First Metabolic Insights into Ex Vivo Cryptosporidium parvum-Infected Bovine Small Intestinal Explants Studied under Physioxic Conditions
Abstract
:Simple Summary
Abstract
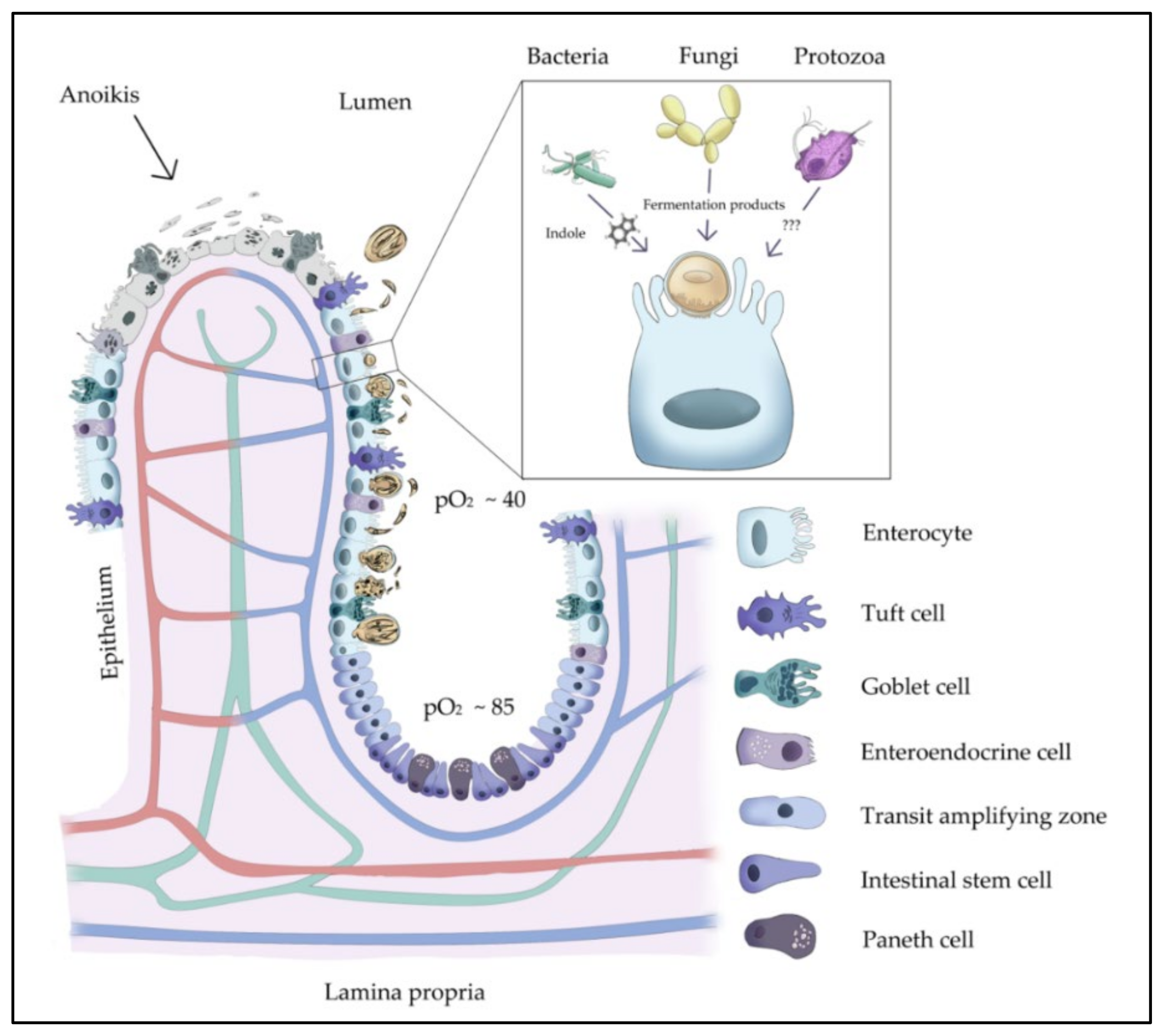
1. Introduction
2. Materials and Methods
2.1. Isolation of Bovine Small Intestinal (BSI) Explants
2.2. Isolation of Bovine Small Intestinal Epithelial Cells (BSIEC) from Intestinal Crypts
2.3. BSI Explant- and BSIEC-Based Host Cell Culture Systems
2.4. Characterization of BSIEC
2.5. Parasites
2.6. BSI Explant and BSIEC Infections with Cryptosporidium parvum Sporozoites
2.7. Scanning Electron Microscopic Analysis of Cryptosporidium parvum-Infected BSI Explants
2.8. Immunofluorescence-Based Quantification of Cryptosporidium parvum Infection Rates in BSIEC
2.9. Tissue DNA Extraction
2.10. qPCR-Based Quantification of Cryptosporidium parvum Replication in BSI Explants
2.11. Quantification of Metabolic Conversion Rates of Cryptosporidium parvum-Infected BSI Explant Supernatants
2.12. Analysis of Glycolytic Responses of Cryptosporidium parvum-Infected BSIEC via Seahorse Technology
2.13. Data Processing and Statistical Analysis
3. Results
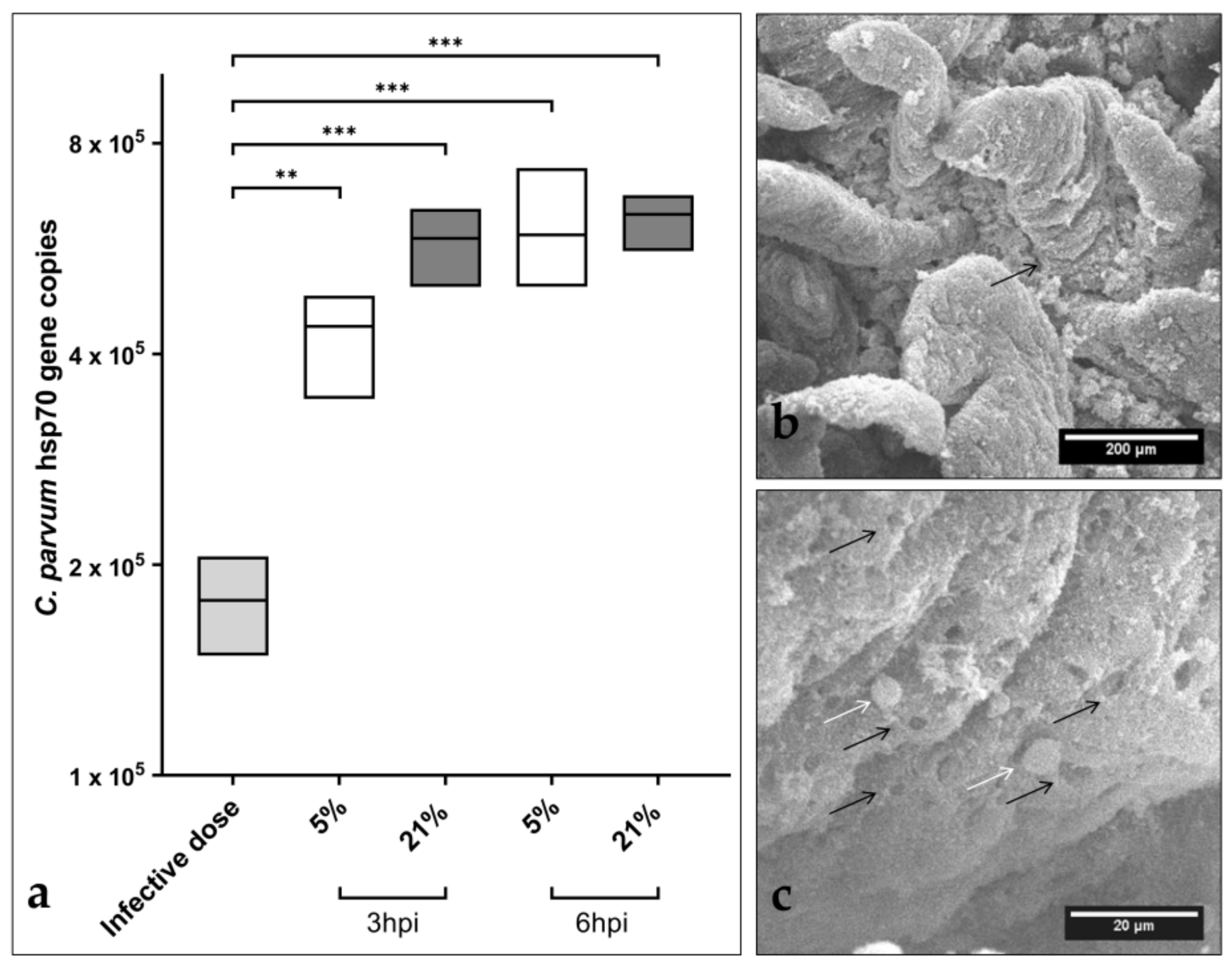
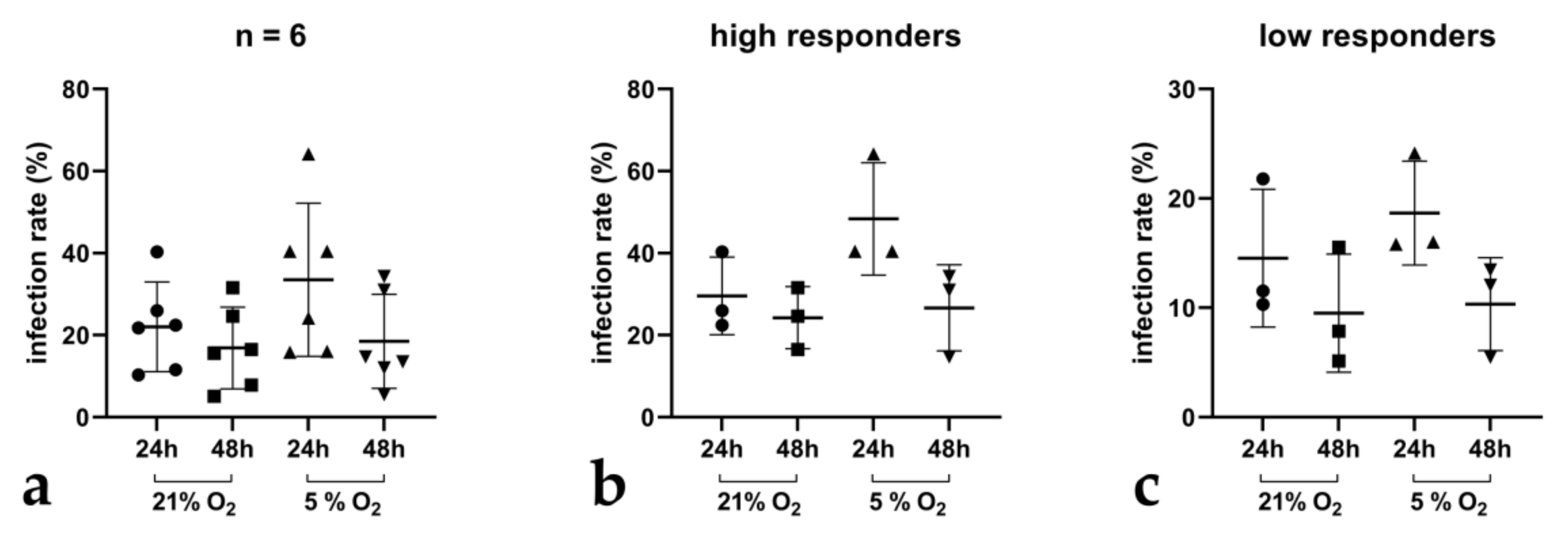
3.1. Cryptosporidium parvum Replication in BSI Explants under Physioxic Oxygen Conditions
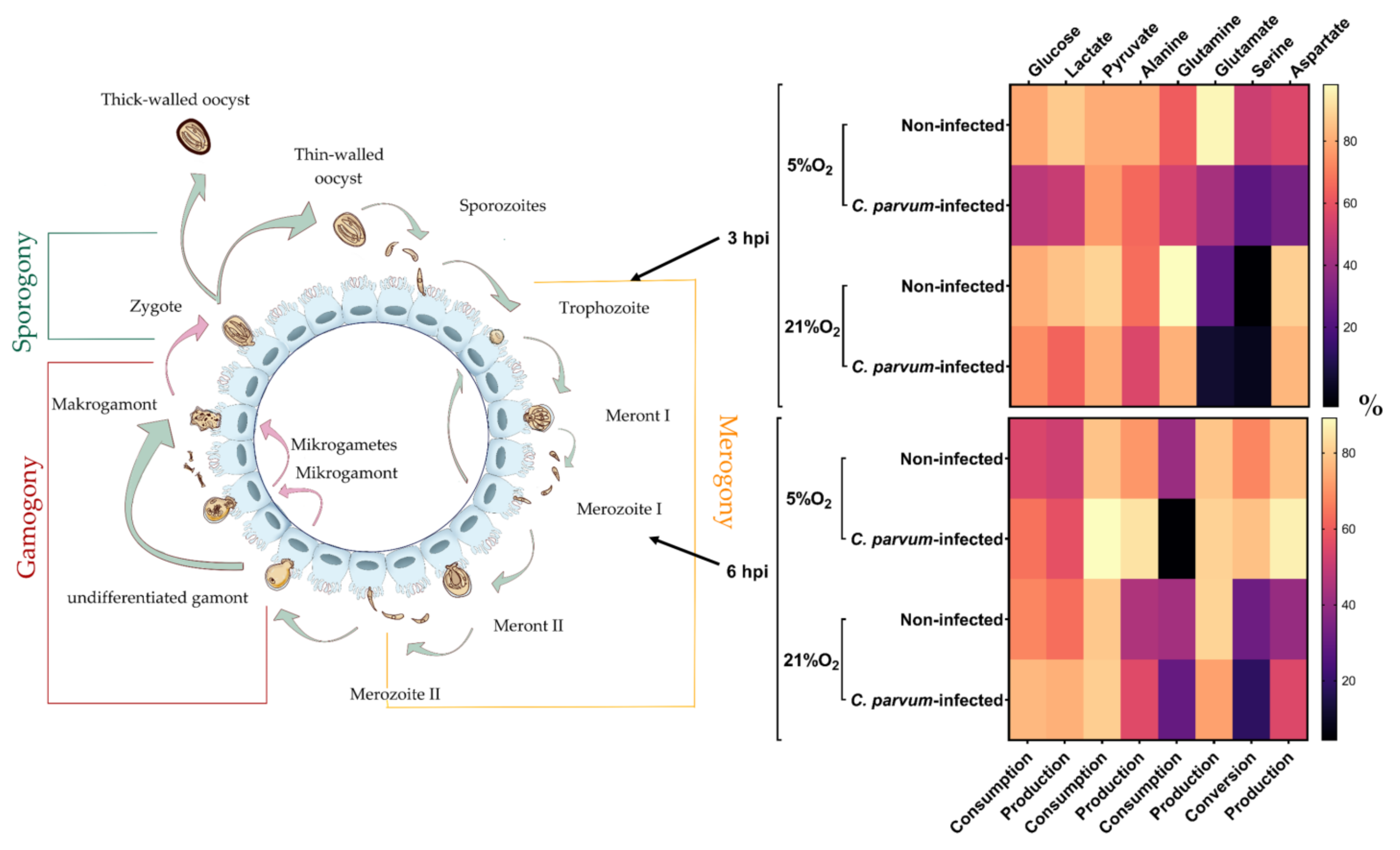
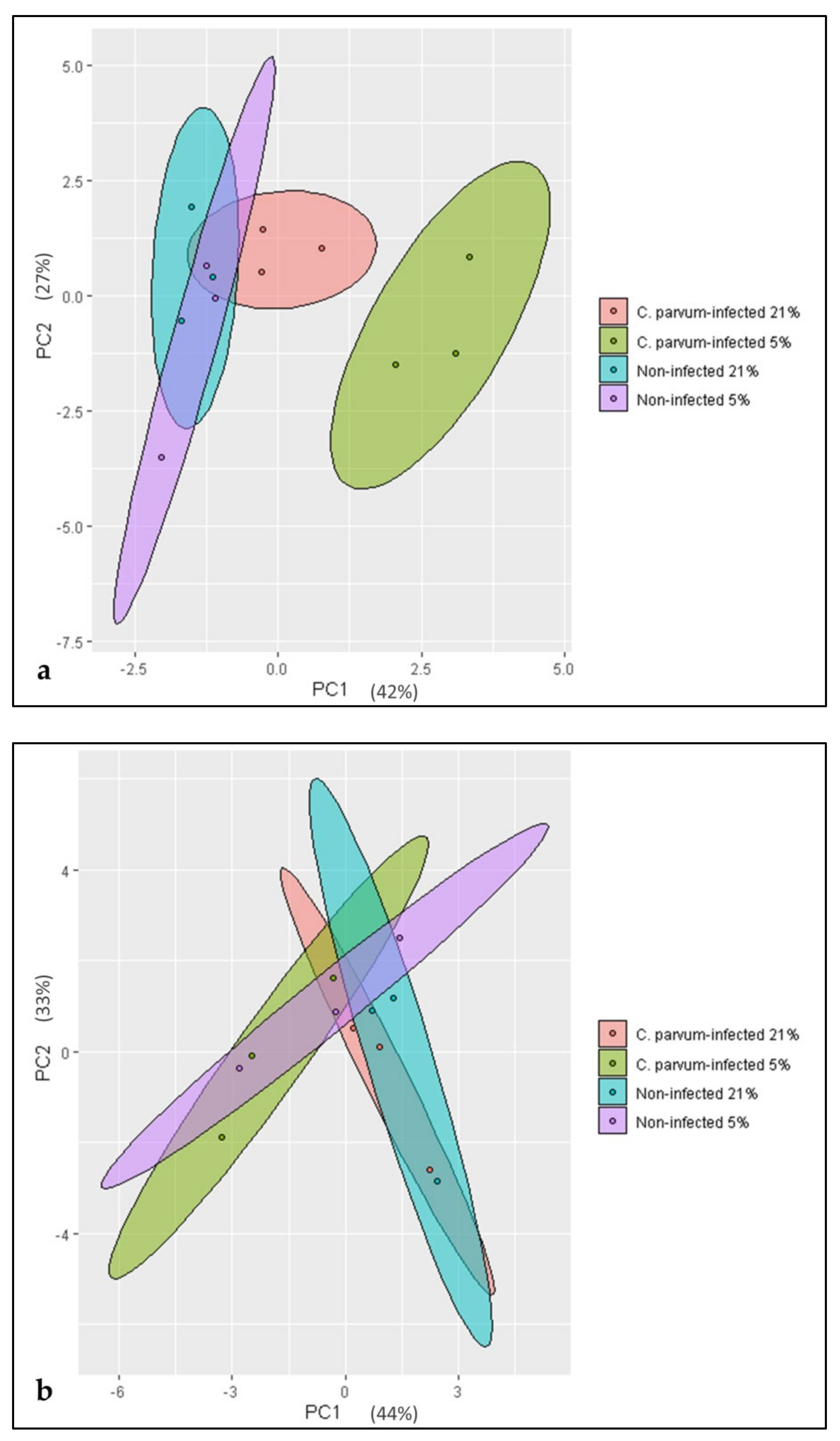
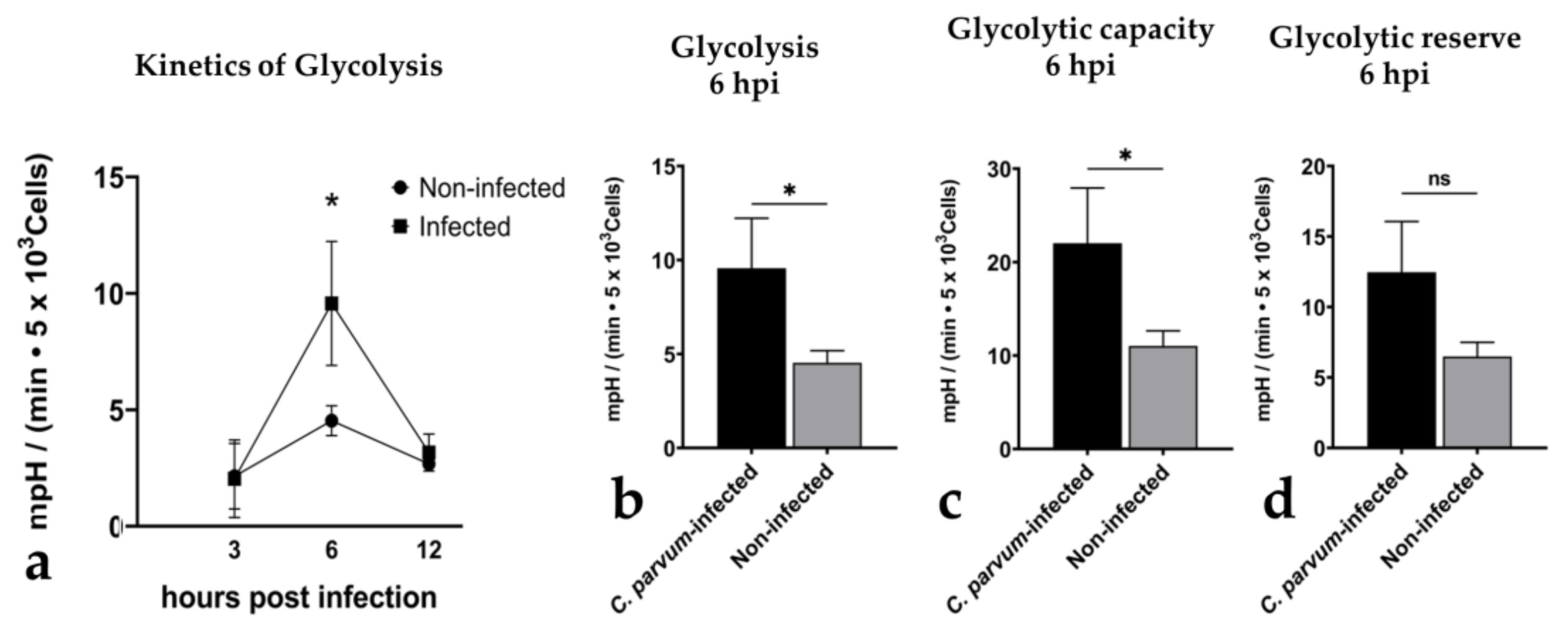
3.2. Metabolic Signatures of Cryptosporidium parvum-Infected BSI Explants Depend on Oxygen Concentrations
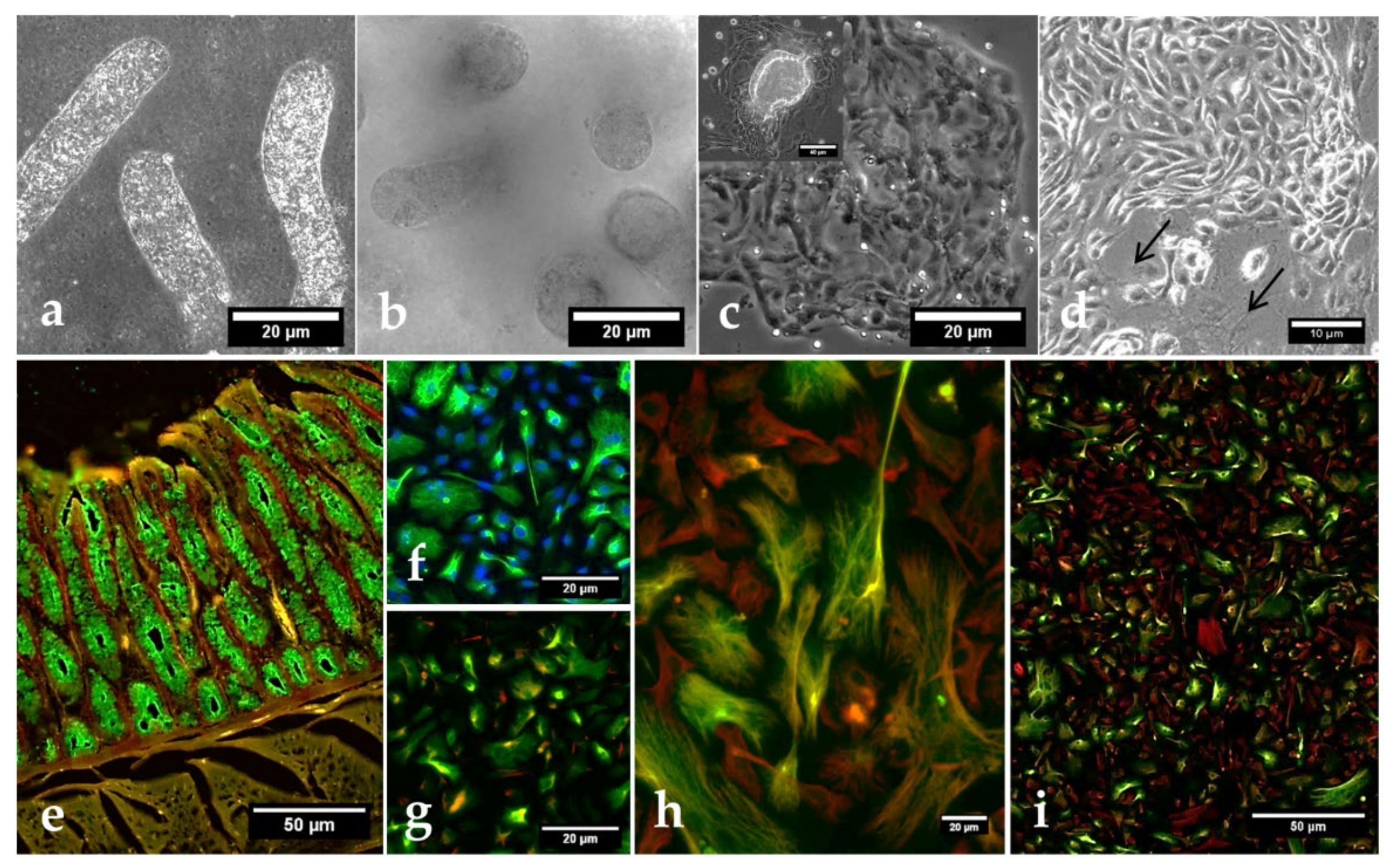
3.3. Characterization and Cryptosporidium parvum-Driven Reactions in Primary BSIEC
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- O’Connor, R.M.; Shaffie, R.; Kang, G.; Ward, H.D. Cryptosporidiosis in Patients with HIV/AIDS. AIDS 2011, 25, 549–560. [Google Scholar] [CrossRef] [PubMed]
- Liu, L.; Johnson, H.L.; Cousens, S.; Perin, J.; Scott, S.; Lawn, J.E.; Rudan, I.; Campbell, H.; Cibulskis, R.; Li, M.; et al. Global, Regional, and National Causes of Child Mortality: An Updated Systematic Analysis for 2010 with Time Trends since 2000. Lancet 2012, 379, 2151–2161. Available online: https://pubmed.ncbi.nlm.nih.gov/22579125/ (accessed on 29 December 2020). [CrossRef]
- Kotloff, K.L.; Nataro, J.P.; Blackwelder, W.C.; Nasrin, D.; Farag, T.H.; Panchalingam, S.; Wu, Y.; Sow, S.O.; Sur, D.; Breiman, R.F.; et al. Burden and Aetiology of Diarrhoeal Disease in Infants and Young Children in Developing Countries (the Global Enteric Multicenter Study, GEMS): A Prospective, Case-Control Study. Lancet 2013, 382, 209–222. [Google Scholar] [CrossRef]
- Vélez, J.; Lange, M.K.; Zieger, P.; Yoon, I.; Failing, K.; Bauer, C. Long-Term Use of Yeast Fermentation Products in Comparison to Halofuginone for the Control of Cryptosporidiosis in Neonatal Calves. Vet. Parasitol. 2019, 269, 57–64. [Google Scholar] [CrossRef] [PubMed]
- WHO. Available online: https://www.who.int (accessed on 18 February 2021).
- Amadi, B.; Mwiya, M.; Sianongo, S.; Payne, L.; Watuka, A.; Katubulushi, M.; Kelly, P. High Dose Prolonged Treatment with Nitazoxanide Is Not Effective for Cryptosporidiosis in HIV Positive Zambian Children: A Randomised Controlled Trial. BMC Infect. Dis. 2009, 9, 195. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Villacorta, I.; Peeters, J.; Vanopdenbosch, E.; Ares-Mazas, E.; Theys, H. Efficacy of Halofuginone Lactate against Cryptosporidium parvum in Calves. Antimicrob. Agents Chemother. 1991, 35, 283–287. [Google Scholar] [CrossRef] [Green Version]
- Abrahamsen, M.S.; Templeton, T.J.; Enomoto, S.; Abrahante, J.E.; Zhu, G.; Lancto, C.A.; Deng, M.; Liu, C.; Widmer, G.; Tzipori, S.; et al. Complete Genome Sequence of the Apicomplexan, Cryptosporidium parvum. Science 2004, 304, 441–445. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, S.; Roellig, D.M.; Guo, Y.; Li, N.; Frace, M.A.; Tang, K.; Zhang, L.; Feng, Y.; Xiao, L. Evolution of Mitosome Metabolism and Invasion-Related Proteins in Cryptosporidium. BMC Genom. 2016, 17, 1006. [Google Scholar] [CrossRef] [Green Version]
- Hublin, J.S.Y.N.; Ryan, U.; Trengove, R.; Maker, G. Metabolomic Profiling of Faecal Extracts from Cryptosporidium parvum Infection in Experimental Mouse Models. PLoS ONE 2013, 8, e77803. [Google Scholar] [CrossRef]
- Xu, T.; Ping, J.; Yu, Y.; Yu, F.; Yu, Y.; Hao, P.; Li, X. Revealing Parasite Influence in Metabolic Pathways in Apicomplexa Infected Patients. BMC Bioinform. 2010, 11, S13. [Google Scholar] [CrossRef] [Green Version]
- Miller, C.N.; Panagos, C.G.; Mosedale, W.R.T.; Kváč, M.; Howard, M.J.; Tsaousis, A.D. NMR Metabolomics Reveals Effects of Cryptosporidium Infections on Host Cell Metabolome. Gut Pathog. 2019, 11, 13. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vélez, J.; Velasquez, Z.; Silva, L.M.R.; Gärtner, U.; Failing, K.; Daugschies, A.; Mazurek, S.; Hermosilla, C.; Taubert, A. Metabolic Signatures of Cryptosporidium parvum-Infected HCT-8 Cells and Impact of Selected Metabolic Inhibitors on C. parvum Infection under Physioxia and Hyperoxia. Biology 2021, 10, 60. [Google Scholar] [CrossRef] [PubMed]
- Saric, J.; Wang, Y.; Li, J.; Coen, M.; Utzinger, J.; Marchesi, J.R.; Keiser, J.; Veselkov, K.; Lindon, J.C.; Nicholson, J.K. Species Variation in the Fecal Metabolome Gives Insight into Differential Gastrointestinal Function. J. Proteome Res. 2008, 7, 352–360. [Google Scholar] [CrossRef] [PubMed]
- Ng, J.S.Y.; Ryan, U.; Trengove, R.D.; Maker, G.L. Development of an Untargeted Metabolomics Method for the Analysis of Human Faecal Samples Using Cryptosporidium-Infected Samples. Mol. Biochem. Parasit. 2012, 185, 145–150. [Google Scholar] [CrossRef]
- Harp, J.A.; Chen, W.; Harmsen, A.G. Resistance of Severe Combined Immunodeficient Mice to Infection with Cryptosporidium parvum: The Importance of Intestinal Microflora. Infect. Immun. 1992, 60, 3509–3512. [Google Scholar] [CrossRef] [Green Version]
- Chappell, C.L.; Darkoh, C.; Shimmin, L.; Farhana, N.; Kim, D.-K.; Okhuysen, P.C.; Hixson, J. Fecal Indole as a Biomarker of Susceptibility to Cryptosporidium Infection. Infect. Immun. 2016, 84, 2299–2306. [Google Scholar] [CrossRef] [Green Version]
- Fuqua, W.C.; Winans, S.C.; Greenberg, E.P. Quorum Sensing in Bacteria: The LuxR-LuxI Family of Cell Density-Responsive Transcriptional Regulators. J. Bacteriol. 1994, 176, 269–275. [Google Scholar] [CrossRef] [Green Version]
- Nadell, C.D.; Xavier, J.B.; Levin, S.A.; Foster, K.R. The Evolution of Quorum Sensing in Bacterial Biofilms. PLoS Biol. 2008, 6, e14. [Google Scholar] [CrossRef] [Green Version]
- Martino, P.D.; Fursy, R.; Bret, L.; Sundararaju, B.; Phillips, R.S. Indole Can Act as an Extracellular Signal to Regulate Biofilm Formation of Escherichia coli and Other Indole-Producing Bacteria. Can. J. Microbiol. 2003, 49, 443–449. [Google Scholar] [CrossRef]
- Muñoz-Caro, T.; Lendner, M.; Daugschies, A.; Hermosilla, C.; Taubert, A. NADPH Oxidase, MPO, NE, ERK1/2, P38 MAPK and Ca2+ Influx Are Essential for Cryptosporidium parvum-Induced NET Formation. Dev. Comp. Immunol. 2015, 52, 245–254. [Google Scholar] [CrossRef]
- Bär, A.-K.; Phukan, N.; Pinheiro, J.; Simoes-Barbosa, A. The Interplay of Host Microbiota and Parasitic Protozoans at Mucosal Interfaces: Implications for the Outcomes of Infections and Diseases. PLoS Neglect. Trop. Dis. 2015, 9, e0004176. [Google Scholar] [CrossRef] [PubMed]
- Zahedi, A.; Ryan, U. Cryptosporidium—An Update with an Emphasis on Foodborne and Waterborne Transmission. Res. Vet. Sci. 2020, 132, 500–512. [Google Scholar] [CrossRef]
- Ryan, U.; Fayer, R.; Xiao, L. Cryptosporidium Species in Humans and Animals: Current Understanding and Research Needs. Parasitology 2014, 141, 1667–1685. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shahiduzzaman, M.; Dyachenko, V.; Obwaller, A.; Unglaube, S.; Daugschies, A. Combination of Cell Culture and Quantitative PCR for Screening of Drugs against Cryptosporidium parvum. Vet. Parasitol. 2009, 162, 271–277. [Google Scholar] [CrossRef] [PubMed]
- Girard, F.; Dziva, F.; van Diemen, P.; Phillips, A.D.; Stevens, M.P.; Frankel, G. Adherence of Enterohemorrhagic Escherichia coli O157, O26, and O111 Strains to Bovine Intestinal Explants. Ex Vivo Appl. Environ. Microbiol. 2007, 73, 3084–3090. [Google Scholar] [CrossRef] [Green Version]
- Castellanos-Gonzalez, A.; Cabada, M.M.; Nichols, J.; Gomez, G.; White, A.C. Human Primary Intestinal Epithelial Cells as an Improved In Vitro Model for Cryptosporidium parvum Infection. Infect. Immun. 2013, 81, 1996–2001. [Google Scholar] [CrossRef] [Green Version]
- Ren, H.J.; Zhang, C.L.; Liu, R.D.; Li, N.; Li, X.G.; Xue, H.K.; Guo, Y.; Wang, Z.Q.; Cui, J.; Ming, L. Primary Cultures of Mouse Small Intestinal Epithelial Cells Using the Dissociating Enzyme Type I Collagenase and Hyaluronidase. Braz. J. Med. Biol. Res. 2017, 50, e5831. [Google Scholar] [CrossRef] [Green Version]
- Zheng, L.; Kelly, C.J.; Colgan, S.P. Physiologic Hypoxia and Oxygen Homeostasis in the Healthy Intestine. A Review in the Theme: Cellular Responses to Hypoxia. Am. J. Physiol. Cell Physiol. 2015, 309, C350–C360. [Google Scholar] [CrossRef] [Green Version]
- Wenger, R.H.; Kurtcuoglu, V.; Scholz, C.C.; Marti, H.H.; Hoogewijs, D. Frequently Asked Questions in Hypoxia Research. Hypoxia 2015, 3, 35–43. [Google Scholar] [CrossRef] [Green Version]
- Rusu, D.; Loret, S.; Peulen, O.; Mainil, J.; Dandrifosse, G. Immunochemical, Biomolecular and Biochemical Characterization of Bovine Epithelial Intestinal Primocultures. BMC Cell Biol. 2005, 6, 42. [Google Scholar] [CrossRef] [Green Version]
- King, B.J.; Keegan, A.R.; Monis, P.T.; Saint, C.P. Environmental Temperature Controls Cryptosporidium Oocyst Metabolic Rate and Associated Retention of Infectivity. Appl. Environ. Microbiol. 2005, 71, 3848–3857. [Google Scholar] [CrossRef] [Green Version]
- Varughese, E.A.; Bennett-Stamper, C.L.; Wymer, L.J.; Yadav, J.S. A New In vitro Model Using Small Intestinal Epithelial Cells to Enhance Infection of Cryptosporidium parvum. J. Microbiol. Methods 2014, 106, 47–54. [Google Scholar] [CrossRef]
- Taubert, A.; Hermosilla, C.; Silva, L.M.R.; Wieck, A.; Failing, K.; Mazurek, S. Metabolic Signatures of Besnoitia besnoiti-Infected Endothelial Host Cells and Blockage of Key Metabolic Pathways Indicate High Glycolytic and Glutaminolytic Needs of the Parasite. Parasitol. Res. 2016, 115, 2023–2034. [Google Scholar] [CrossRef]
- Ammon, H.P.T. Methoden Der Enzymatischen Analyse. Von H. U. Bergmeyer (Hrsg. Unter Mitarb. von K. Gawehn). 3., Neu Bearb. Und Erw. Aufl., 2 Bde., Insg. 2513 S., Verlag Chemie GmbH Weinheim/Bergstr. 1974, DM 460,—. Pharmazie Zeit 1975, 4, 92–93. [Google Scholar] [CrossRef]
- Mazurek, S.; Eigenbrodt, E.; Failing, K.; Steinberg, P. Alterations in the Glycolytic and Glutaminolytic Pathways after Malignant Transformation of Rat Liver Oval Cells. J. Cell. Physiol. 1999, 181, 136–146. [Google Scholar] [CrossRef]
- Taubert, A.; Zahner, H.; Hermosilla, C. Dynamics of Transcription of Immunomodulatory Genes in Endothelial Cells Infected with Different Coccidian Parasites. Vet. Parasitol. 2006, 142, 214–222. [Google Scholar] [CrossRef] [PubMed]
- RStudio. Open Source & Professional Software for Data Science Teams. Available online: https://rstudio.com/ (accessed on 21 February 2021).
- R: The R Project for Statistical Computing. Available online: https://www.r-project.org/ (accessed on 21 February 2021).
- Dragulescu, A.; Arendt, C. Xlsx: Read, Write, Format Excel 2007 and Excel 97/2000/XP/2003 Files. Available online: https://CRAN.R-project.org/package=xlsx (accessed on 21 February 2021).
- Angus, K.W.; Tzipori, S.; Gray, E.W. Intestinal Lesions in Specific-Pathogen-Free Lambs Associated with a Cryptosporidium from Calves with Diarrhea. Vet. Pathol. 1982, 19, 67–78. [Google Scholar] [CrossRef] [Green Version]
- Mahmoud, A.H.A.; Slate, J.R.; Hong, S.; Yoon, I.; McGill, J.L. Supplementing a Saccharomyces cerevisiae Fermentation Product Modulates Innate Immune Function and Ameliorates Bovine Respiratory Syncytial Virus Infection in Neonatal Calves. J. Anim. Sci. 2020, 98, skaa252. [Google Scholar] [CrossRef] [PubMed]
- Vázquez Flores, S.; de Jesús Guerrero Carrillo, M.; Scott, M.F.; Hamann, J.; Almanza, S.B.; Guizar Bravo, C.; Quintana, A.P.B.; Vargasb, P.J.A. 1472 Effects of Saccharomyces cerevisiae Fermentation Products on Intestinal Villi Integrity in Neonatal Calves Naturally Infected with Cryptosporidium spp. J. Anim. Sci. 2016, 94, 714–715. [Google Scholar] [CrossRef]
- Becattini, S.; Sorbara, M.T.; Kim, S.G.; Littmann, E.L.; Dong, Q.; Walsh, G.; Wright, R.; Amoretti, L.; Fontana, E.; Hohl, T.M.; et al. Rapid Transcriptional and Metabolic Adaptation of Intestinal Microbes to Host Immune Activation. Cell Host Microbe 2021, 29, 378–393. [Google Scholar] [CrossRef]
- Fischbach, M.A.; Sonnenburg, J.L. Eating For Two: How Metabolism Establishes Interspecies Interactions in the Gut. Cell Host Microbe 2011, 10, 336–347. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Weinstein, N.; Garten, B.; Vainer, J.; Minaya, D.; Czaja, K. Managing the Microbiome: How the Gut Influences Development and Disease. Nutrients 2021, 13, 74. [Google Scholar] [CrossRef]
- Ryan, R.P.; Dow, J.M. Diffusible Signals and Interspecies Communication in Bacteria. Microbiology 2008, 154, 1845–1858. [Google Scholar] [CrossRef] [Green Version]
- McCole, D.F.; Eckmann, L.; Laurent, F.; Kagnoff, M.F. Intestinal Epithelial Cell Apoptosis Following Cryptosporidium parvum Infection. Infect. Immun. 2000, 68, 1710–1713. [Google Scholar] [CrossRef] [Green Version]
- Laurent, F.; Lacroix-Lamandé, S. Innate Immune Responses Play a Key Role in Controlling Infection of the Intestinal Epithelium by Cryptosporidium. Int. J. Parasitol. 2017, 47, 711–721. [Google Scholar] [CrossRef] [PubMed]
- Isaacs, D.; Hunt, G.; Phillips, A.; Price, E.; Raafat, F.; Walker-Smith, J. Cryptosporidiosis in Immunocompetent Children. J. Clin. Pathol. 1985, 38, 76–81. [Google Scholar] [CrossRef] [Green Version]
- Gookin, J.L.; Nordone, S.K.; Argenzio, R.A. Host Responses to Cryptosporidium Infection. J. Vet. Intern. Med. 2002, 16, 12–21. [Google Scholar] [CrossRef]
- Woodmansee, D.; Pohlenz, J.F.L. Development of Cryptosporidium sp. in a Human Rectal Tumor Cell Line. In Proceedings of the Fourth International Symposium on Neonatal Diarrhea, Saskatoon, SK, Canada, 3–5 October 1983; pp. 306–319. [Google Scholar]
- Current, W.L.; Haynes, T.B. Complete Development of Cryptosporidium in Cell Culture. Science 1984, 224, 603–605. [Google Scholar] [CrossRef]
- Alcantara Warren, C.; Destura, R.V.; Sevilleja, J.E.A.D.; Barroso, L.F.; Carvalho, H.; Barrett, L.J.; O’Brien, A.D.; Guerrant, R.L. Detection of Epithelial-Cell Injury, and Quantification of Infection, in the HCT-8 Organoid Model of Cryptosporidiosis. J. Infect. Dis. 2008, 198, 143–149. [Google Scholar] [CrossRef] [PubMed]
- RePass, M.A.D.; Chen, Y.; Lin, Y.; Zhou, W.; Kaplan, D.L.; Ward, H.D. Novel Bioengineered Three-Dimensional Human Intestinal Model for Long-Term Infection of Cryptosporidium parvum. Infect. Immun. 2017, 85. [Google Scholar] [CrossRef] [Green Version]
- Miller, C.N.; Jossé, L.; Brown, I.; Blakeman, B.; Povey, J. A cell culture platform for Cryptosporidium that enables long-term cultivation and new tools for the systematic investigation of its biology. Int. J. Parasitol. 2018, 48, 197–201. [Google Scholar] [CrossRef]
- Wilke, G.; Funkhouser-Jones, L.J.; Wang, Y.; Ravindran, S.; Wang, Q.; Beatty, W.L.; Baldridge, M.T.; VanDussen, K.L.; Shen, B.; Kuhlenschmidt, M.S.; et al. A Stem-Cell-Derived Platform Enables Complete Cryptosporidium Development In Vitro and Genetic Tractability. Cell Host Microbe 2019, 26, 123–134.e8. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Carreau, A.; Hafny-Rahbi, B.E.; Matejuk, A.; Grillon, C.; Kieda, C. Why Is the Partial Oxygen Pressure of Human Tissues a Crucial Parameter? Small Molecules and Hypoxia. J. Cell Mol. Med. 2011, 15, 1239–1253. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Papandreou, I.; Cairns, R.A.; Fontana, L.; Lim, A.L.; Denko, N.C. HIF-1 Mediates Adaptation to Hypoxia by Actively Downregulating Mitochondrial Oxygen Consumption. Cell Metab. 2006, 3, 187–197. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lindmark, D.G.; Müller, M. Hydrogenosome, a Cytoplasmic Organelle of the Anaerobic Flagellate Tritrichomonas foetus, and Its Role in Pyruvate Metabolism. J. Biol. Chem. 1973, 248, 7724–7728. [Google Scholar] [CrossRef]
- Zimorski, V.; Mentel, M.; Tielens, A.G.M.; Martin, W.F. Energy Metabolism in Anaerobic Eukaryotes and Earth’s Late Oxygenation. Free Radic. Biol. Med. 2019, 140, 279–294. [Google Scholar] [CrossRef] [PubMed]
- Cacciò, S.M.; Widmer, G. Cryptosporidium: Parasite and Disease; Springer: Berlin/Heidelberg, Germany, 2013; ISBN 3-7091-1562-0. [Google Scholar]
- Radwanski, E.R.; Last, R.L. Tryptophan Biosynthesis and Metabolism: Biochemical and Molecular Genetics. Plant. Cell 1995, 7, 921–934. [Google Scholar] [PubMed] [Green Version]
- Shanmugasundram, A.; Gonzalez-Galarza, F.F.; Wastling, J.M.; Vasieva, O.; Jones, A.R. Library of Apicomplexan Metabolic Pathways: A Manually Curated Database for Metabolic Pathways of Apicomplexan Parasites. Nucleic Acids Res. 2013, 41, D706–D713. [Google Scholar] [CrossRef] [Green Version]
- Burgess, S.L.; Gilchrist, C.A.; Lynn, T.C.; Petri, W.A. Parasitic Protozoa and Interactions with the Host Intestinal Microbiota. Infect. Immun. 2017, 85, e00101-17. [Google Scholar] [CrossRef] [Green Version]
- Bassler, B.L. How Bacteria Talk to Each Other: Regulation of Gene Expression by Quorum Sensing. Curr. Opin. Microbiol. 1999, 2, 582–587. [Google Scholar] [CrossRef]
- Wang, W.; Huang, J.; Zhang, J.; Wang, Z.; Li, H.; Amenyogbe, E.; Chen, G. Effects of Hypoxia Stress on the Intestinal Microflora of Juvenile of Cobia (Rachycentron Canadum). Aquaculture 2021, 536, 736419. [Google Scholar] [CrossRef]
- Lee, J.-H.; Lee, J. Indole as an Intercellular Signal in Microbial Communities. FEMS Microbiol. Rev. 2010, 34, 426–444. [Google Scholar] [CrossRef] [PubMed]
- Xu, J.; Li, Y.; Yang, Z.; Li, C.; Liang, H.; Wu, Z.; Pu, W. Yeast probiotics shape the gut microbiome and improve the health of early-weaned piglets. Front. Microbiol. 2018, 9, 2011. [Google Scholar] [CrossRef]
- Chou, W.K.; Park, J.; Carey, J.B.; McIntyre, D.R.; Berghman, L.R. Immunomodulatory Effects of Saccharomyces cerevisiae Fermentation Product Supplementation on Immune Gene Expression and Lymphocyte Distribution in Immune Organs in Broilers. Front. Vet. Sci. 2017, 4, 37. [Google Scholar] [CrossRef] [Green Version]
- Adams, C.A. The Probiotic Paradox: Live and Dead Cells Are Biological Response Modifiers. Nutr. Res. Rev. 2010, 23, 37–46. [Google Scholar] [CrossRef] [Green Version]
- Bouzid, M.; Hunter, P.R.; Chalmers, R.M.; Tyler, K.M. Cryptosporidium Pathogenicity and Virulence. Clin. Microbiol. Rev. 2013, 26, 115–134. [Google Scholar] [CrossRef] [Green Version]
- Naghavi, M.; Abajobir, A.A.; Abbafati, C.; Abbas, K.M.; Abd-Allah, F.; Abera, S.F.; Aboyans, V.; Adetokunboh, O.; Afshin, A.; Agrawal, A.; et al. Global, Regional, and National Age-Sex Specific Mortality for 264 Causes of Death, 1980–2016: A Systematic Analysis for the Global Burden of Disease Study 2016. Lancet 2017, 390, 1151–1210. [Google Scholar] [CrossRef] [Green Version]
- Roth, G.A.; Abate, D.; Abate, K.H.; Abay, S.M.; Abbafati, C.; Abbasi, N.; Abbastabar, H.; Abd-Allah, F.; Abdela, J.; Abdelalim, A.; et al. Global, Regional, and National Age-Sex-Specific Mortality for 282 Causes of Death in 195 Countries and Territories, 1980–2017: A Systematic Analysis for the Global Burden of Disease Study 2017. Lancet 2018, 392, 1736–1788. [Google Scholar] [CrossRef] [Green Version]
- Karpe, A.V.; Hutton, M.L.; Mileto, S.J.; James, M.L.; Evans, C.; Shah, R.M.; Ghodke, A.B.; Hillyer, K.E.; Metcalfe, S.S.; Liu, J.-W.; et al. Cryptosporidiosis Modulates the Gut Microbiome and Metabolism in a Murine Infection Model. Metabolites 2021, 11, 380. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Vélez, J.; Silva, L.M.R.; Gärtner, U.; Daugschies, A.; Mazurek, S.; Hermosilla, C.; Taubert, A. First Metabolic Insights into Ex Vivo Cryptosporidium parvum-Infected Bovine Small Intestinal Explants Studied under Physioxic Conditions. Biology 2021, 10, 963. https://doi.org/10.3390/biology10100963
Vélez J, Silva LMR, Gärtner U, Daugschies A, Mazurek S, Hermosilla C, Taubert A. First Metabolic Insights into Ex Vivo Cryptosporidium parvum-Infected Bovine Small Intestinal Explants Studied under Physioxic Conditions. Biology. 2021; 10(10):963. https://doi.org/10.3390/biology10100963
Chicago/Turabian StyleVélez, Juan, Liliana M. R. Silva, Ulrich Gärtner, Arwid Daugschies, Sybille Mazurek, Carlos Hermosilla, and Anja Taubert. 2021. "First Metabolic Insights into Ex Vivo Cryptosporidium parvum-Infected Bovine Small Intestinal Explants Studied under Physioxic Conditions" Biology 10, no. 10: 963. https://doi.org/10.3390/biology10100963
APA StyleVélez, J., Silva, L. M. R., Gärtner, U., Daugschies, A., Mazurek, S., Hermosilla, C., & Taubert, A. (2021). First Metabolic Insights into Ex Vivo Cryptosporidium parvum-Infected Bovine Small Intestinal Explants Studied under Physioxic Conditions. Biology, 10(10), 963. https://doi.org/10.3390/biology10100963






