Coexpression of Fungal Cell Wall-Modifying Enzymes Reveals Their Additive Impact on Arabidopsis Resistance to the Fungal Pathogen, Botrytis cinerea
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Arabidopsis Growth Conditions
2.2. Generation of Transgenic Plants
2.3. RNA Extraction, cDNA Synthesis, and Real-Time qPCR
2.4. CW Composition Analysis
2.5. Infection of Arabidopsis Genotypes with Botrytis Cinerea
2.6. Determination of H2O2 Accumulation
2.7. Statistical Analysis
3. Results
3.1. Generation of Transgenic Arabidopsis Plants Expressing A. nidulans Acetylesterases (AnAXE and AnRAE) and Feruloylesterase (AnFAE)
3.2. Expression of Fungal Acetylesterases Reduced the Degree of CW Acetylation in Transgenic Arabidopsis
3.3. Transgenic Arabidopsis Expressing Fungal Feruloylesterase Showed Reduction in CW Feruloylation
3.4. Acetylesterase Expressing Plants Have Increased Resistance to Necrotrophic Fungal Pathogen
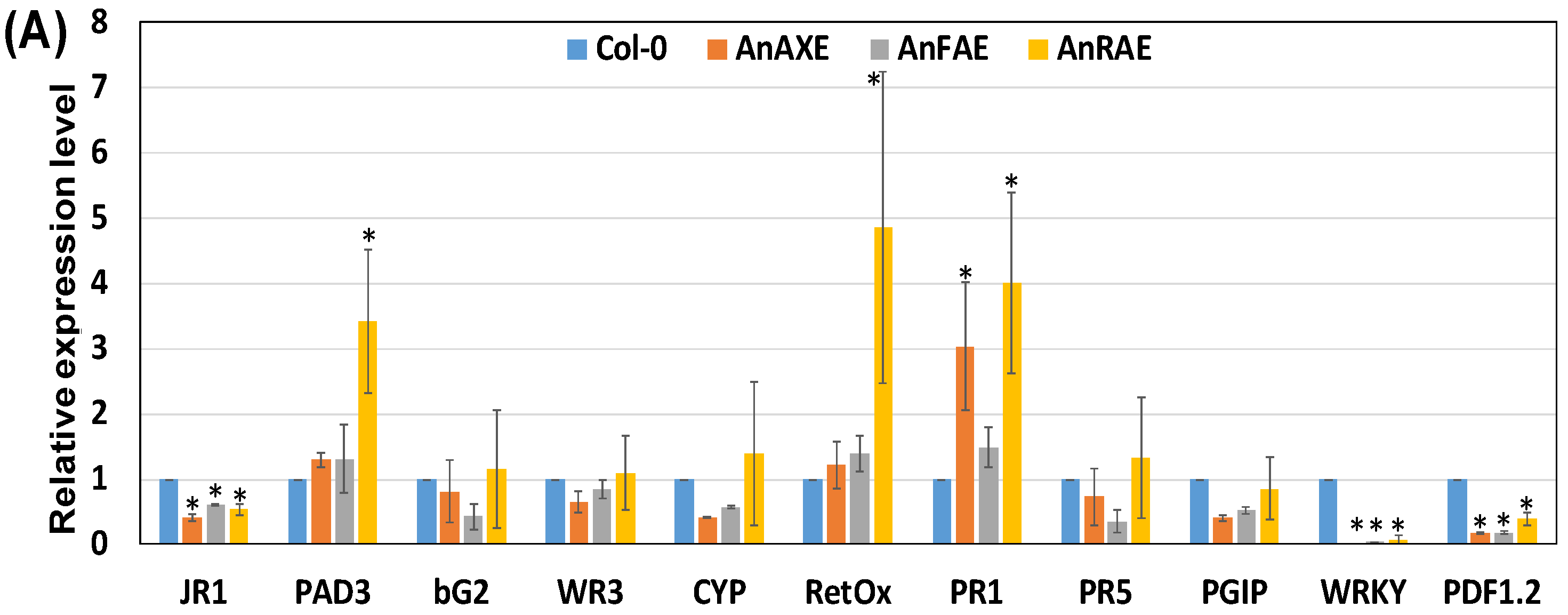
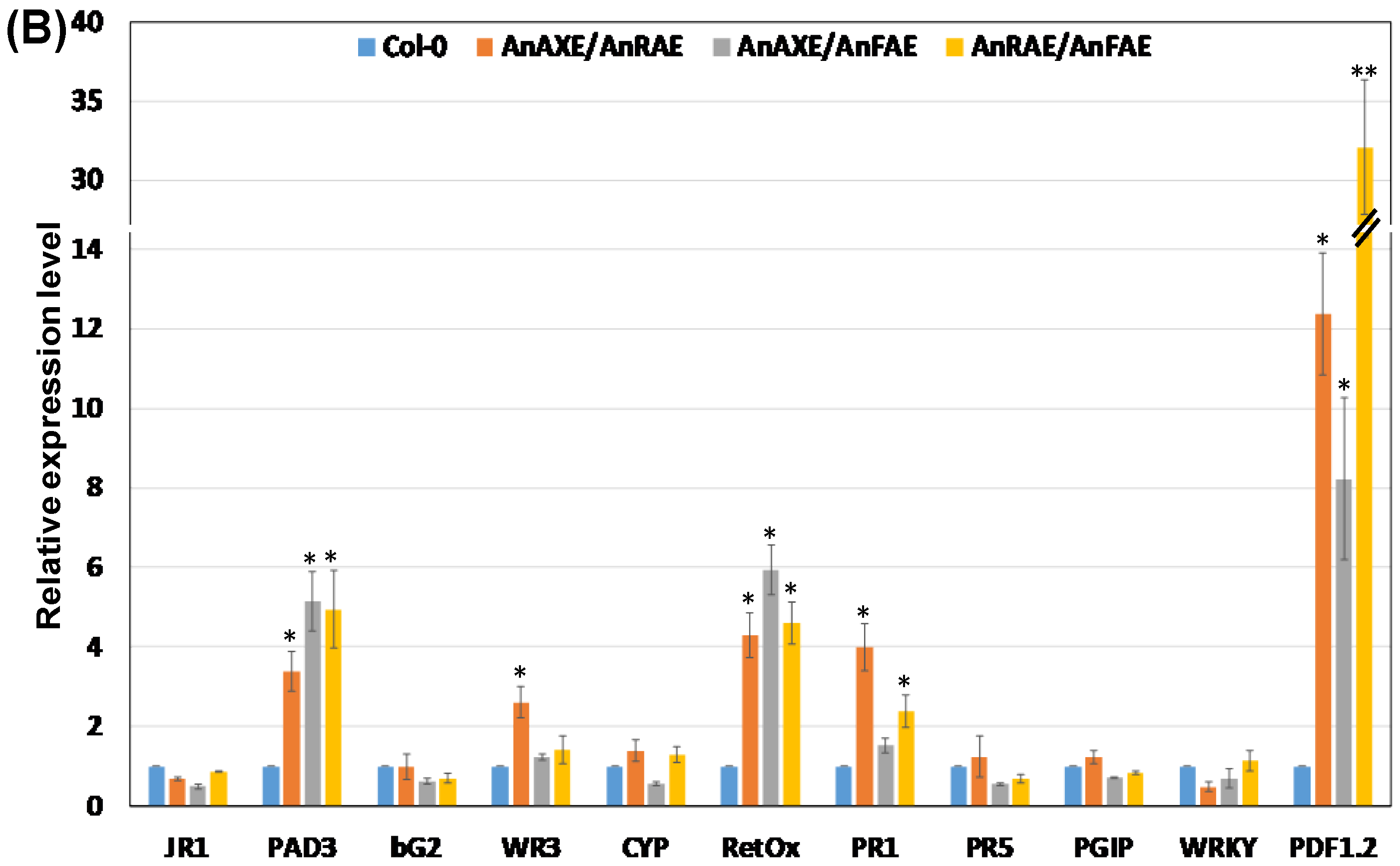
3.5. Deacetylation and Deferuloylation of CW Resulted in Expression of Pathogen-Responsive Genes
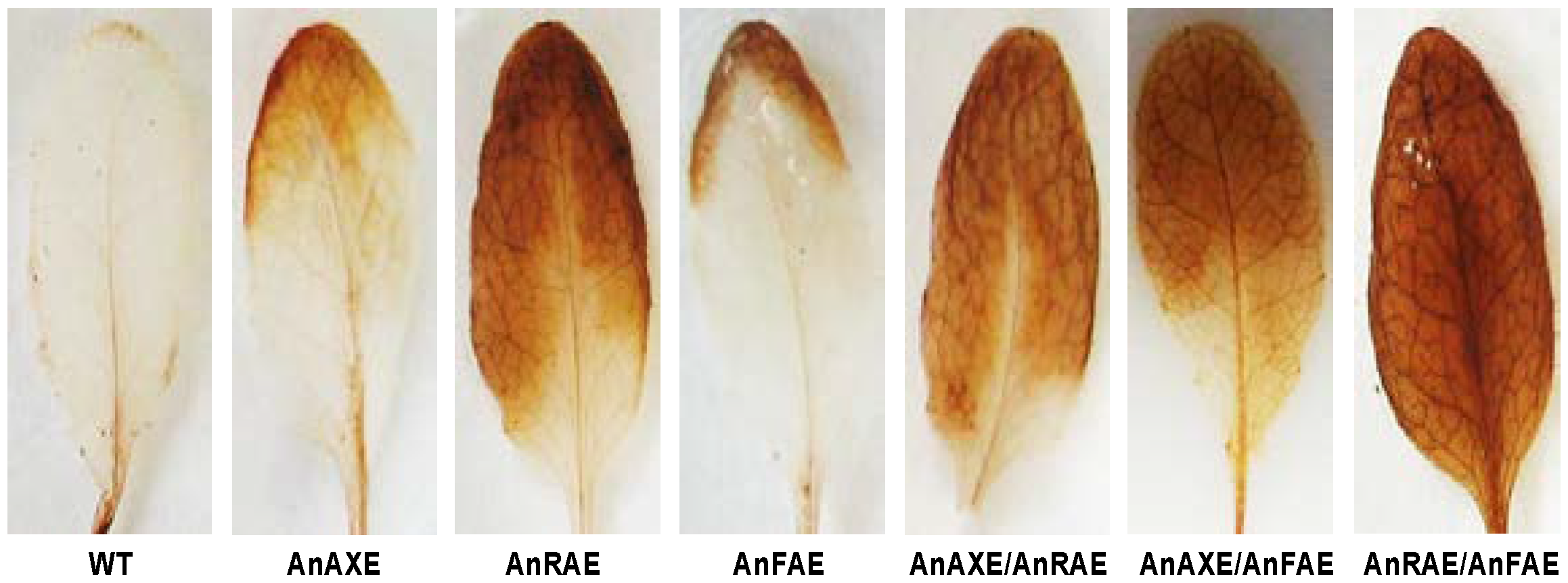
3.6. Acetylesterase Expressing Plants Have Enhanced H2O2 Accumulation
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Srivastava, V.; McKee, L.S.; Bulone, V. Plant Cell Walls. In eLS; John Wiley & Sons Ltd.: Chichester, UK, 2017; Volume 154, pp. 1–17. [Google Scholar]
- Cosgrove, D.J. Growth of the plant cell wall. Nat. Rev. Mol. Cell Biol. 2005, 6, 850–861. [Google Scholar] [CrossRef] [PubMed]
- Lampugnani, E.R.; Khan, G.A.; Somssich, M.; Persson, S. Building a plant cell wall at a glance. J. Cell Sci. 2018, 131, jcs207373. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Caffall, K.H.; Mohnen, D. The structure, function, and biosynthesis of plant cell wall pectic polysaccharides. Carbohydr. Res. 2009, 344, 1879–1900. [Google Scholar] [CrossRef] [PubMed]
- Cosgrove, D.J. Plant cell wall extensibility: Connecting plant cell growth with cell wall structure, mechanics, and the action of wall-modifying enzymes. J. Exp. Bot. 2016, 67, 463–476. [Google Scholar] [CrossRef] [PubMed]
- Gou, J.-Y.; Miller, L.M.; Hou, G.; Yu, X.-H.; Chen, X.-Y.; Liu, C.-J. Acetylesterase-mediated deacetylation of pectin impairs cell elongation, pollen germination, and plant reproduction. Plant Cell 2012, 24, 50–65. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lionetti, V.; Metraux, J.P. Plant cell wall in pathogenesis, parasitism and symbiosis. Front. Plant Sci. 2014, 612, 5. [Google Scholar] [CrossRef] [Green Version]
- Bellincampi, D.; Cervone, F.; Lionetti, V. Plant cell wall dynamics and wall-related susceptibility in plant-pathogen interactions. Front. Plant Sci. 2014, 5, 228. [Google Scholar] [CrossRef] [Green Version]
- Kesten, C.; Menna, A.; Sanchez-Rodriguez, C. Regulation of cellulose synthesis in response to stress. Curr. Opin. Plant Biol. 2017, 40, 106–113. [Google Scholar] [CrossRef]
- Malinovsky, F.G.; Fangel, J.U.; Willats, W.G.T. The role of the cell wall in plant immunity. Front. Plant Sci. 2014, 5, 178. [Google Scholar] [CrossRef] [Green Version]
- Miedes, E.; Vanholme, R.; Boerjan, W.; Molina, A. The role of the secondary cell wall in plant resistance to pathogens. Front. Plant Sci. 2014, 5, 358. [Google Scholar] [CrossRef] [Green Version]
- Bacete, L.; Melida, H.; Lopez, G.; Dabos, P.; Tremousaygue, D.; Denance, N.; Miedes, E.; Bulone, V.; Goffner, D.; Molina, A. Arabidopsis Response Regulator 6 (ARR6) modulates plant cell-wall composition and disease resistance. Mol. Plant-Microbe Interact. 2020, 33, 767–780. [Google Scholar] [CrossRef]
- Houston, K.; Tucker, M.R.; Chowdhury, J.; Shirley, N.; Little, A. The plant cell wall: A complex and dynamic structure as revealed by the responses of genes under stress conditions. Front. Plant Sci. 2016, 7, 984. [Google Scholar] [CrossRef] [Green Version]
- Molina, A.; Miedes, E.; Bacete, L.; Rodriguez, T.; Melida, H.; Denance, N.; Sanchez-Vallet, A.; Riviere, M.P.; Lopez, G.; Freydier, A.; et al. Arabidopsis cell wall composition determines disease resistance specificity and fitness. Proc. Natl. Acad. Sci. USA 2021, 118, 5. [Google Scholar] [CrossRef] [PubMed]
- Nafisi, M.; Fimognari, L.; Sakuragi, Y. Interplays between the cell wall and phytohormones in interaction between plants and necrotrophic pathogens. Phytochemistry 2015, 112, 63–71. [Google Scholar] [CrossRef] [Green Version]
- Bacete, L.; Melida, H.; Miedes, E.; Molina, A. Plant cell wall-mediated immunity: Cell wall changes trigger disease resistance responses. Plant J. 2018, 93, 614–636. [Google Scholar] [CrossRef]
- Lionetti, V.; Raiola, A.; Cervone, F.; Bellincampi, D. Transgenic expression of pectin methylesterase inhibitors limits tobamovirus spread in tobacco and Arabidopsis. Mol. Plant Pathol. 2014, 15, 265–274. [Google Scholar] [CrossRef]
- Boller, T.; Felix, G. A Renaissance of elicitors: Perception of microbe-associated molecular patterns and danger signals by pattern-recognition receptors. Annu. Rev. Plant Biol. 2009, 60, 379–406. [Google Scholar] [CrossRef] [PubMed]
- Boutrot, F.; Zipfel, C. Function, discovery, and exploitation of plant pattern recognition receptors for broad-spectrum disease resistance. Annu. Rev. Phytopathol. 2017, 55, 257–286. [Google Scholar] [CrossRef] [PubMed]
- Hamann, T. Plant cell wall integrity maintenance as an essential component of biotic stress response mechanisms. Front. Plant Sci. 2012, 3, 77. [Google Scholar] [CrossRef] [Green Version]
- Lionetti, V.; Fabri, E.; De Caroli, M.; Hansen, A.R.; Willats, W.G.T.; Piro, G.; Bellincampi, D. Three pectin methylesterase inhibitors protect cell wall integrity for Arabidopsis immunity to Botrytis. Plant Physiol. 2017, 173, 1844–1863. [Google Scholar] [CrossRef] [Green Version]
- Kubicek, C.P.; Starr, T.L.; Glass, N.L. Plant cell wall-degrading enzymes and their secretion in plant-pathogenic fungi. Annu. Rev. Phytopathol. 2014, 52, 427–451. [Google Scholar] [CrossRef]
- Nuehse, T.S. Cell wall integrity signaling and innate immunity in plants. Front. Plant Sci. 2012, 3, 280. [Google Scholar] [CrossRef] [Green Version]
- Zhang, H.; Hong, Y.; Huang, L.; Li, D.; Song, F. Arabidopsis AtERF014 acts as a dual regulator that differentially modulates immunity against Pseudomonas syringae pv. tomato and Botrytis cinerea. Sci. Rep. 2016, 6, 30251. [Google Scholar] [CrossRef]
- Bethke, G.; Thao, A.; Xiong, G.; Li, B.; Soltis, N.E.; Hatsugai, N.; Hillmer, R.A.; Katagiri, F.; Kliebenstein, D.J.; Pauly, M.; et al. Pectin biosynthesis is critical for cell wall integrity and immunity in Arabidopsis thaliana. Plant Cell 2016, 28, 537–556. [Google Scholar] [CrossRef] [Green Version]
- Engelsdorf, T.; Will, C.; Hofmann, J.; Schmitt, C.; Merritt, B.B.; Rieger, L.; Frenger, M.S.; Marschall, A.; Franke, R.B.; Pattathil, S.; et al. Cell wall composition and penetration resistance against the fungal pathogen Colletotrichum higginsianum are affected by impaired starch turnover in Arabidopsis mutants. J. Exp. Bot. 2017, 68, 701–713. [Google Scholar]
- Vogel, J.P.; Raab, T.K.; Schiff, C.; Somerville, S.C. PMR6, a pectate lyase-like gene required for powdery mildew susceptibility in Arabidopsis. Plant Cell 2002, 14, 2095–2106. [Google Scholar] [CrossRef] [Green Version]
- Vogel, J.P.; Raab, T.K.; Somerville, C.R.; Somerville, S.C. Mutations in PMR5 result in powdery mildew resistance and altered cell wall composition. Plant J. 2004, 40, 968–978. [Google Scholar] [CrossRef] [PubMed]
- Atmodjo, M.A.; Hao, Z.; Mohnen, D. Evolving views of pectin biosynthesis. Annu. Rev. Plant Biol. 2013, 64, 747–779. [Google Scholar] [CrossRef] [Green Version]
- Lionetti, V.; Cervone, F.; Bellincampi, D. Methyl esterification of pectin plays a role during plant-pathogen interactions and affects plant resistance to diseases. J. Plant Physiol. 2012, 169, 1623–1630. [Google Scholar] [CrossRef] [PubMed]
- Del Corpo, D.; Fullone, M.R.; Miele, R.; Lafond, M.; Pontiggia, D.; Grisel, S.; Kieffer-Jaquinod, S.; Giardina, T.; Bellincampi, D.; Lionetti, V. AtPME17 is a functional Arabidopsis thaliana pectin methylesterase regulated by its PRO region that triggers PME activity in the resistance to Botrytis cinerea. Mol. Plant Pathol. 2020, 21, 1620–1633. [Google Scholar] [CrossRef] [PubMed]
- Lionetti, V.; Raiola, A.; Camardella, L.; Giovane, A.; Obel, N.; Pauly, M.; Favaron, F.; Cervone, F.; Bellincampi, D. Overexpression of pectin methylesterase inhibitors in Arabidopsis restricts fungal infection by Botrytis cinerea. Plant Physiol. 2007, 143, 1871–1880. [Google Scholar] [CrossRef] [Green Version]
- Volpi, C.; Janni, M.; Lionetti, V.; Bellincampi, D.; Favaron, F.; D’Ovidio, R. The ectopic expression of a pectin methyl esterase inhibitor increases pectin methyl esterification and limits fungal diseases in wheat. Mol. Plant-Microbe Interact. 2011, 24, 1012–1019. [Google Scholar] [CrossRef] [Green Version]
- Liu, N.; Sun, Y.; Pei, Y.; Zhang, X.; Wang, P.; Li, X.; Li, F.; Hou, Y. A pectin methylesterase inhibitor enhances resistance to Verticillium wilt. Plant Physiol. 2018, 176, 2202–2220. [Google Scholar] [CrossRef] [Green Version]
- Aguero, C.B.; Uratsu, S.L.; Greve, C.; Powell, A.L.T.; Labavitch, J.M.; Meredith, C.P.; Dandekar, A.M. Evaluation of tolerance to Pierce’s disease and Botrytis in transgenic plants of Vitis vinifera L. expressing the pear PGIP gene. Mol. Plant Pathol. 2005, 6, 43–51. [Google Scholar] [CrossRef] [PubMed]
- Ferrari, S.; Sella, L.; Janni, M.; De Lorenzo, G.; Favaron, F.; D’Ovidio, R. Transgenic expression of polygalacturonase-inhibiting proteins in Arabidopsis and wheat increases resistance to the flower pathogen Fusarium graminearum. Plant Biol. 2012, 14, 31–38. [Google Scholar] [CrossRef]
- Pogorelko, G.; Lionetti, V.; Fursova, O.; Sundaram, R.M.; Qi, M.; Whitham, S.A.; Bogdanove, A.J.; Bellincampi, D.; Zabotina, O.A. Arabidopsis and Brachypodium distachyon transgenic plants expressing Aspergillus nidulans acetylesterases have decreased degree of polysaccharide acetylation and increased resistance to pathogens. Plant Physiol. 2013, 162, 9–23. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rui, Y.; Dinneny, J.R. A wall with integrity: Surveillance and maintenance of the plant cell wall under stress. New Phytol. 2020, 225, 1428–1439. [Google Scholar] [CrossRef] [Green Version]
- Brutus, A.; Sicilia, F.; Macone, A.; Cervone, F.; De Lorenzo, G. A domain swap approach reveals a role of the plant wall-associated kinase 1 (WAK1) as a receptor of oligogalacturonides. Proc. Natl. Acad. Sci. USA 2010, 107, 9452–9457. [Google Scholar] [CrossRef] [Green Version]
- Davidson, P.; Broberg, M.; Kariola, T.; Sipari, N.; Pirhonen, M.; Palva, E.T. Short oligogalacturonides induce pathogen resistance-associated gene expression in Arabidopsis thaliana. BMC Plant Biol. 2017, 17, 19. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ferrari, S.; Savatin, D.V.; Sicilia, F.; Gramegna, G.; Cervone, F.; De Lorenzo, G. Oligogalacturonides: Plant damage-associated molecular patterns and regulators of growth and development. Front. Plant Sci. 2013, 4, 49. [Google Scholar] [CrossRef] [Green Version]
- Savatin, D.V.; Bisceglia, N.G.; Marti, L.; Fabbri, C.; Cervone, F.; De Lorenzo, G. The Arabidopsis nucleus- and phragmoplast-localized kinase1-related protein kinases are required for elicitor-induced oxidative burst and immunity. Plant Physiol. 2014, 165, 1188–1202. [Google Scholar] [CrossRef] [Green Version]
- Wagner, T.A.; Kohorn, B.D. Wall-associated kinases are expressed throughout plant development and are required for cell expansion. Plant Cell 2001, 13, 303–318. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Brown, D.M.; Zeef, L.A.H.; Ellis, J.; Goodacre, R.; Turner, S.R. Identification of novel genes in Arabidopsis involved in secondary cell wall formation using expression profiling and reverse genetics. Plant Cell 2005, 17, 2281–2295. [Google Scholar] [CrossRef] [PubMed]
- Rogers, L.A.; Dubos, C.; Surman, C.; Willment, J.; Cullis, I.F.; Mansfield, S.D.; Campbell, M.M. Comparison of lignin deposition in three ectopic lignification mutants. New Phytol. 2005, 168, 123–140. [Google Scholar] [CrossRef] [PubMed]
- Sampedro, J.; Pardo, B.; Gianzo, C.; Guitian, E.; Revilla, G.; Zarra, I. Lack of alpha-xylosidase activity in Arabidopsis alters xyloglucan composition and results in growth defects. Plant Physiol. 2010, 154, 1105–1115. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Delgado-Cerezo, M.; Sanchez-Rodriguez, C.; Escudero, V.; Miedes, E.; Virginia Fernandez, P.; Jorda, L.; Hernandez-Blanco, C.; Sanchez-Vallet, A.; Bednarek, P.; Schulze-Lefert, P.; et al. Arabidopsis heterotrimeric G-protein regulates cell wall defense and resistance to necrotrophic fungi. Mol. Plant 2012, 5, 98–114. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Llorente, F.; Alonso-Blanco, C.; Sanchez-Rodriguez, C.; Jorda, L.; Molina, A. ERECTA receptor-like kinase and heterotrimeric G protein from Arabidopsis are required for resistance to the necrotrophic fungus Plectosphaerella cucumerina. Plant J. 2005, 43, 165–180. [Google Scholar] [CrossRef]
- Sanchez-Rodriguez, C.; Estevez, J.M.; Llorente, F.; Hernandez-Blanco, C.; Jorda, L.; Pagan, I.; Berrocal, M.; Marco, Y.; Somerville, S.; Molina, A. The ERECTA receptor-like kinase regulates cell wall-mediated resistance to pathogens in Arabidopsis thaliana. Mol. Plant-Microbe Interact. 2009, 22, 953–963. [Google Scholar] [CrossRef] [Green Version]
- Manabe, Y.; Nafisi, M.; Verhertbruggen, Y.; Orfila, C.; Gille, S.; Rautengarten, C.; Cherk, C.; Marcus, S.E.; Somerville, S.; Pauly, M.; et al. Loss-of-function mutation of reduced wall acetylation2 in Arabidopsis leads to reduced cell wall acetylation and increased resistance to Botrytis cinerea. Plant Physiol. 2011, 155, 1068–1078. [Google Scholar] [CrossRef] [Green Version]
- Pawar, P.M.-A.; Derba-Maceluch, M.; Chong, S.-L.; Gomez, L.D.; Miedes, E.; Banasiak, A.; Ratke, C.; Gaertner, C.; Mouille, G.; McQueen-Mason, S.J.; et al. Expression of fungal acetyl xylan esterase in Arabidopsis thaliana improves saccharification of stem lignocellulose. Plant Biotechnol. J. 2016, 14, 387–397. [Google Scholar] [CrossRef] [Green Version]
- Gille, S.; Pauly, M. O-acetylation of plant cell wall polysaccharides. Front. Plant Sci. 2012, 3, 12. [Google Scholar] [CrossRef] [Green Version]
- Escudero, V.; Jorda, L.; Sopena-Torres, S.; Melida, H.; Miedes, E.; Munoz-Barrios, A.; Swami, S.; Alexander, D.; McKee, L.S.; Sanchez-Vallet, A.; et al. Alteration of cell wall xylan acetylation triggers defense responses that counterbalance the immune deficiencies of plants impaired in the beta-subunit of the heterotrimeric G-protein. Plant J. 2017, 92, 386–399. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lugan, R.; Niogret, M.-F.; Kervazo, L.; Larher, F.R.; Kopka, J.; Bouchereau, A. Metabolome and water status phenotyping of Arabidopsis under abiotic stress cues reveals new insight into ESK1 function. Plant Cell Environ. 2009, 32, 95–108. [Google Scholar] [CrossRef] [PubMed]
- Xin, Z.; Mandaokar, A.; Chen, J.; Last, R.L.; Browse, J. Arabidopsis ESK1 encodes a novel regulator of freezing tolerance. Plant J. 2007, 49, 786–799. [Google Scholar] [CrossRef]
- Xu, F.; Liu, Z.; Xie, H.; Zhu, J.; Zhang, J.; Kraus, J.; Blaschnig, T.; Nehls, R.; Wang, H. Increased drought tolerance through the suppression of ESKMO1 gene and overexpression of CBF-related genes in Arabidopsis. PLoS ONE 2014, 9, e106509. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Buanafina, M.M.d.O.; Fescemyer, H.W. Modification of esterified cell wall phenolics increases vulnerability of tall fescue to herbivory by the fall armyworm. Planta 2012, 236, 513–523. [Google Scholar] [CrossRef] [PubMed]
- Santiago, R.; Malvar, R.A. Role of dehydrodiferulates in maize resistance to pests and diseases. Int. J. Mol. Sci. 2010, 11, 691–703. [Google Scholar] [CrossRef] [Green Version]
- Buanafina, M.M.d.O. Feruloylation in grasses: Current and future perspectives. Mol. Plant 2009, 2, 861–872. [Google Scholar] [CrossRef]
- Bily, A.C.; Reid, L.M.; Taylor, J.H.; Johnston, D.; Malouin, C.; Burt, A.J.; Bakan, B.; Regnault-Roger, C.; Pauls, K.P.; Arnason, J.T.; et al. Dehydrodimers of ferulic acid in maize grain pericarp and aleurone: Resistance factors to Fusarium graminearum. Phytopathology 2003, 93, 712–719. [Google Scholar] [CrossRef] [Green Version]
- Reem, N.T.; Pogorelko, G.; Lionetti, V.; Chambers, L.; Held, M.A.; Bellincampi, D.; Zabotina, O.A. Decreased polysaccharide feruloylation compromises plant cell wall integrity and increases susceptibility to necrotrophic fungal pathogens. Front. Plant Sci. 2016, 7, 630. [Google Scholar] [CrossRef] [Green Version]
- Pogorelko, G.; Fursova, O.; Lin, M.; Pyle, E.; Jass, J.; Zabotina, O.A. Post-synthetic modification of plant cell walls by expression of microbial hydrolases in the apoplast. Plant Mol. Biol. 2011, 77, 433–445. [Google Scholar] [CrossRef]
- Norris, S.R.; Meyer, S.E.; Callis, J. The intron of Arabidopsis thaliana polyubiquitin genes is conserved in location and is a quantitative determinant of chimeric gene expression. Plant Mol. Biol. 1993, 21, 895–906. [Google Scholar] [CrossRef]
- Grefen, C.; Donald, N.; Hashimoto, K.; Kudla, J.; Schumacher, K.; Blatt, M.R. A ubiquitin-10 promoter-based vector set for fluorescent protein tagging facilitates temporal stability and native protein distribution in transient and stable expression studies. Plant J. 2010, 64, 355–365. [Google Scholar] [CrossRef]
- Clough, S.J.; Bent, A.F. Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 1998, 16, 735–743. [Google Scholar] [CrossRef] [Green Version]
- Gibson, D.G.; Young, L.; Chuang, R.-Y.; Venter, J.C.; Hutchison, C.A., III; Smith, H.O. Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat. Methods 2009, 6, 343–345. [Google Scholar] [CrossRef] [PubMed]
- Schmittgen, T.D.; Livak, K.J. Analyzing real-time PCR data by the comparative C-T method. Nat. Protoc. 2008, 3, 1101–1108. [Google Scholar] [CrossRef] [PubMed]
- McComb, E.A.; McCready, R.M. Determination of acetyl in pectin and in acetylated carbohydrate polymers. Anal. Chem. 1957, 29, 819–821. [Google Scholar] [CrossRef]
- Ferrari, S.; Galletti, R.; Pontiggia, D.; Manfredini, C.; Lionetti, V.; Bellincampi, D.; Cervone, F.; De Lorenzo, G. Transgenic expression of a fungal endo-polygalacturonase increases plant resistance to pathogens and reduces auxin sensitivity. Plant Physiol. 2008, 146, 669–681. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Abràmoff, M.D.; Magalhães, P.J.; Ram, S.J. Image processing with ImageJ. Biophotonics Int. 2004, 11, 36–42. [Google Scholar]
- Daudi, A.; O’Brien, J.A. Detection of hydrogen peroxide by DAB staining in Arabidopsis leaves. Bio-Protocol 2012, 2, 18. [Google Scholar] [CrossRef] [Green Version]
- Dormatey, R.; Sun, C.; Ali, K.; Coulter, J.A.; Bi, Z.; Bai, J. Gene pyramiding for sustainable crop improvement against biotic and abiotic stresses. Agronomy 2020, 10, 1255. [Google Scholar] [CrossRef]
- Mundt, C.C. Durable resistance: A key to sustainable management of pathogens and pests. Infect. Genet. Evol. 2014, 27, 446–455. [Google Scholar] [CrossRef] [PubMed]
- Pilet-Nayel, M.-L.; Moury, B.; Caffier, V.; Montarry, J.; Kerlan, M.-C.; Fournet, S.; Durel, C.-E.; Delourme, R. Quantitative resistance to plant pathogens in pyramiding strategies for durable crop protection. Front. Plant Sci. 2017, 8, 1838. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wan, B. Transgenic pyramiding for crop improvement. In Advances in Plant Breeding Strategies: Breeding, Biotechnology and Molecular Tools; Al-Khayri, J.M., Jain, S.M., Johnson, D.V., Eds.; Springer: Berlin, Germany, 2015; Volume 1, pp. 369–396. [Google Scholar]
- Buanafina, M.M.D.; Langdon, T.; Hauck, B.; Dalton, S.; Timms-Taravella, E.; Morris, P. Targeting expression of a fungal ferulic acid esterase to the apoplast, endoplasmic reticulum or golgi can disrupt feruloylation of the growing cell wall and increase the biodegradability of tall fescue (Festuca arundinacea). Plant Biotechnol. J. 2010, 8, 316–331. [Google Scholar] [CrossRef]
- Harholt, J.; Bach, I.C.; Lind-Bouquin, S.; Nunan, K.J.; Madrid, S.M.; Brinch-Pedersen, H.; Holm, P.B.; Scheller, H.V. Generation of transgenic wheat (Triticum aestivum L.) accumulating heterologous endo-xylanase or ferulic acid esterase in the endosperm. Plant Biotechnol. J. 2010, 8, 351–362. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tundo, S.; Kalunke, R.; Janni, M.; Volpi, C.; Lionetti, V.; Bellincampi, D.; Favaron, F.; D’Ovidio, R. Pyramiding PvPGIP2 and TAXI-III but not PvPGIP2 and PMEI enhances resistance against Fusarium graminearum. Mol. Plant-Microbe Interact. 2016, 29, 629–639. [Google Scholar] [CrossRef] [Green Version]
- Seong, S.Y.; Matzinger, P. Hydrophobicity: An ancient damage-associated molecular pattern that initiates innate immune responses. Nat. Rev. Immunol. 2004, 4, 469–478. [Google Scholar] [CrossRef] [PubMed]
- Ferrari, S.; Galletti, R.; Denoux, C.; De Lorenzo, G.; Ausubel, F.M.; Dewdney, J. Resistance to Botrytis cinerea induced in Arabidopsis by elicitors is independent of salicylic acid, ethylene, or jasmonate signaling but requires PHYTOALEXIN DEFICIENT3. Plant Physiol. 2007, 144, 367–379. [Google Scholar] [CrossRef] [Green Version]
- Denoux, C.; Galletti, R.; Mammarella, N.; Gopalan, S.; Werck, D.; De Lorenzo, G.; Ferrari, S.; Ausubel, F.M.; Dewdney, J. Activation of defense response pathways by OGs and Flg22 elicitors in Arabidopsis seedlings. Mol. Plant 2008, 1, 423–445. [Google Scholar] [CrossRef] [Green Version]
- Schuhegger, R.; Nafisi, M.; Mansourova, M.; Petersen, B.L.; Olsen, C.E.; Svatos, A.; Halkier, B.A.; Glawischnig, E. CYP71B15 (PAD3) catalyzes the final step in camalexin biosynthesis. Plant Physiol. 2006, 141, 1248–1254. [Google Scholar] [CrossRef] [Green Version]
- Rojo, E.; Leon, J.; Sanchez-Serrano, J.J. Cross-talk between wound signalling pathways determines local versus systemic gene expression in Arabidopsis thaliana. Plant J. 1999, 20, 135–142. [Google Scholar] [CrossRef] [Green Version]
- Benedetti, M.; Verrascina, I.; Pontiggia, D.; Locci, F.; Mattei, B.; De Lorenzo, G.; Cervone, F. Four Arabidopsis berberine bridge enzyme-like proteins are specific oxidases that inactivate the elicitor-active oligogalacturonides. Plant J. 2018, 94, 260–273. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Windram, O.; Madhou, P.; McHattie, S.; Hill, C.; Hickman, R.; Cooke, E.; Jenkins, D.J.; Penfold, C.A.; Baxter, L.; Breeze, E.; et al. Arabidopsis defense against Botrytis cinerea: Chronology and regulation deciphered by high-resolution temporal transcriptomic analysis. Plant Cell 2012, 24, 3530–3557. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zander, M.; La Camera, S.; Lamotte, O.; Metraux, J.-P.; Gatz, C. Arabidopsis thaliana class-II TGA transcription factors are essential activators of jasmonic acid/ethylene-induced defense responses. Plant J. 2010, 61, 200–210. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Breen, S.; Williams, S.J.; Outram, M.; Kobe, B.; Solomon, P.S. Emerging insights into the functions of pathogenesis-related protein 1. Trends Plant Sci. 2017, 22, 871–879. [Google Scholar] [CrossRef] [PubMed]
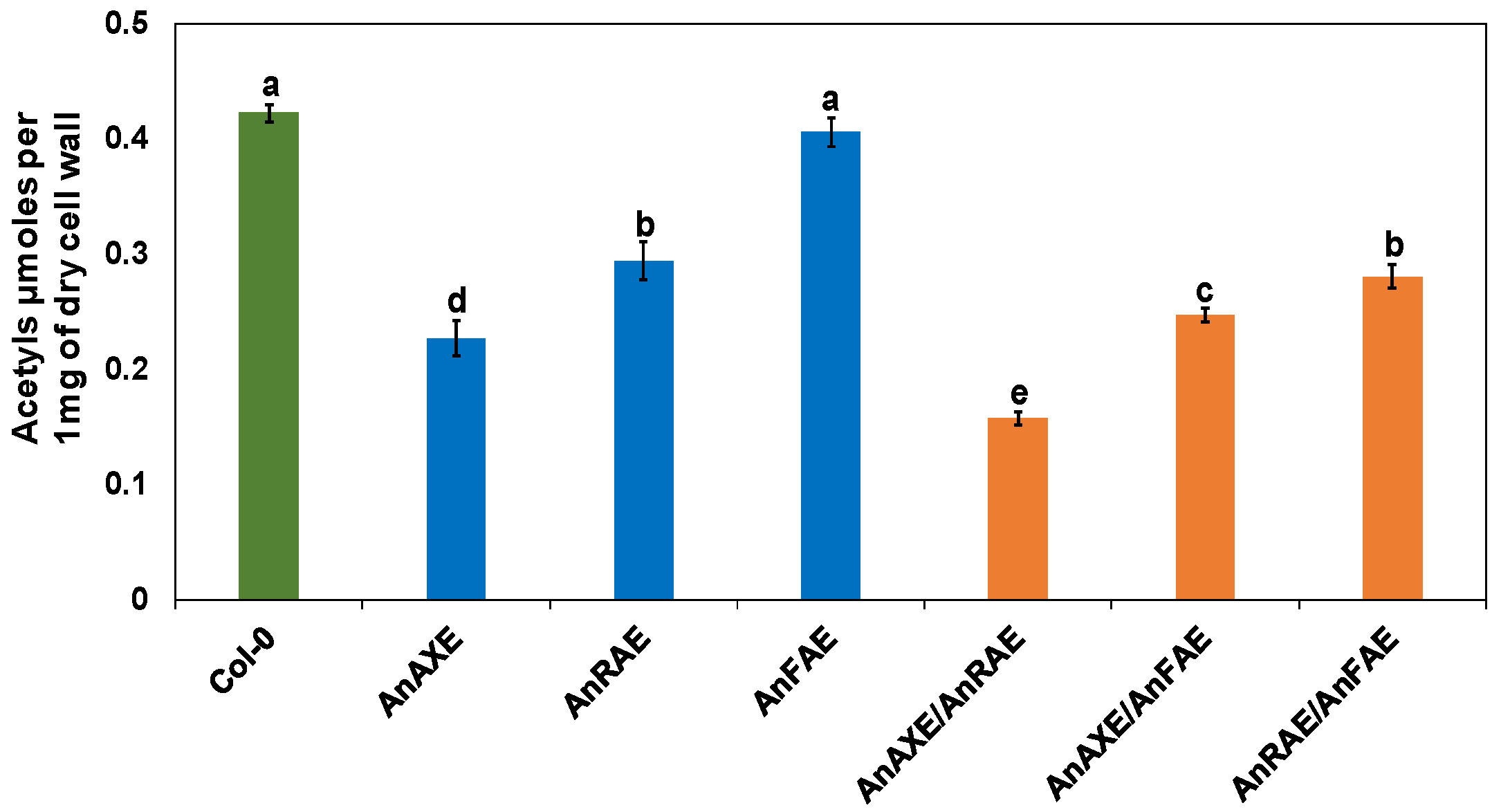
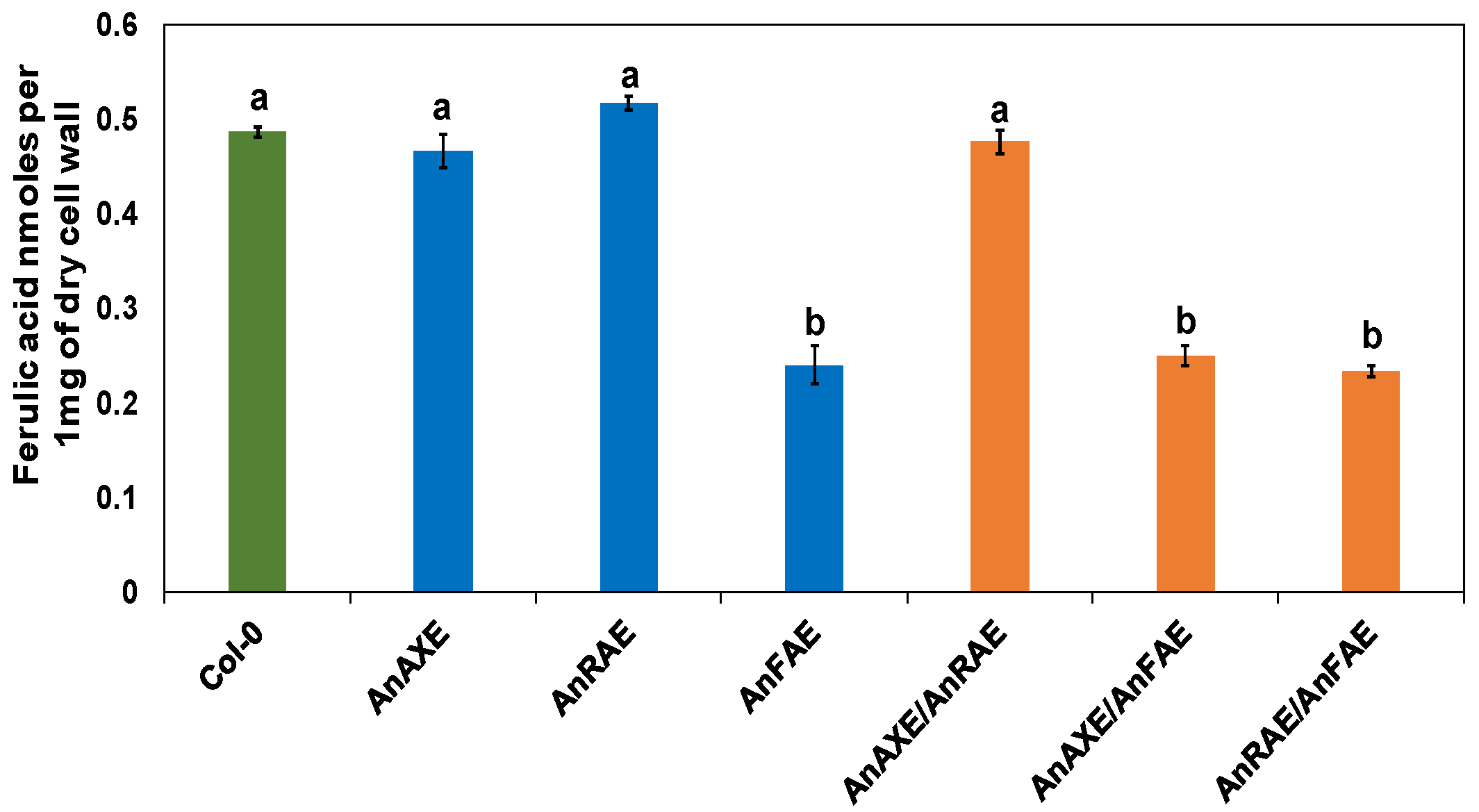

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Swaminathan, S.; Reem, N.T.; Lionetti, V.; Zabotina, O.A. Coexpression of Fungal Cell Wall-Modifying Enzymes Reveals Their Additive Impact on Arabidopsis Resistance to the Fungal Pathogen, Botrytis cinerea. Biology 2021, 10, 1070. https://doi.org/10.3390/biology10101070
Swaminathan S, Reem NT, Lionetti V, Zabotina OA. Coexpression of Fungal Cell Wall-Modifying Enzymes Reveals Their Additive Impact on Arabidopsis Resistance to the Fungal Pathogen, Botrytis cinerea. Biology. 2021; 10(10):1070. https://doi.org/10.3390/biology10101070
Chicago/Turabian StyleSwaminathan, Sivakumar, Nathan T. Reem, Vincenzo Lionetti, and Olga A. Zabotina. 2021. "Coexpression of Fungal Cell Wall-Modifying Enzymes Reveals Their Additive Impact on Arabidopsis Resistance to the Fungal Pathogen, Botrytis cinerea" Biology 10, no. 10: 1070. https://doi.org/10.3390/biology10101070
APA StyleSwaminathan, S., Reem, N. T., Lionetti, V., & Zabotina, O. A. (2021). Coexpression of Fungal Cell Wall-Modifying Enzymes Reveals Their Additive Impact on Arabidopsis Resistance to the Fungal Pathogen, Botrytis cinerea. Biology, 10(10), 1070. https://doi.org/10.3390/biology10101070







