 0.913 ** | −0.604 ** | 0.913 ** |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hou, Y.; Zeng, W.; Hou, M.; Wang, Z.; Luo, Y.; Lei, G.; Zhou, B.; Huang, J. Responses of the Soil Microbial Community to Salinity Stress in Maize Fields. Biology 2021, 10, 1114. https://doi.org/10.3390/biology10111114
Hou Y, Zeng W, Hou M, Wang Z, Luo Y, Lei G, Zhou B, Huang J. Responses of the Soil Microbial Community to Salinity Stress in Maize Fields. Biology. 2021; 10(11):1114. https://doi.org/10.3390/biology10111114
Chicago/Turabian StyleHou, Yaling, Wenzhi Zeng, Menglu Hou, Zhao Wang, Ying Luo, Guoqing Lei, Bo Zhou, and Jiesheng Huang. 2021. "Responses of the Soil Microbial Community to Salinity Stress in Maize Fields" Biology 10, no. 11: 1114. https://doi.org/10.3390/biology10111114
APA StyleHou, Y., Zeng, W., Hou, M., Wang, Z., Luo, Y., Lei, G., Zhou, B., & Huang, J. (2021). Responses of the Soil Microbial Community to Salinity Stress in Maize Fields. Biology, 10(11), 1114. https://doi.org/10.3390/biology10111114







