The Possible Role of Microbial Proteases in Facilitating SARS-CoV-2 Brain Invasion
Abstract
:Simple Summary
Abstract
1. Introduction
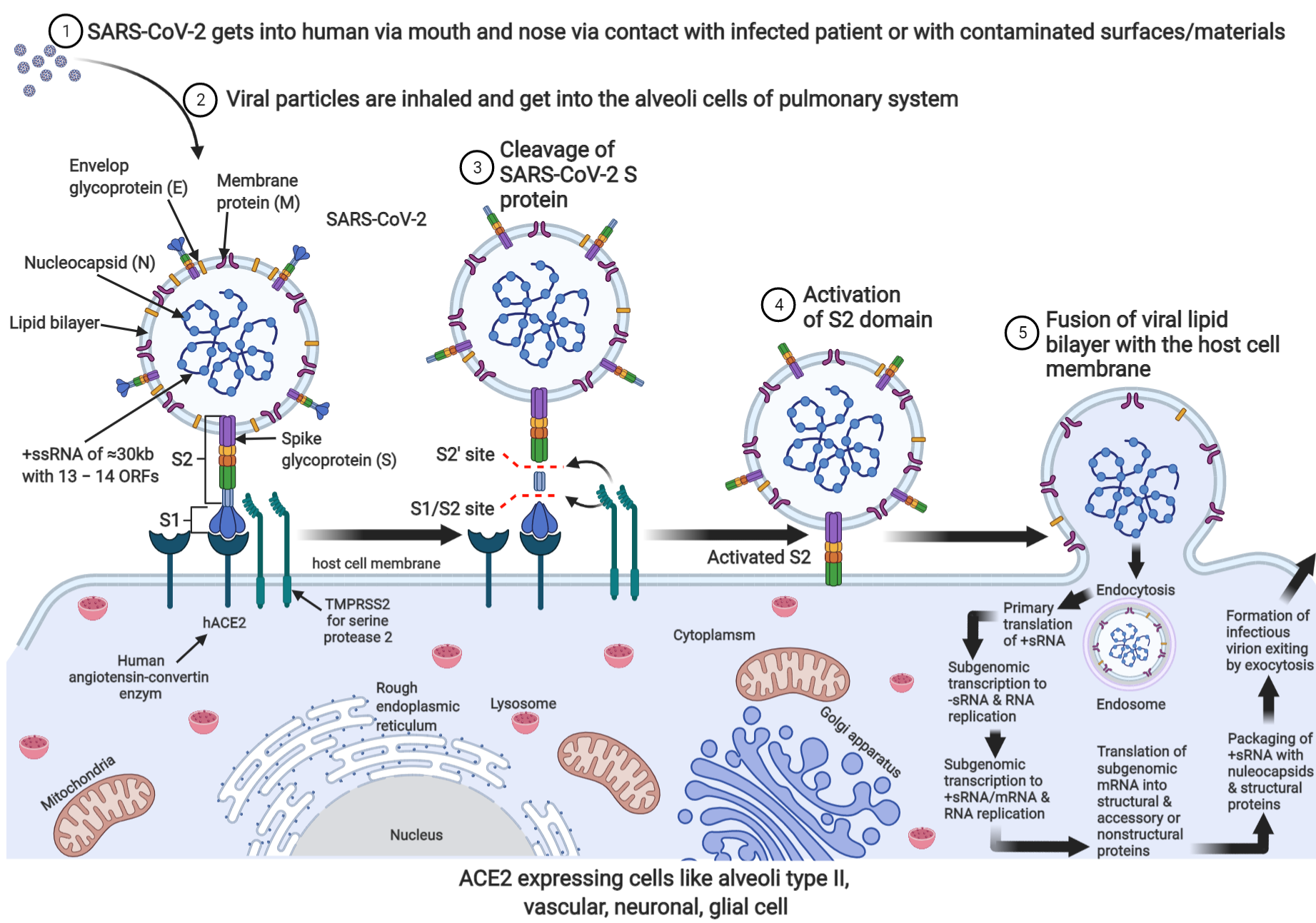
2. SARS-CoV-2: Current Understanding of Its Possible Brain Invasion
3. Possible Role of Cryptococcal Proteases in Promoting Brain Invasion in the Context of Co-Infection with SARS-COV-2
| Sequence | Type | Source | MW (kDa) | pH | Catalytic Residue | Zinc Chelating | Traditional Function | Purported Function in the Context of SARS-CoV-2 Infection | References |
|---|---|---|---|---|---|---|---|---|---|
| 1 | Serine-based protease (uncharacterised) | Cryptococcus neoformans | 75 | 7.2 | Ser | No | Promotes fungal brain invasion | May activate the viral S protein or promote viral transcellular migration | [72,77,78] |
| 2 | Fungalysin (metalloprotease, Mpr1) | Cryptococcus neoformans | 42 | 7.5–8.0 | Glu, Tyr, Asp | Yes | Promotes fungal brain invasion | May activate the viral S protein or promote viral transcellular migration | [79,80,81] |
| 3 | Major aspartyl peptidase (May1) | Cryptococcus neoformans | 45 | 5 | Asp | No | Promotes fungal brain invasion | May activate the viral S protein or promote viral transcellular migration | [71,82] |
| 4 | Furin (serine-based) | Homo sapiens (Human) | 104 | 5–8 | Asp, His, Ser | No (calcium-chelating) | Activates functionally important protein precursors | Activates the viral S protein for membrane fusion into cells | [83,84,85] |
| 5 | Transmembrane serine protease 2 (TMPRSS2) (serine-based) | Homo sapiens (Human) | 58 | 8 | His, Asp, Ser | No (calcium-chelating) | Promotes spike protein cleavage and activation for membrane fusion and viral uptake | Activates the viral S protein for membrane fusion into cells | [86,87,88] |
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Zhu, X.; Ge, Y.; Wu, T.; Zhao, K.; Chen, Y.; Wu, B.; Zhu, F.; Zhu, B.; Cui, L. Co-infection with respiratory pathogens among COVID-2019 cases. Virus Res. 2020, 285, 1–6. [Google Scholar] [CrossRef]
- Bruce, E.A.; Huang, M.L.; Perchetti, G.A.; Tighe, S.; Laaguiby, P.; Hoffman, J.J.; Gerrard, D.L.; Nalla, A.K.; Wei, Y.; Greninger, A.L.; et al. Direct RT-qPCR detection of SARS-CoV-2 RNA from patient nasopharyngeal swabs without an RNA extraction step. PLoS Biol. 2020, 18, e3000896. [Google Scholar] [CrossRef]
- Iwasaki, A.; Yang, Y. The potential danger of suboptimal antibody responses in COVID-19. Nat. Rev. Immunol. 2020, 20, 339–341. [Google Scholar] [CrossRef] [Green Version]
- Procop, G.W.; Shrestha, N.K.; Vogel, S.; van Sickle, K.; Harrington, S.; Rhoads, D.D.; Rubin, B.P.; Terpeluk, P. A Direct Comparison of Enhanced Saliva to Nasopharyngeal Swab for the Detection of SARS-CoV-2 in Symptomatic Patients. J. Clin. Microbiol. 2020, 58, 1–6. [Google Scholar] [CrossRef]
- Wyllie, A.L.; Fournier, J.; Casanovas-Massana, A.; Campbell, M.; Tokuyama, M.; Vijayakumar, P.; Geng, B.; Muenker, M.C.; Moore, A.J.; Vogels, C.B.F.; et al. Saliva is more sensitive for SARS-CoV-2 detection in COVID-19 patients than nasopharyngeal swabs. medRxiv 2020, 1–12. [Google Scholar] [CrossRef] [Green Version]
- Sidonie, L.N.; Alexis, C.; Samuel, L.P.; Christelle, V.F.; Laurence, M.J.; Anne-Marie, R.A.; Le, G.J.; Delaugerre, C. Evalua-tion of a Rapid Diagnostic Assay for Detection of SARS-CoV-2 Antigen in Nasopharyngeal Swabs. J. Clin. Microbiol. 2021, 58, 1814–1820. [Google Scholar] [CrossRef]
- Wei, Z.; Ryhana, M.; Elizabeth, S.J.; BG, J.M.A. Comparison of Four Molecular In Vitro Diagnostic Assays for the Detec-tion of SARS-CoV-2 in Nasopharyngeal Specimens. J. Clin. Microbiol. 2021, 58, 1–8. [Google Scholar] [CrossRef]
- Routley, N. Visualizing What COVID-19 Does to Your Body. Available online: https://www.visualcapitalist.com/visualizing-what-covid-19-does-to-your-body/ (accessed on 26 June 2021).
- Hameid, R.A.; Cormet-Boyaka, E.; Kuebler, W.M.; Uddin, M.; Berdiev, B.K. SARS-CoV-2 may hijack GPCR signaling pathways to dysregulate lung ion and fluid transport. Am. J. Physiol. Lung Cell Mol. Physiol. 2021, 320, L430–L435. [Google Scholar] [CrossRef]
- Zaki, A.M.; Van Boheemen, S.; Bestebroer, T.M.; Osterhaus, A.D.M.E.; Fouchier, R.A.M. Isolation of a novel coronavirus from a man with pneumonia in Saudi Arabia. N. Engl. J. Med. 2012, 367, 1814–1820. [Google Scholar] [CrossRef]
- Chen, J.; Qi, T.; Liu, L.; Ling, Y.; Qian, Z.; Li, T.; Li, F.; Xu, Q.; Zhang, Y.; Xu, S. Clinical progression of patients with COVID-19 in Shanghai, China. J. Infect. 2020, 80, e1–e6. [Google Scholar] [CrossRef] [PubMed]
- Huang, A.; Lu, M.; Ling, E.; Li, P.; Wang, C. A M35 family metalloprotease is required for fungal virulence against in-sects by inactivating host prophenoloxidases and beyond. Virulence 2020, 11, 222–237. [Google Scholar] [CrossRef] [Green Version]
- Chu, D.K.W.; Pan, Y.; Cheng, S.M.S.; Hui, K.P.Y.; Krishnan, P.; Liu, Y.; Ng, D.Y.M.; Wan, C.K.C.; Yang, P.; Wang, Q.; et al. Molecular diagnosis of a novel coronavirus (2019-nCoV) causing an outbreak of pneumonia. Clin. Chem. 2020, 66, 549–555. [Google Scholar] [CrossRef] [Green Version]
- Lescure, F.X.; Bouadma, L.; Nguyen, D.; Parisey, M.; Wicky, P.H.; Behillil, S.; Gaymard, A.; Bouscambert-Duchamp, M.; Donati, F.; Le, H.Q. Clinical and virological data of the first cases of COVID-19 in Europe: A case series. Lancet Infect. Dis. 2020, 20, 697–706. [Google Scholar] [CrossRef] [Green Version]
- Machhi, J.; Herskovitz, J.; Senan, A.M.; Dutta, D.; Nath, B.; Oleynikov, M.D.; Blomberg, W.R.; Meigs, D.D.; Hasan, M.; Patel, M. The natural history, pathobiology, and clinical manifestations of SARS-CoV-2 infections. J. Neuroimmune Pharmacol. 2020, 15, 359–386. [Google Scholar] [CrossRef]
- Wang, T.; Du, Z.; Zhu, F.; Cao, Z.; An, Y.; Gao, Y.; Jiang, B. Comorbidities and multi-organ injuries in the treatment of COVID-19. Lancet 2020, 395, e52. [Google Scholar] [CrossRef]
- Lau, S.K.P.; Luk, H.K.H.; Wong, A.C.P.; Li, K.S.M.; Zhu, L.; He, Z.; Fung, J.; Chan, T.T.Y.; Fung, K.S.C.; Woo, P.C.Y. Pos-sible bat origin of severe acute respiratory syndrome coronavirus 2. Emerg. Infect. Dis. 2020, 26, 1542–1547. [Google Scholar] [CrossRef] [PubMed]
- Lam, T.T.Y.; Jia, N.; Zhang, Y.W.; Shum, M.H.H.; Jiang, J.F.; Zhu, H.C.; Tong, Y.G.; Shi, Y.X.; Ni, X.B.; Liao, Y.S. Identify-ing SARS-CoV-2-related coronaviruses in Malayan pangolins. Nature 2020, 583, 282–285. [Google Scholar] [CrossRef] [Green Version]
- Xiao, K.; Zhai, J.; Feng, Y.; Zhou, N.; Zhang, X.; Zou, J.J.; Li, N.; Guo, Y.; Li, X.; Shen, X. Isolation of SARS-CoV-2-related coronavirus from Malayan pangolins. Nature 2020, 583, 286–289. [Google Scholar] [CrossRef] [PubMed]
- Chan, J.F.W.; Kok, K.H.; Zhu, Z.; Chu, H.; To, K.K.W.; Yuan, S.; Yuen, K.Y. Genomic characterisation of the 2019 novel human pathogenic coronavirus isolated from a patient with atypical pneumonia after visiting Wuhan. Emerg. Microbes Infect. 2020, 9, 221–236. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, Q.; Guan, X.; Wu, P.; Wang, X.; Zhou, L.; Tong, Y.; Ren, R.; Leung, K.S.M.; Lau, E.H.Y.; Wong, J.Y.; et al. Early Transmission Dynamics in Wuhan, China, of Novel Coronavirus–Infected Pneumonia. N. Engl. J. Med. 2020, 382, 1199–1207. [Google Scholar] [CrossRef]
- Phan, L.T.; Nguyen, T.V.; Luong, Q.C.; Nguyen, T.V.; Nguyen, H.T.; Le, H.Q.; Nguyen, T.T.; Cao, T.M.; Pham, Q.D. Importation and human-to-human transmission of a novel coronavirus in Vietnam. N. Engl. J. Med. 2020, 382, 872–874. [Google Scholar] [CrossRef] [Green Version]
- Petersen, E.; Koopmans, M.; Go, U.; Hamer, D.H.; Petrosillo, N.; Castelli, F.; Storgaard, M.; Al Khalili, S.; Simonsen, L. Comparing SARS-CoV-2 with SARS-CoV and influenza pandemics. Lancet Infect. Dis. 2020, 20, e238–e244. [Google Scholar] [CrossRef]
- WHO. Report of the WHO-China Joint Mission on Coronavirus Disease 2019 (COVID-19)—16–24 Feburary 2020. Available online: https://www.who.int/docs/default-source/coronaviruse/who-china-joint-mission-on-covid-19-final-report.pdf (accessed on 24 May 2021).
- Bhatraju, P.K.; Ghassemieh, B.J.; Nichols, M.; Kim, R.; Jerome, K.R.; Nalla, A.K.; Greninger, A.L.; Pipavath, S.; Wurfel, M.M.; Evans, L. Covid-19 in critically ill patients in the Seattle region—Case series. N. Engl. J. Med. 2020, 382, 2012–2022. [Google Scholar] [CrossRef]
- Chow, N.; Fleming-Dutra, K.; Gierke, R.; Hall, A.; Hughes, M.; Pilishvili, T.; CDC COVID-19 Response Team. Prelimi-nary estimates of the prevalence of selected underlying health conditions among patients with coronavirus disease 2019—United States, 12 February–28 March 2020. MMWR Morb. Mortal. Wkly. Rep. 2020, 69, 382–386. [Google Scholar] [CrossRef]
- Ejaz, H.; Alsrhani, A.; Zafar, A.; Javed, H.; Junaid, K.; Abdalla, A.E.; Abosalif, K.O.A.; Ahmed, Z.; Younas, S. COVID-19 and comorbidities: Deleterious impact on infected patients. J. Infect. Public Health 2020, 13, 1833–1839. [Google Scholar] [CrossRef] [PubMed]
- Guan, W.-J.; Ni, Z.-Y.; Hu, Y.; Liang, W.-H.; Ou, C.-Q.; He, J.-X.; Liu, L.; Shan, H.; Lei, C.-L.; Hui, D.S.; et al. Clinical characteristics of coro-navirus disease 2019 in China. N. Engl. J. Med. 2020, 382, 1708–1720. [Google Scholar] [CrossRef]
- Li, Z.; Wu, M.; Yao, J.; Guo, J.; Liao, X.; Song, S.; Li, J.; Duan, G.; Zhou, Y.; Wu, X. Caution on kidney dysfunctions of COVID-19 patients. medRxiv 2020, 1–25. [Google Scholar] [CrossRef]
- Simonnet, A.; Chetboun, M.; Poissy, J.; Raverdy, V.; Noulette, J.; Duhamel, A.; Labreuche, J.; Mathieu, D.; Pattou, F.; Jourdain, M. High prevalence of obesity in severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2) requiring invasive mechanical ventilation. Obesity 2020, 28, 1195–1199. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.; Yu, Y.; Xu, J.; Shu, H.; Liu, H.; Wu, Y.; Zhang, L.; Yu, Z.; Fang, M.; Yu, T. Clinical course and outcomes of criti-cally ill patients with SARS-CoV-2 pneumonia in Wuhan, China: A single-centered, retrospective, observational study. Lancet Respir. Med. 2020, 8, 475–481. [Google Scholar] [CrossRef] [Green Version]
- Zheng, Y.Y.; Ma, Y.T.; Zhang, J.Y.; Xie, X. COVID-19 and the cardiovascular system. Nat. Rev. Cardiol. 2020, 17, 259–260. [Google Scholar] [CrossRef] [Green Version]
- Zhu, N.; Zhang, D.; Wang, W.; Li, X.; Yang, B.; Song, J.; Zhao, X.; Huang, B.; Shi, W.; Lu, R.; et al. A Novel Coronavirus from Patients with Pneumonia in China, 2019. N. Engl. J. Med. 2020, 382, 727–733. [Google Scholar] [CrossRef] [PubMed]
- Hung, E.C.W.; Chim, S.S.C.; Chan, P.K.S.; Tong, Y.K.; Ng, E.K.O.; Chiu, R.W.K.; Leung, C.B.; Sung, J.J.Y.; Tam, J.S.; Lo, Y.M.D. Detection of SARS coronavirus RNA in the cerebrospinal fluid of a patient with severe acute respiratory syndrome. Clin. Chem. 2003, 49, 2108–2109. [Google Scholar] [CrossRef] [Green Version]
- Gu, J.; Gong, E.; Zhang, B.; Zheng, J.; Gao, Z.; Zhong, Y.; Zou, W.; Zhan, J.; Wang, S.; Xie, Z. Multiple organ infection and the pathogenesis of SARS. J. Exp. Med. 2005, 202, 415–424. [Google Scholar] [CrossRef] [PubMed]
- Saad, M.; Omrani, A.S.; Baig, K.; Bahloul, A.; Elzein, F.; Matin, M.A.; Selim, M.A.A.; Al Mutairi, M.; Al Nakhli, D.; Al Aidaroos, A.Y. Clinical aspects and outcomes of 70 patients with Middle East respiratory syndrome coronavirus infection: A single-center experience in Saudi Arabia. Int. J. Infect. Dis. 2014, 29, 301–306. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Helms, J.; Kremer, S.; Merdji, H.; Clere-Jehl, R.; Schenck, M.; Kummerlen, C.; Collange, O.; Boulay, C.; Fafi-Kremer, S.; Ohana, M. Neurologic features in severe SARS-CoV-2 infection. N. Engl. J. Med. 2020, 382, 2268–2270. [Google Scholar] [CrossRef] [PubMed]
- Mao, L.; Jin, H.; Wang, M.; Hu, Y.; Chen, S.; He, Q.; Chang, J.; Hong, C.; Zhou, Y.D.; Wang, X.M. Neurologic manifestations of hospitalised patients with coronavirus disease 2019 in Wuhan, China. JAMA Neurol. 2020, 77, 683–690. [Google Scholar] [CrossRef] [Green Version]
- Moriguchi, T.; Harii, N.; Goto, J.; Harada, D.; Sugawara, H.; Takamino, J.; Ueno, M.; Sakata, H.; Kondo, K.; Myose, N. A first case of meningitis/encephalitis associated with SARS-Coronavirus-2. Int. J. Infect. Dis. 2020, 94, 55–58. [Google Scholar] [CrossRef] [PubMed]
- Zhou, L.; Zhang, M.; Wang, J.; Gao, J. Sars-Cov-2: Underestimated damage to nervous system. Travel Med. Infect. Dis. 2020, 36, 101642. [Google Scholar] [CrossRef]
- Netland, J.; Meyerholz, D.K.; Moore, S.; Cassell, M.; Perlman, S. Severe Acute Respiratory Syndrome Coronavirus Infection Causes Neuronal Death in the Absence of Encephalitis in Mice Transgenic for Human ACE2. J. Virol. 2008, 82, 7264–7275. [Google Scholar] [CrossRef] [Green Version]
- Hamming, I.; Timens, W.; Bulthuis, M.L.C.; Lely, A.T.; Navis, G.J.; van Goor, H. Tissue distribution of ACE2 protein, the functional receptor for SARS coronavirus. A first step in understanding SARS pathogenesis. J. Pathol. 2004, 203, 631–637. [Google Scholar] [CrossRef]
- Li, Y.; Bai, W.; Hashikawa, T. The neuroinvasive potential of SARS-CoV2 may play a role in the respiratory failure of COVID-19 patients. J. Med. Virol. 2020, 92, 552–555. [Google Scholar] [CrossRef]
- Wu, Y.; Xu, X.; Chen, Z.; Duan, J.; Hashimoto, K.; Yang, L.; Liu, C.; Yang, C. Nervous system involvement after infection with COVID-19 and other coronaviruses. Brain Behav. Immun. 2020, 87, 18–22. [Google Scholar] [CrossRef]
- Zhou, P.; Yang, X.L.; Wang, X.G.; Hu, B.; Zhang, L.; Zhang, W.; Si, H.R.; Zhu, Y.; Li, B.; Huang, C.L. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature 2020, 579, 270–273. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hartenian, E.; Nandakumar, D.; Lari, A.; Ly, M.; Tucker, J.M.; Glaunsinger, B.A. The molecular virology of coronaviruses. J. Biol. Chem. 2020, 295, 12910–12934. [Google Scholar] [CrossRef] [PubMed]
- Andersen, K.G.; Rambaut, A.; Lipkin, W.I.; Holmes, E.C.; Garry, R.F. The proximal origin of SARS-CoV-2. Nat. Med. 2020, 26, 450–452. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wrobel, A.G.; Benton, D.J.; Xu, P.; Roustan, C.; Martin, S.R.; Rosenthal, P.B.; Skehel, J.J.; Gamblin, S.J. SARS-CoV-2 and bat RaTG13 spike glycoprotein structures inform on virus evolution and furin-cleavage effects. Nat. Struct. Mol. Biol. 2020, 27, 763–767. [Google Scholar] [CrossRef]
- Al Saiegh, F.; Ghosh, R.; Leibold, A.; Avery, M.B.; Schmidt, R.F.; Theofanis, T.; Mouchtouris, N.; Philipp, L.; Peiper, S.C.; Wang, Z.X. Status of SARS-CoV-2 in cerebrospinal fluid of patients with COVID-19 and stroke. J. Neurol. Neurosurg. Psychiatry 2020, 91, 846–848. [Google Scholar] [CrossRef]
- Baig, A.M.; Khaleeq, A.; Ali, U.; Syeda, H. Evidence of the COVID-19 virus targeting the CNS: Tissue distribution, host–virus interaction, and proposed neurotropic mechanisms. ACS Chem. Neurosci. 2020, 11, 995–998. [Google Scholar] [CrossRef] [Green Version]
- Chen, T.; Wu, D.; Chen, H.; Yan, W.; Yang, D.; Chen, G.; Ma, K.; Xu, D.; Yu, H.; Wang, H.; et al. Clinical characteristics of 113 deceased patients with coronavirus disease 2019: Retrospective study. BMJ 2020, 368, m1091. [Google Scholar] [CrossRef] [Green Version]
- Chen, N.; Zhou, M.; Dong, X.; Qu, J.; Gong, F.; Han, Y.; Qiu, Y.; Wang, J.; Liu, Y.; Wei, Y. Epidemiological and clinical characteristics of 99 cases of 2019 novel coronavirus pneumonia in Wuhan, China: A descriptive study. Lancet 2020, 395, 507–513. [Google Scholar] [CrossRef] [Green Version]
- Duong, L.; Xu, P.; Liu, A. Meningoencephalitis without respiratory failure in a young female patient with COVID-19 infection in Downtown Los Angeles, early April 2020. Brain Behav. Immun. 2020, 87, 1–33. [Google Scholar] [CrossRef] [PubMed]
- Eliezer, M.; Hautefort, C.; Hamel, A.-L.; Verillaud, B.; Herman, P.; Houdart, E.; Eloit, C. Sudden and complete olfactory loss of function as a possible symptom of COVID-19. JAMA Otolaryngol. Neck Surg. 2020, 146, 674–675. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Oxley, T.J.; Mocco, J.; Majidi, S.; Kellner, C.P.; Shoirah, H.; Singh, I.P.; Singh, I.P.; De Leacy, R.A.; Shigematsu, T.; Ladner, T.R.; et al. Large-vessel stroke as a presenting feature of Covid-19 in the young. N. Engl. J. Med. 2020, 382, e60(1)–e60(3). [Google Scholar] [CrossRef]
- Rogers, J.P.; Chesney, E.; Oliver, D.; Pollak, T.A.; McGuire, P.; Fusar-Poli, P.; Zandi, M.S.; Lewis, G.; David, A.S. Psychiatric and neuropsychiatric presentations associated with severe coronavirus infections: A systematic review and meta-analysis with comparison to the COVID-19 pandemic. Lancet Psychiatry 2020, 7, 611–627. [Google Scholar] [CrossRef]
- Zhao, H.; Shen, D.; Zhou, H.; Liu, J.; Chen, S. Guillain-Barré syndrome associated with SARS-CoV-2 infection: Causality or coincidence? Lancet Neurol. 2020, 19, 383–384. [Google Scholar] [CrossRef]
- Mehta, P.; McAuley, D.F.; Brown, M.; Sanchez, E.; Tattersall, R.S.; Manson, J.J. COVID-19: Consider cytokine storm syndromes and immunosuppression. Lancet 2020, 395, 1033–1034. [Google Scholar] [CrossRef]
- Poyiadji, N.; Shahin, G.; Noujaim, D.; Stone, M.; Patel, S.; Griffith, B. COVID-19–associated Acute Hemorrhagic Necrotizin Encephalopathy: Imaging Features. Radiology 2020, 296, E119–E120. [Google Scholar] [CrossRef] [Green Version]
- Grifoni, A.; Weiskopf, D.; Ramirez, S.I.; Mateus, J.; Dan, J.M.; Moderbacher, C.R.; Rawlings, S.A.; Sutherland, A.; Premkumar, L.; Jadi, R.S. Targets of T cell responses to SARS-CoV-2 coronavirus in humans with COVID-19 disease and unexposed individuals. Cell 2020, 181, 1489–1501. [Google Scholar] [CrossRef]
- Zhang, H.; Kang, Z.; Gong, H.; Xu, D.; Wang, J.; Li, Z.; Cui, X.; Xiao, J.; Meng, T.; Zhou, W. The digestive system is a potential route of 2019-nCov infection: A bioinformatics analysis based on single-cell transcriptomes. BioRxiv 2020, 1–20. [Google Scholar] [CrossRef]
- Park, M.D. Macrophages: A Trojan horse in COVID-19? Nat. Rev. Immunol. 2020, 20, 351. [Google Scholar] [CrossRef]
- Elkington, P.T.G.; O’kane, C.M.; Friedland, J.S. The paradox of matrix metalloproteinases in infectious disease. Clin. Exp. Immunol. 2005, 142, 12–20. [Google Scholar] [CrossRef]
- Vilcinskas, A.; Wedde, M. Insect inhibitors of metalloproteinases. IUBMB Life 2002, 54, 339–343. [Google Scholar] [CrossRef]
- Master, S.S.; Rampini, S.K.; Davis, A.S.; Keller, C.; Ehlers, S.; Springer, B.; Timmins, G.S.; Sander, P.; Deretic, V. Mycobacterium tuberculosis Prevents Inflammasome Activation. Cell Host Microbe 2008, 3, 224–232. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Marathe, K.R.; Patil, R.H.; Vishwakarma, K.S.; Chaudhari, A.B.; Maheshwari, V.L. Protease inhibitors and their applications: An overview. In Studies in Natural Products Chemistry; Elsevier: Rahman, The Netherlands, 2019; pp. 211–242. [Google Scholar] [CrossRef]
- Igarashi, Y.; Heureux, E.; Doctor, K.S.; Talwar, P.; Gramatikova, S.; Gramatikoff, K.; Zhang, Y.; Blinov, M.; Ibragimova, S.S.; Boyd, S.; et al. PMAP: Databases for analysing proteolytic events and pathways. Nucleic Acids Res. 2008, 37, D611–D618. [Google Scholar] [CrossRef] [PubMed]
- Quesada, V.; Ordóñez, G.R.; Sánchez, L.M.; Puente, X.S.; López-Otín, C. The degradome database: Mammalian proteases and diseases of proteolysis. Nucleic Acids Res. 2009, 37, D239–D243. [Google Scholar] [CrossRef] [Green Version]
- Rawlings, N.D.; Barrett, A.J.; Thomas, P.D.; Huang, X.; Bateman, A.; Finn, R.D. The MEROPS database of proteolytic enzymes, their substrates and inhibitors in 2017 and a comparison with peptidases in the PANTHER database. Nucleic Acids Res. 2018, 46, D624–D632. [Google Scholar] [CrossRef] [PubMed]
- Llano, E.; Pendás, A.M.; Aza-Blanc, P.; Kornberg, T.B.; Lopez-Otin, C. Dm1-MMP, a matrix metalloproteinase from Drosophila with a potential role in extracellular matrix remodeling during neural development. J. Biol. Chem. 2000, 275, 35978–35985. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kryštůfek, R.; Šácha, P.; Starková, J.; Brynda, J.; Hradilek, M.; Tloušt’ová, E.G.J.; Rut, W.; Boucher, M.J.; Drąg, M.; Majer, P.; et al. Re-emerging Aspartic Protease Targets: Examining Cryptococcus neoformans Major Aspartyl Peptidase 1 as a Target for Antifungal Drug Discovery. J. Med. Chem. 2021, 64, 6706–6719. [Google Scholar] [CrossRef]
- Strickland, A.B.; Shi, M. Mechanisms of fungal dissemination. Cell Mol. Life Sci. 2021, 78, 3219–3238. [Google Scholar] [CrossRef] [PubMed]
- Esher, S.K.; Zaragoza, O.; Alspaugh, J.A. Cryptococcal pathogenic mechanisms: A dangerous trip from the environment to the brain. Mem. Inst. Oswaldo Cruz 2018, 113, 1–15. [Google Scholar] [CrossRef] [Green Version]
- García-Rodas, R.; Zaragoza, O. Catch me if you can: Phagocytosis and killing avoidance by Cryptococcus neoformans. FEMS Immunol. Med. Microbiol. 2012, 64, 147–161. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gaylord, E.A.; Choy, H.L.; Doering, T.L. Dangerous liaisons: Interactions of Cryptococcus neoformans with host phagocytes. Pathogens 2020, 9, 891. [Google Scholar] [CrossRef]
- Liu, T.B.; Perlin, D.S.; Xue, C. Molecular mechanisms of cryptococcal meningitis. Virulence 2012, 3, 173–181. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rodrigues, M.L.; Dos, R.F.C.G.; Puccia, R.; Travassos, L.R.; Alviano, C.S. Cleavage of human fibronectin and other basement membrane-associated proteins by a Cryptococcus neoformans serine proteinase. Microb. Pathog. 2003, 34, 65–71. [Google Scholar] [CrossRef]
- Vu, K.; Tham, R.; Uhrig, J.P.; Thompson, G.R.; Pombejra, S.N.; Jamklang, M.; Bautos, J.M.; Gelli, A. Invasion of the central nervous system by Cryptococcus neoformans requires a secreted fungal metalloprotease. MBio 2014, 5, 1–13. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xu, C.Y.; Zhu, H.M.; Wu, J.H.; Wen, H.; Liu, C.J. Increased permeability of blood-brain barrier is mediated by serine protease during Cryptococcus meningitis. J. Int. Med. Res. 2012, 42, 85–92. [Google Scholar] [CrossRef] [PubMed]
- Pombejra, N.S.; Jamklang, M.; Uhrig, J.P.; Vu, K.; Gelli, A. The structure-function analysis of the Mpr1 metalloprotease determinants of activity during migration of fungal cells across the blood-brain barrier. PLoS ONE 2018, 13, e0203020. [Google Scholar] [CrossRef] [PubMed]
- Vu, K.; Garcia, J.A.; Gelli, A. Cryptococcal meningitis and anti-virulence therapeutic strategies. Front. Microbiol. 2019, 10, 1–7. [Google Scholar] [CrossRef] [Green Version]
- Clarke, S.C.; Dumesic, P.A.; Homer, C.M.; O’Donoghue, A.J.; La Greca, F.; Pallova, L.; Majer, P.; Madhani, H.D.; Craik, C.S. Integrated Activity and Genetic Profiling of Secreted Peptidases in Cryptococcus neoformans Reveals an Aspartyl Peptidase Required for Low pH Survival and Virulence. PLoS Pathog. 2016, 12, e1006051. [Google Scholar] [CrossRef]
- Creemers, J.W.M.; Siezen, R.J.; Roebroek, A.J.M.; Ayoubi, T.A.Y.; Huylebroeck, D.; Van de Ven, W.J.M. Modulation of furin-mediated proprotein processing activity by site-directed mutagenesis. J. Biol. Chem. 1993, 268, 21826–21834. [Google Scholar] [CrossRef]
- Cieplik, M.; Klenk, H.W.G. Identification and characterisation of spodoptera frugiperda furin: A thermostable subtilisin-like endopeptidase. Biol. Chem. 1998, 379, 1433–1440. [Google Scholar] [CrossRef] [PubMed]
- Solovyeva, N.I.; Gureeva, T.A.; Timoshenko, O.S.; Moskvitina, T.A.; Kugaevskaya, E.V. Furin as Proprotein Convertase and Its Role in Normal and Pathological Biological Processes. Biomed. Khim. 2016, 11, 609–621. [Google Scholar] [CrossRef] [Green Version]
- Meng, T.; Cao, H.; Zhang, H.; Kang, Z.; Xu, D.; Gong, H.; Wang, J.; Li, Z.; Cui, X.; Xu, H.; et al. The insert sequence in SARS-CoV-2 enhances spike protein cleavage by TMPRSS. bioRxiv 2020, 1–36. [Google Scholar] [CrossRef]
- Thunders, M.; Delahunt, B. Gene of the month: TMPRSS2 (transmembrane serine protease 2). J. Clin. Pathol. 2020, 73, 773–776. [Google Scholar] [CrossRef]
- Sonawane, K.D.; Barale, S.S.; Dhanavade, M.J.; Waghmare, S.R.; Nadaf, N.H.; Kamble, S.A.; Mohammed, A.A.; Makandar, A.M.; Fandilolu, P.M.; Dound, A.S.; et al. Structural insights and inhibition mechanism of TMPRSS2 by experimentally known inhibitors Camostat mesylate, Nafamostat and Bromhexine hydrochloride to control SARS-coronavirus-2: A molecular modeling approach. Inform. Med. Unlocked 2021, 24, 1–15. [Google Scholar] [CrossRef]
- Belouzard, S.; Chu, V.C.; Whittaker, G.R. Activation of the SARS coronavirus spike protein via sequential proteolytic cleavage at two distinct sites. Proc. Natl. Acad. Sci. USA 2009, 106, 5871–5876. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Levitz, S.M.; Harrison, T.S.; Tabuni, A.; Liu, X. Chloroquine induces human mononuclear phagocytes to inhibit and kill Cryptococcus neoformans by a mechanism independent of iron deprivation. J. Clin. Investig. 1997, 100, 1640–1646. [Google Scholar] [CrossRef] [Green Version]
- van Weert, A.W.; Geuze, H.J.; Groothuis, B.; Stoorvogel, W. Primaquine interferes with membrane recycling from endosomes to the plasma membrane through a direct interaction with endosomes which does not involve neutralisation of endosomal pH nor osmotic swelling of endosomes. Eur. J. Cell Biol. 2000, 79, 394–399. [Google Scholar] [CrossRef] [PubMed]
- Seglen, P.O. Inhibitors of lysosomal function. Methods Enzymol. 1983, 96, 737–764. [Google Scholar] [CrossRef]
- Geary, T.G.; Jensen, J.B.; Ginsburg, H. Uptake of [3H] chloroquine by drug-sensitive and -resistant strains of the human malaria parasite Plasmodium falciparum. Biochem. Pharmacol. 1986, 35, 3805–3812. [Google Scholar] [CrossRef]
- Newman, S.L.; Gootee, L.; Brunner, G.; Deepe, G.S. Chloroquine induces human macrophage killing of Histoplasma capsulatum by limiting the availability of intracellular iron and is therapeutic in a murine model of histoplasmosis. J. Clin. Investig. 1994, 93, 1422–1429. [Google Scholar] [CrossRef]
- López-Otín, C.; Bond, J.S. Proteases: Multifunctional enzymes in life and disease. J. Biol. Chem. 2008, 283, 30433–30437. [Google Scholar] [CrossRef] [Green Version]
- Blume, C.; Jackson, C.L.; Spalluto, C.M.; Legebeke, J.; Nazlamova, L.; Conforti, F.; Perotin, J.M.; Frank, M.; Butler, J.; Crispin, M.; et al. A novel ACE2 isoform is expressed in human respiratory epithelia and is upregulated in response to interferons and RNA respiratory virus infection. Nat. Genet. 2021, 53, 205–214. [Google Scholar] [CrossRef] [PubMed]
- Chandran, K.; Sullivan, N.J.; Felbor, U.; Whelan, S.P.; Cunningham, J.M. Endosomal proteolysis of the Ebola virus glycoprotein is necessary for infection. Science 2005, 308, 1643–1645. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Barrientos, L.G.; Rollin, P.E. Release of cellular proteases into the acidic extracellular milieu exacerbates Ebola virus-induced cell damage. Virology 2007, 358, 1–9. [Google Scholar] [CrossRef] [Green Version]
- Powers, J.C.; Asgian, J.L.; Ekici, O.D.; James, K.E. Irreversible inhibitors of serine, cysteine, and threonine proteases. Chem. Rev. 2002, 102, 4639–4750. [Google Scholar] [CrossRef]
- Otto, H.H.; Schirmeister, T. Cysteine Proteases and Their Inhibitors. Chem. Rev. 1997, 97, 133–172. [Google Scholar] [CrossRef]
- Kawase, M.; Shirato, K.; van der Hoek, L.; Taguchi, F.; Matsuyama, S. Simultaneous Treatment of Human Bronchial Epithelial Cells with Serine and Cysteine Protease Inhibitors Prevents Severe Acute Respiratory Syndrome Coronavirus Entry. J. Virol. 2012, 86, 6537–6545. [Google Scholar] [CrossRef] [Green Version]
- Hoffmann, M.; Kleine-Weber, H.; Schroeder, S.; Krüger, N.; Herrler, T.; Erichsen, S.; Erichsen, S.; Schiergens, T.S.; Herrler, G.; Wu, N.H.; et al. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell 2020, 181, 271–280.e8. [Google Scholar] [CrossRef]
- Djomkam, A.L.Z.; Olwal, C.O.; Sala, T.B.; Paemka, L. Commentary: SARS-CoV-2 Cell Entry Depends on ACE2 and TMPRSS2 and Is Blocked by a Clinically Proven Protease Inhibitor. Front. Oncol. 2020, 10, e1448. [Google Scholar] [CrossRef]
- Kim, J.C.; Spence, R.A.; Currier, P.F.; Lu, X.; Denison, M.R. Coronavirus protein processing and RNA synthesis is inhibited by the cysteine proteinase inhibitor E64d. Virology 1995, 208, 1–8. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lv, Z.; Chu, Y.; Wang, Y. HIV protease inhibitors: A review of molecular selectivity and toxicity. HIV/AIDS 2015, 7, 95–104. [Google Scholar] [CrossRef] [Green Version]
- Pfizer Inc. Available online: https://www.pfizer.com/news/press-release/press-release-detail/pfizer-initiates-phase-1-study-novel-oral-antiviral (accessed on 7 September 2021).

| The Co-Occurrence of SARS-CoV-2 with Other Infections | |||||
|---|---|---|---|---|---|
| Non-Infectious Conditions | Aetiological Agents | ||||
| Scopus | PubMed | Scopus | PubMed | ||
| Meningitis-causing agents | |||||
| Cancer | 42,719 | 12,180 | M. tuberculosis | 8440 | 1206 |
| Diabetes | 26,288 | 6663 | S. pneumoniae | 1065 | 122 |
| Hypertension | 16,623 | 4393 | P. aeruginosa | 1773 | 107 |
| Obesity | 14,643 | 2923 | H. capsulatum | 49 | 8 |
| Cerebrovascular disease | 4351 | 1628 | C. neoformans | 160 | 11 |
| Asthma | 8238 | 1358 | Mucoralean spp. | 4 | 26 |
| Non-meningitis-causing agents | |||||
| Renal failure | 6098 | 1656 | A. fumigatus | 435 | 44 |
| Chronic obstructive pulmonary disease | 4613 | 779 | Ca. albicans | 840 | 36 |
| Hepatic dysfunction | 1698 | 1956 | Ca. auris | 187 | 107 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mjokane, N.; Folorunso, O.S.; Ogundeji, A.O.; Sebolai, O.M. The Possible Role of Microbial Proteases in Facilitating SARS-CoV-2 Brain Invasion. Biology 2021, 10, 966. https://doi.org/10.3390/biology10100966
Mjokane N, Folorunso OS, Ogundeji AO, Sebolai OM. The Possible Role of Microbial Proteases in Facilitating SARS-CoV-2 Brain Invasion. Biology. 2021; 10(10):966. https://doi.org/10.3390/biology10100966
Chicago/Turabian StyleMjokane, Nozethu, Olufemi S. Folorunso, Adepemi O. Ogundeji, and Olihile M. Sebolai. 2021. "The Possible Role of Microbial Proteases in Facilitating SARS-CoV-2 Brain Invasion" Biology 10, no. 10: 966. https://doi.org/10.3390/biology10100966






