A Universal Model for the Log-Normal Distribution of Elasticity in Polymeric Gels and Its Relevance to Mechanical Signature of Biological Tissues
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Collagen Gel
2.2. Cell Culture
2.3. Atomic Force Microscopy
3. Results
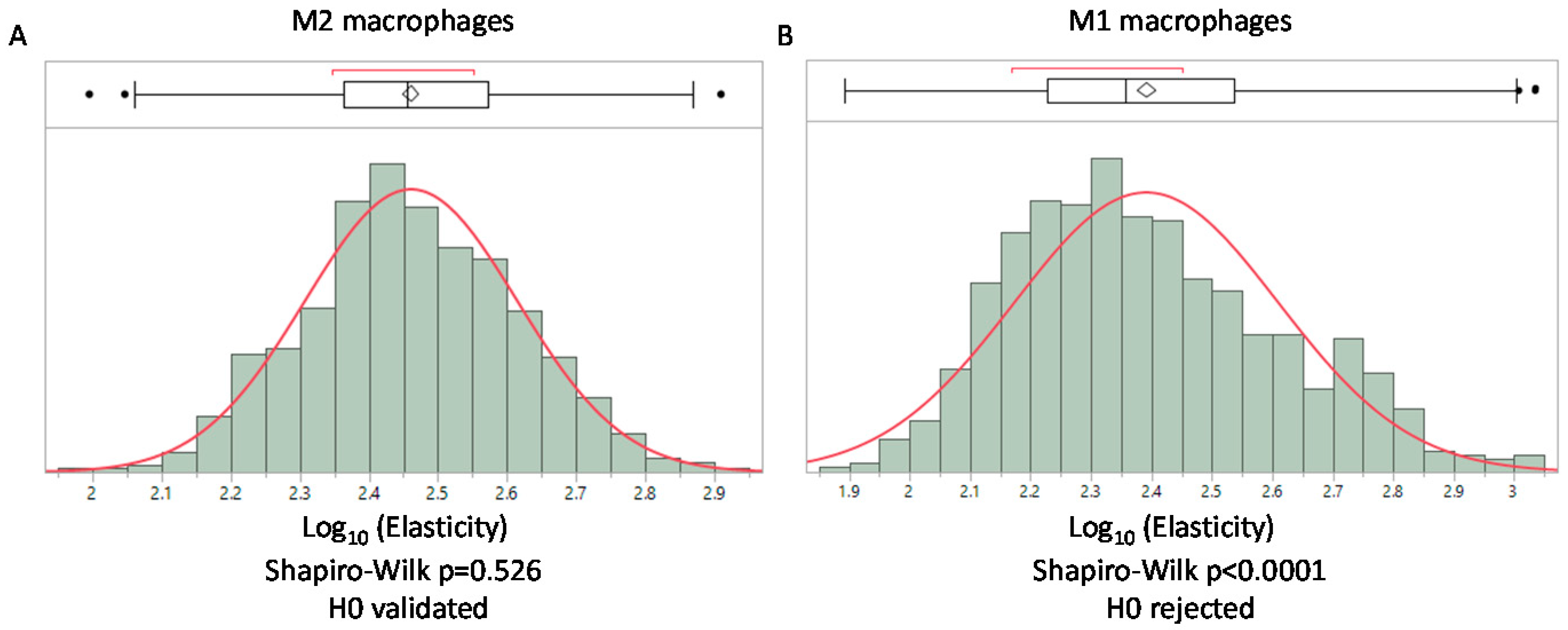
3.1. Elastic Moduli of Collagen Based Scaffolds Displayed a Log-Normal Distribution
3.2. Log-Normal and Normal Distribution Laws
3.3. Log-Normal Distribution in Biology: A Heuristic Argument
3.4. The Heuristic Argument is Unable to Explain the Log-Normal Distribution for Gels
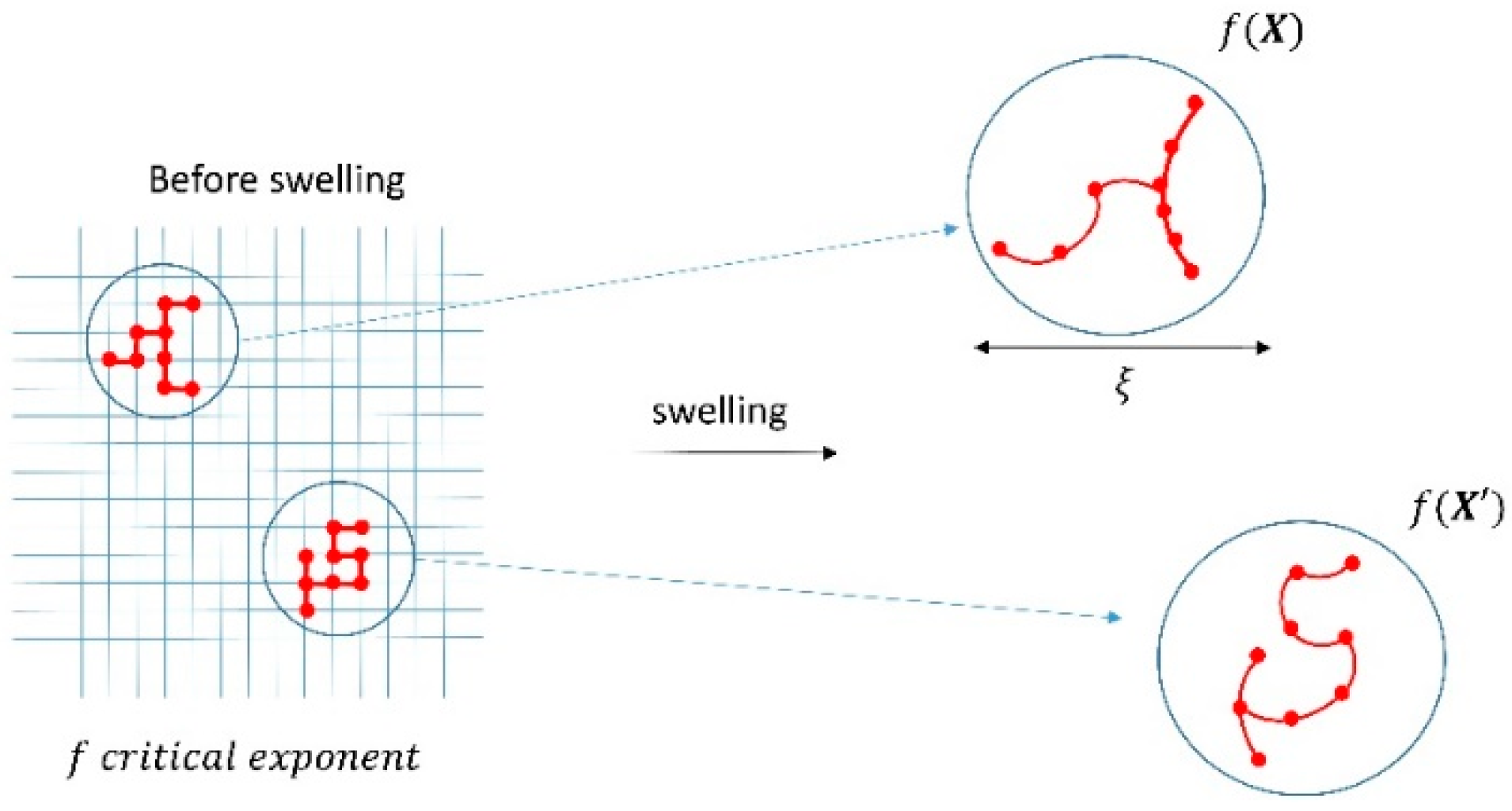
3.5. Percolation Model of the Elastic Modulus of A Polymeric Gel
3.6. AFM Analysis of A Simplified Biological Tissue
4. Discussion
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Hanahan, D.; Weinberg, R.A. Hallmarks of Cancer: The Next Generation. Cell 2011, 144, 646–674. [Google Scholar] [CrossRef] [PubMed]
- Mierke, C.T. The fundamental role of mechanical properties in the progression of cancer disease and inflammation. Rep. Prog. Phys. 2014, 77, 076602. [Google Scholar] [CrossRef] [PubMed]
- Katira, P.; Bonnecaze, R.T.; Zaman, M.H. Modeling the Mechanics of Cancer: Effect of Changes in Cellular and Extra-Cellular Mechanical Properties. Front. Oncol. 2013, 3, 145. [Google Scholar] [CrossRef] [PubMed]
- Butcher, D.T.; Alliston, T.; Weaver, V.M. A tense situation: Forcing tumour progression. Nat. Rev. Cancer 2009, 9, 108–122. [Google Scholar] [CrossRef] [PubMed]
- Hochmuth, R.M. Micropipette aspiration of living cells. J. Biomech. 2000, 33, 15–22. [Google Scholar] [CrossRef]
- Lincoln, B.; Wottawah, F.; Schinkinger, S.; Ebert, S.; Guck, J. High-Throughput Rheological Measurements with an Optical Stretcher. In Methods in Cell Biology; Cell Mechanics; Academic Press: Cambridge, MA, USA, 2007; Volume 83, pp. 397–432. [Google Scholar] [CrossRef]
- Kollmannsberger, P.; Fabry, B. BaHigh-force magnetic tweezers with force feedback for biological applications. Rev. Sci. Instrum. 2007, 78, 114301. [Google Scholar] [CrossRef] [PubMed]
- Plodinec, M.; Loparic, M.; Monnier, C.A.; Obermann, E.C.; Zanetti-Dallenbach, R.; Oertle, P.; Hyotyla, J.T.; Aebi, U.; Bentires-Alj, M.; Lim, R.Y.H.; et al. The nanomechanical signature of breast cancer. Nat. Nanotechnol. 2012, 7, 757–765. [Google Scholar] [CrossRef]
- Hecht, F.M.; Rheinlaender, J.; Schierbaum, N.; Goldmann, W.H.; Fabry, B.; Schäffer, T.E. Imaging viscoelastic properties of live cells by AFM: Power-law rheology on the nanoscale. Soft Matter 2015, 11, 4584–4591. [Google Scholar] [CrossRef]
- Limpert, E.; Stahel, W.A.; Abbt, M. Log-normal Distributions across the Sciences: Keys and CluesOn the charms of statistics, and how mechanical models resembling gambling machines offer a link to a handy way to characterize log-normal distributions, which can provide deeper insight into variability and probability—normal or log-normal: That is the question. BioScience 2001, 51, 341–352. [Google Scholar] [CrossRef]
- Court, M.; Malier, M.; Millet, A. 3D type I collagen environment leads up to a reassessment of the classification of human macrophage polarizations. Biomaterials 2019, 208, 98–109. [Google Scholar] [CrossRef]
- Sader, J.E.; Borgani, R.; Gibson, C.T.; Haviland, D.B.; Higgins, M.J.; Kilpatrick, J.I.; Lu, J.; Mulvaney, P.; Shearer, C.J.; Slattery, A.D.; et al. A virtual instrument to standardise the calibration of atomic force microscope cantilevers. Rev. Sci. Instrum. 2016, 87, 093711. [Google Scholar] [CrossRef] [PubMed]
- Hermanowicz, P.; Sarna, M.; Burda, K.; Gabryś, H. AtomicJ: An open source software for analysis of force curves. Rev. Sci. Instrum. 2014, 85, 063703. [Google Scholar] [CrossRef] [PubMed]
- Jacod, J.; Protter, P. Probability Essentials, 2nd ed.; Springer: New York, NY, USA, 2004. [Google Scholar]
- Koch, A.L. The logarithm in biology 1. Mechanisms generating the log-normal distribution exactly. J. Theor. Biol. 1966, 12, 276–290. [Google Scholar] [CrossRef]
- Furusawa, C.; Suzuki, T.; Kashiwagi, A.; Yomo, T.; Kaneko, K. Ubiquity of log-normal distributions in intra-cellular reaction dynamics. Biophysics 2005, 1, 25–31. [Google Scholar] [CrossRef]
- Vernerey, F.J.; Bryant, S. The role of percolation in hydrogel-based tissue engineering and bioprinting. Curr. Opin. Biomed. Eng. 2020, 15, 68–74. [Google Scholar] [CrossRef]
- De Gennes, P.-G. On a relation between percolation theory and the elasticity of gels. J. Phys. Lett. 1976, 37, 1–2. [Google Scholar] [CrossRef]
- Tokita, M.; Niki, R.; Hikichi, K. Percolation Theory and Elastic Modulus of Gel. J. Phys. Soc. Jpn. 1984, 53, 480–482. [Google Scholar] [CrossRef]
- Kantor, Y.; Webman, I. Elastic Properties of Random Percolating Systems. Phys. Rev. Lett. 1984, 52, 1891–1894. [Google Scholar] [CrossRef]
- Arbabi, S.; Sahimi, M. Critical properties of viscoelasticity of gels and elastic percolation networks. Phys. Rev. Lett. 1990, 65, 725–728. [Google Scholar] [CrossRef]
- Wynn, T.A.; Chawla, A.; Pollard, J.W. Macrophage biology in development, homeostasis and disease. Nature 2013, 496, 445–455. [Google Scholar] [CrossRef]
- Binnewies, M.; Roberts, E.W.; Kersten, K.; Chan, V.; Fearon, D.F.; Merad, M.; Coussens, L.M.; Gabrilovich, D.I.; Ostrand-Rosenberg, S.; Hedrick, C.C.; et al. Understanding the tumor immune microenvironment (TIME) for effective therapy. Nat. Med. 2018, 24, 541–550. [Google Scholar] [CrossRef] [PubMed]
- Yang, M.; McKay, D.; Pollard, J.W.; Lewis, C.E. Diverse Functions of Macrophages in Different Tumor Microenvironments. Cancer Res. 2018, 78, 5492–5503. [Google Scholar] [CrossRef] [PubMed]
- Aras, S.; Zaidi, M.R. TAMeless traitors: Macrophages in cancer progression and metastasis. Br. J. Cancer 2017, 117, 1583–1591. [Google Scholar] [CrossRef] [PubMed]
- Olson, O.C.; Kim, H.; Quail, D.F.; Foley, E.A.; Joyce, J.A. Tumor-Associated Macrophages Suppress the Cytotoxic Activity of Antimitotic Agents. Cell Rep. 2017, 19, 101–113. [Google Scholar] [CrossRef] [PubMed]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Millet, A. A Universal Model for the Log-Normal Distribution of Elasticity in Polymeric Gels and Its Relevance to Mechanical Signature of Biological Tissues. Biology 2021, 10, 64. https://doi.org/10.3390/biology10010064
Millet A. A Universal Model for the Log-Normal Distribution of Elasticity in Polymeric Gels and Its Relevance to Mechanical Signature of Biological Tissues. Biology. 2021; 10(1):64. https://doi.org/10.3390/biology10010064
Chicago/Turabian StyleMillet, Arnaud. 2021. "A Universal Model for the Log-Normal Distribution of Elasticity in Polymeric Gels and Its Relevance to Mechanical Signature of Biological Tissues" Biology 10, no. 1: 64. https://doi.org/10.3390/biology10010064
APA StyleMillet, A. (2021). A Universal Model for the Log-Normal Distribution of Elasticity in Polymeric Gels and Its Relevance to Mechanical Signature of Biological Tissues. Biology, 10(1), 64. https://doi.org/10.3390/biology10010064




