Using Unstated Cases to Correct for COVID-19 Pandemic Outbreak and Its Impact on Easing the Intervention for Qatar
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Parameters of the Model
2.2. Fitting the Model to the Data
2.3. Self Starting Function for the Cumulative Function
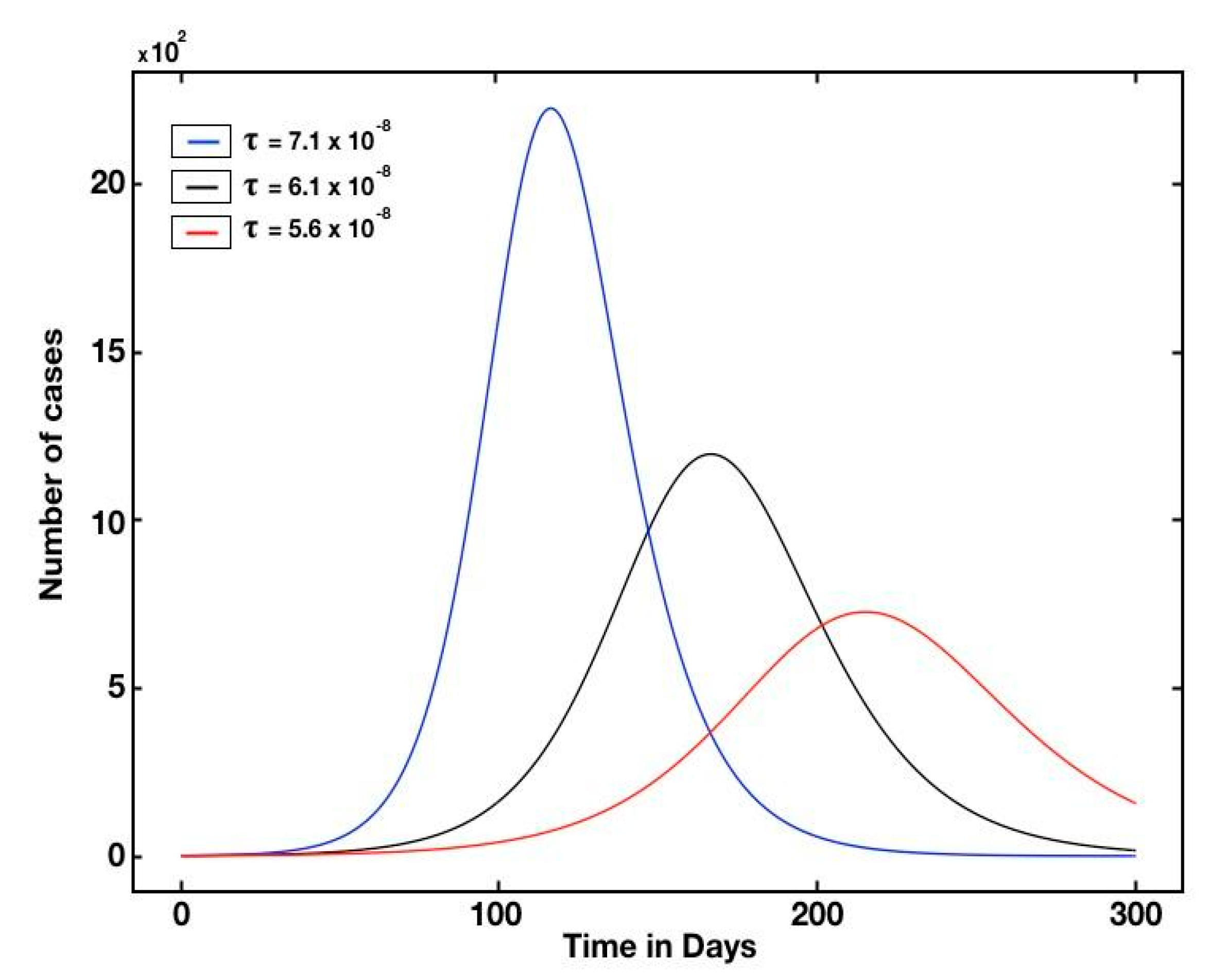
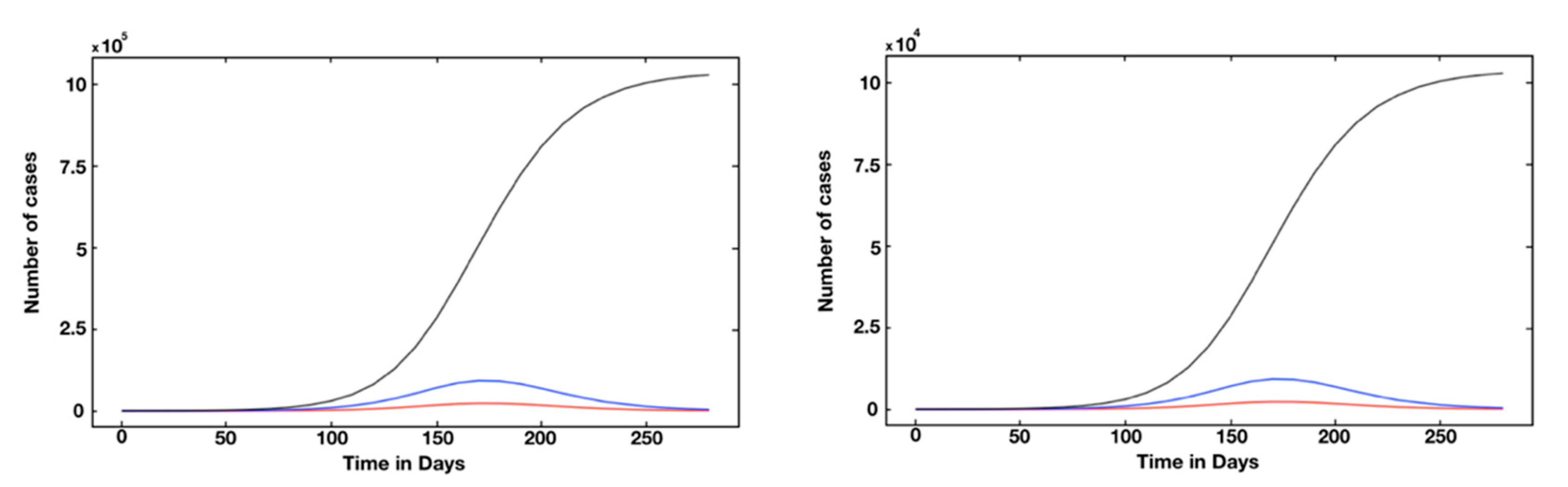
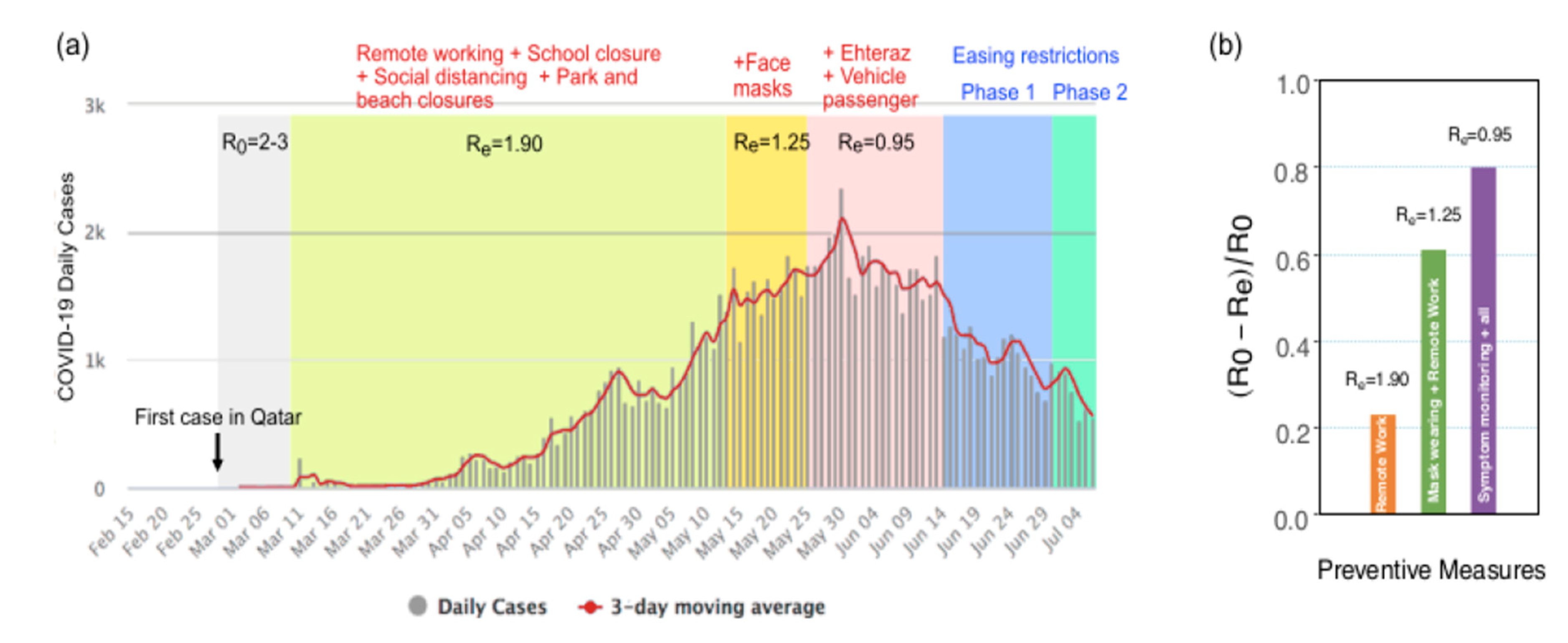
3. Results
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Appendix A
Appendix A.1
Appendix A.2
References
- World Health Organization. WHO Director-General’s Opening Remarks at the Media Briefing on COVID-19. Available online: https://www.who.int/director-general/speeches/detail/who-director-general-s-opening-remarks-at-the-media-briefing-on-covid-19---11-march-2020 (accessed on 11 March 2020).
- Ministry of Public Health, Qatar, Government Entity. Available online: https://www.moph.gov.qa/english/Pages/default.aspx (accessed on 1 April 2020).
- Alabdulkarim, N.; Alsultan, F.; Bashir, S. Gulf Countries Responding to COVID-19. Dubai Med. J. 2020, 3, 58–60. [Google Scholar] [CrossRef]
- New England Journal of Medicine, Letter to the Editor. Available online: https://www.nejm.org/doi/full/10.1056/NEJMc2001468 (accessed on 30 January 2020).
- Malkov, E. Simulation of coronavirus disease 2019 (COVID-19) scenarios with possibility of reinfection. Chaos Solitons Fractals 2020, 139, 110296. [Google Scholar] [CrossRef]
- Liu, Z.; Magal, P.; Seydi, O.; Webb, G. Understanding unreported cases in the COVID-19 epidemic outbreak in Wuhan, China, and the importance of major public health interventions. Biology 2020, 9, 50. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.; Magal, P.; Seydi, O.; Webb, G. A COVID-19 epidemic model with latency period. Infect. Dis. Model. 2020, 5, 323–337. [Google Scholar] [CrossRef]
- Alvarez, F.E.; Argente, D.; Lippi, F. A Simple Planning Problem for Covid-19 Lockdown; National Bureau of Economic Research: Cambridge, MA, USA, 2020. [Google Scholar]
- Eichenbaum, M.S.; Rebelo, S.; Trabandt, M. The Macroeconomics of Epidemics; National Bureau of Economic Research: Cambridge, MA, USA, 2020. [Google Scholar]
- Hortaçsu, A.; Liu, J.; Schwieg, T. Estimating the fraction of unreported infections in epidemics with a known epicenter: An application to COVID-19. J. Econom. 2021, 220, 106–129. [Google Scholar] [CrossRef] [PubMed]
- Kermack, W.O.; McKendrick, A.G. A contribution to the mathematical theory of epidemics. Proc. R. Soc. Lond. Ser. A Contain. Pap. Math. Phys. Character 1927, 115, 700–721. [Google Scholar]
- Sahafizadeh, E.; Sartoli, S. Estimating the reproduction number of COVID-19 in Iran using epidemic modeling. J. Travel Med. 2020, 27. [Google Scholar] [CrossRef] [PubMed]
- Zhuang, Z.; Zhao, S.; Lin, Q.; Cao, P.; Lou, Y.; Yang, L.; Yang, S.; He, D.; Xiao, L. Preliminary estimates of the reproduction number of the coronavirus disease (COVID-19) outbreak in Republic of Korea and Italy by 5 March 2020. Int. J. Infect. Dis. 2020, 95, 308–310. [Google Scholar] [CrossRef] [PubMed]
- Chanilkul, G.; Tongkhonburi, P.; Meesubthong, C. The reproductive index from SEIR model of Covid-19 epidemic in Asean. medRxiv 2020. [Google Scholar] [CrossRef]
- Last, M. The First Wave of COVID-19 in Israel-Initial Analysis of Publicly Available Data. PLoS ONE 2020, 15, e0240393. [Google Scholar] [CrossRef]
- Hamidouche, M. COVID-19 Outbreak in Algeria: A Model to Predict Cumulative Cases. J. Contemp. Stud. Epidemiol. Public Health 2020, 1, 1–6. [Google Scholar] [CrossRef]
- Gunzler, D.; Sehgal, A.R. Time-Varying COVID-19 Reproduction Number in the United States. medRxiv 2020. PMID: 32511659 PMCID: PMC7277015. [Google Scholar] [CrossRef]
- da Luz Scherf, E.; da Silva, M.V.V.; Fachini, J.S. The Management (or Lack Thereof) of COVID-19 in Brazil: Implications for Human Rights and Public Health. Health Hum. Rights J. Forthcom. 2020, 1–24. [Google Scholar] [CrossRef]
- Jung, S.M.; Endo, A.; Kinoshita, R.; Nishiura, H. Projecting a second wave of COVID-19 in Japan with variable interventions in high-risk settings. R. Soc. Open Sci. 2021, 8, 202169. [Google Scholar] [CrossRef]
- Li, Y.; Wang, L.W.; Peng, Z.H.; Shen, H.B. Basic reproduction number and predicted trends of coronavirus disease 2019 epidemic in the mainland of China. Infect. Dis. Poverty 2020, 9, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Saxena, A. Here are the Key Takeaways from Delhi’s Serological Survey. In The Indian Express; Indian Express Group: Uttar Pradesh, India, 2020. [Google Scholar]
- Wu, S.L.; Mertens, A.N.; Crider, Y.S.; Nguyen, A.; Pokpongkiat, N.N.; Djajadi, S.; Seth, A.; Hsiang, M.S.; Colford, J.M., Jr.; Reingold, A.; et al. Substantial underestimation of SARS-CoV-2 infection in the United States. Nat. Commun. 2020, 11, 4507. [Google Scholar] [CrossRef]
- Deo, V.; Grover, G. A new extension of state-space SIR model to account for Underreporting—An application to the COVID-19 transmission in California and Florida. J. Results Phys. 2021, 24, 104182. [Google Scholar] [CrossRef]
- Bernoulli, D. Essai d’une Nouvelle Analyse de la Mortalité Causée par la Petite Vérole, et des Avantages de L’inoculation Pour la Prévenir; Mémoire Académie Royale des Sciences: Paris, France, 1760. [Google Scholar]
- Saeedian, M.; Khalighi, M.; Azimi-Tafreshi, N.; Jafari, G.R.; Ausloos, M. Memory effects on epidemic evolution: The susceptible-infected-recovered epidemic model. Phys. Rev. E 2017, 95, 022409. [Google Scholar] [CrossRef]
- Materassi, M. Some fractal thoughts about the COVID-19 infection outbreak. Chaos Solitons Fractals 2019, 4, 100032. [Google Scholar] [CrossRef]
- Jahanshahi, H.; Munoz-Pacheco, J.M.; Bekiros, S.; Alotaibi, N.D. A fractional-order SIRD model with time-dependent memory indexes for encompassing the multi-fractional characteristics of the COVID-19. Chaos Solitons Fractals 2021, 143, 110632. [Google Scholar] [CrossRef]
- Ziff, A.L.; Ziff, R.M. Fractal kinetics of COVID-19 pandemic. medRxiv 2020. [Google Scholar]
- Gowrisankar, A.; Rondoni, L.; Banerjee, S. Can India develop herd immunity against COVID-19? Eur. Phys. J. Plus 2020, 135, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Eftekhari, H.; Mukherjee, D.; Banerjee, M.; Ritov, Y. Markovian and Non-Markovian Processes with Active Decision Making Strategies for Addressing the COVID-19 Pandemic. arXiv 2008, arXiv:2008.00375. [Google Scholar]
- Demongeot, J.; Gaudart, J.; Mintsa, J.; Rachdi, M. Demography in epidemics modelling. Commun. Pure Appl. Anal. 2012, 11, 61. [Google Scholar] [CrossRef]
- Weiss, H. The SIR model and the foundations of Public Health. Mater. Mat. 2013, 3, 1–17. [Google Scholar]
- Farrington, C.P.; Kanaan, M.N.; Gay, N.J. Estimation of the basic reproduction number for infectious diseases from age-stratified serological survey data. J. R. Stat. Soc. Ser. C (Applied Stat.) 2001, 50, 251–292. [Google Scholar] [CrossRef]
- Van den Driessche, P. Reproduction numbers of infections disease models. Infect. Dis. Model. 2017, 2, 288–303. [Google Scholar]
- Rahman, B.; Aziz, I.A.; Khdhr, F.W.; Mahmood, D.F. Preliminary estimation of the basic reproduction number of SARS-CoV-2 in the Middle East. Bull. World Health Organ 2020. E-Pub, 2020/5. Preprint, WHO. [Google Scholar] [CrossRef]
- Al-Shammari, A.A.; Ali, H.; Al-Ahmad, B.; Al-Refaei, F.H.; Al-Sabah, S.; Jamal, M.H.; Alshukry, A.; Al-Duwairi, Q.; Al-Mulla, F. Real-time tracking and forecasting of the of COVID-19 outbreak in Kuwait: A mathematical modeling study. MedRxiv 2020. [Google Scholar] [CrossRef]
- Al Wahaibi, A.; Al Manji, A.; Al Maani, A.; Al Rawahi, B.; Al Harthy, K.; Alyaquobi, F.; Al-Jardani, A.; Petersen, E.; Al Abri, S. COVID-19 epidemic monitoring after non-pharmaceutical interventions: The use of time-varying reproduction number in a country with a large migrant population. Int. J. Infect. Dis. 2020, 99, 466–472. [Google Scholar] [CrossRef]
- Billah, A.; Miah, M.; Khan, N. Reproductive number of coronavirus: A systematic review and meta-analysis based on global level evidence. PLoS ONE 2020, 15, e0242128. [Google Scholar] [CrossRef] [PubMed]
- Rahman, B.; Sadraddin, E.; Porreca, A. The basic reproduction number of SARS-CoV-2 in Wuhan is about to die out, how about the rest of the World? Rev. Med. Virol. 2020, 20, e2111. [Google Scholar] [CrossRef] [PubMed]
- Ritz, C.; Jensen, S.M.; Gerhard, D.; Streibig, J.C. Dose-Response Analysis Using R.; CRC Press: Boca Raton, FL, USA, 2019. [Google Scholar]
- Demongeot, J.; Hansen, O.; Jannot, A.S.; Taramasco, C. Random Modelling of Contagious (Social and Infectious) Diseases: Examples of Obesity and HIV and Perspectives Using Social Networks. In Proceedings of the 2012 26th International Conference on Advanced Information Networking and Applications Workshops, Fukuoka-shi, Japan, 26–29 March 2012; pp. 1153–1160. [Google Scholar]
- Magal, P.; Webb, G. The parameter identification problem for SIR epidemic models: Identifying unreported cases. J. Math. Biol. 2018, 77, 1629–1648. [Google Scholar] [CrossRef]
- Dolbeault, J.; Turinici, G. Heterogeneous social interactions and the COVID-19 lockdown outcome in a multi-group SEIR model. Math. Model. Nat. Phenom. 2020, 15, 36. [Google Scholar] [CrossRef]
- Martcheva, M. An Introduction to Mathematical Epidemiology; Springer: New York, NY, USA, 2015; Volume 61. [Google Scholar]
- Jazeera, A. Qatar Announces Closure of Schools, Universities over Coronavirus; Al Jazeera: Doha, Qatar, 2020. [Google Scholar]
- Park, H.; Bentria, E.T.; Arredouani, A.; Bensmail, H.; Mellouhi, F.E. Artificial Intelligence accelerated design of antimicrobial/antiviral surfaces: A promising route to mitigate SARS-CoV-2 fomite and aerosol transmission (Accepted). ACS Pharmacol. Transl. Sci. 2020. [Google Scholar]
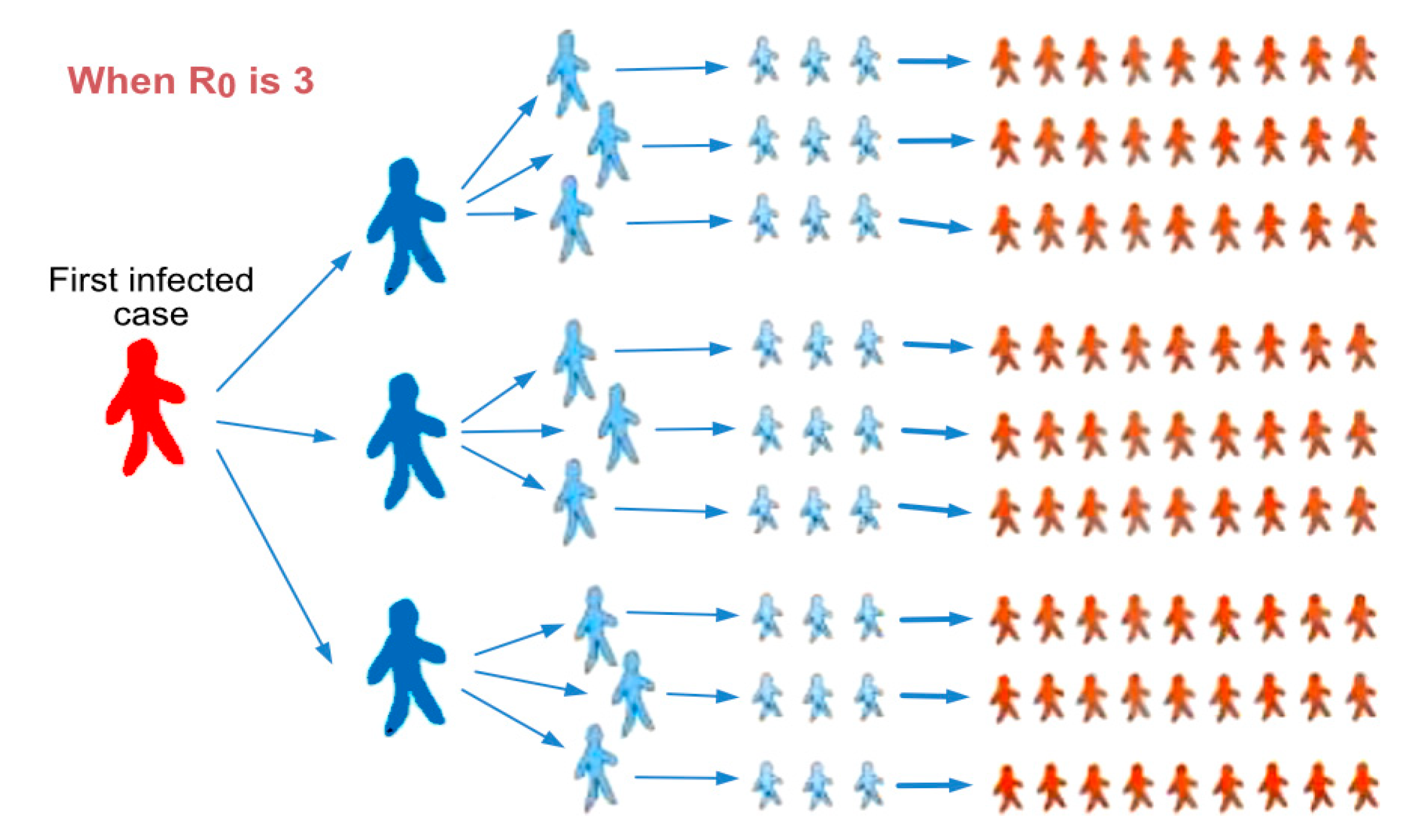
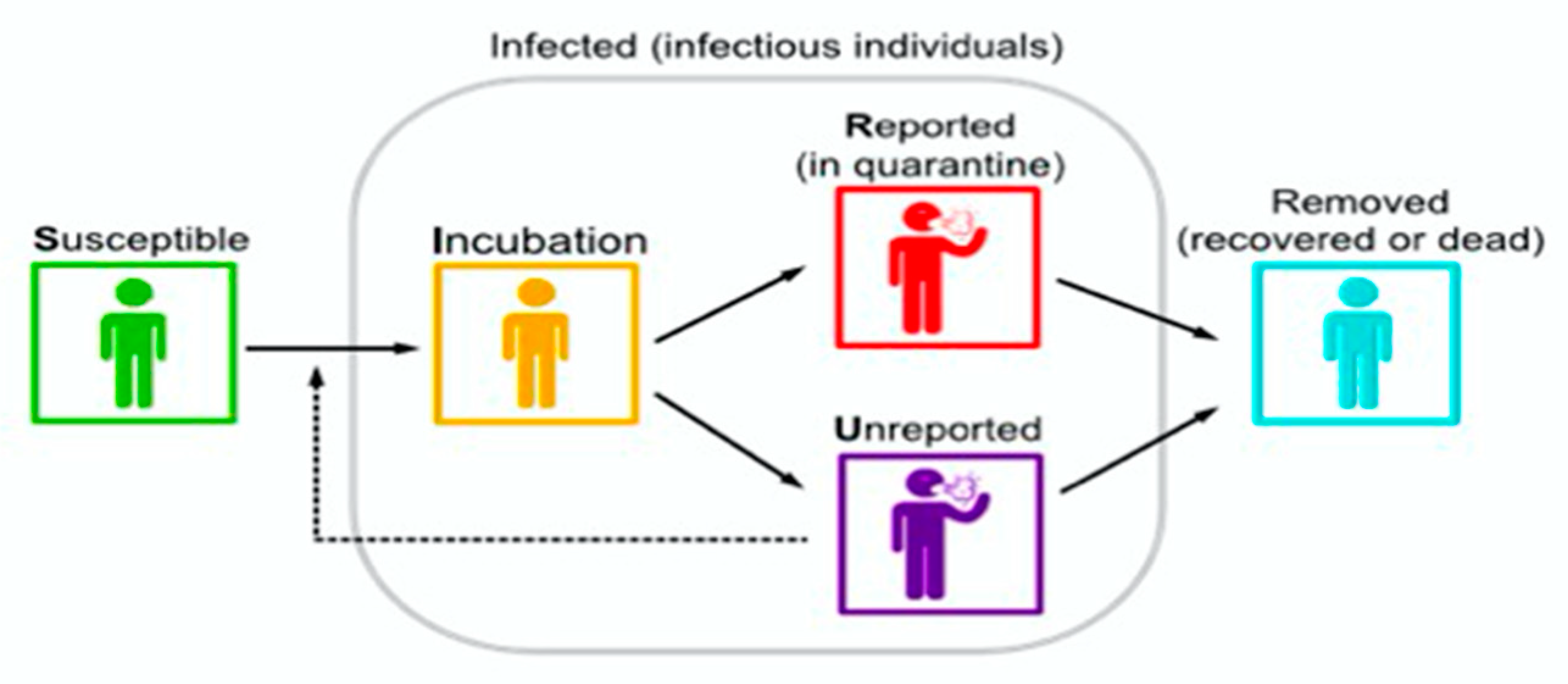
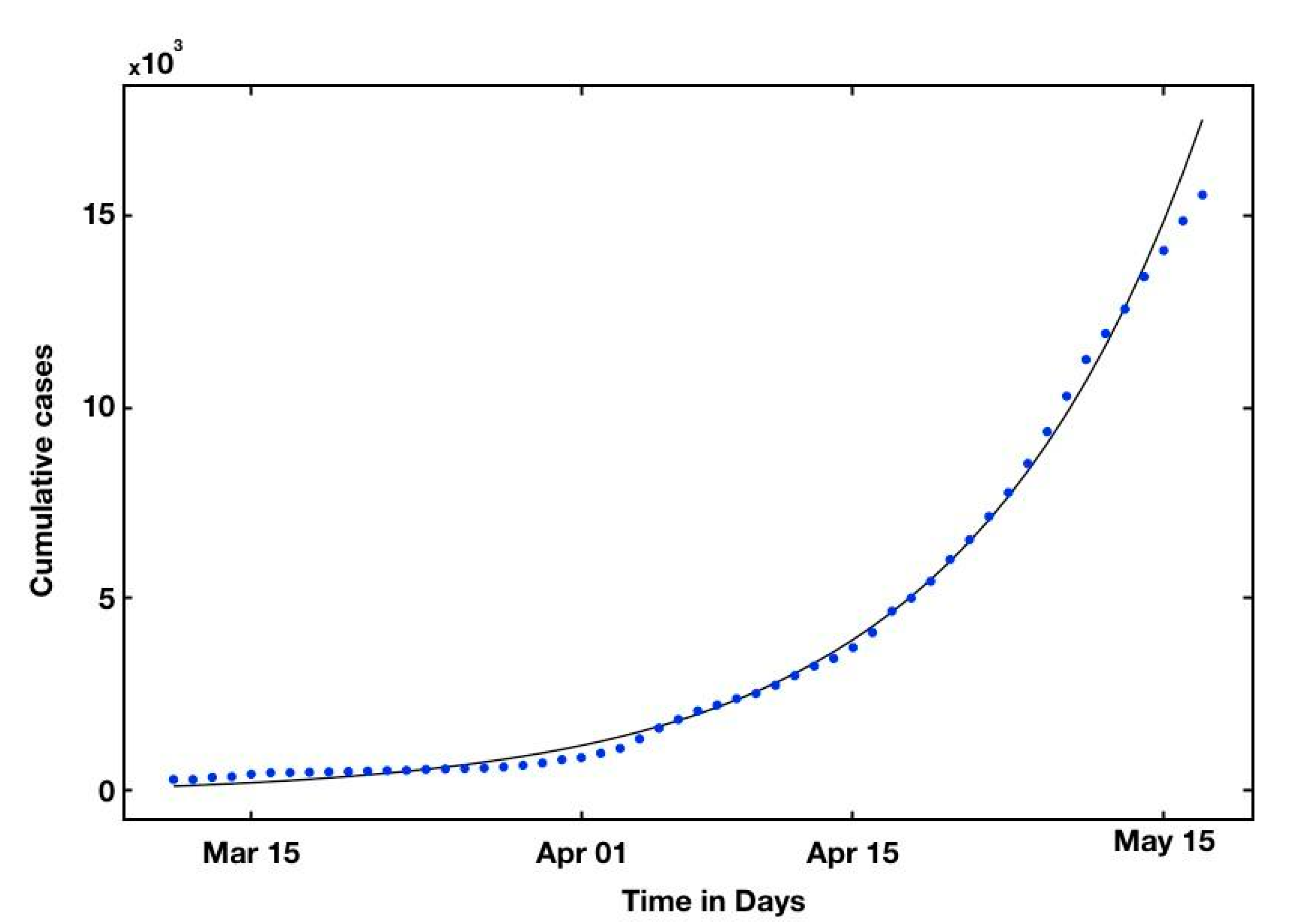
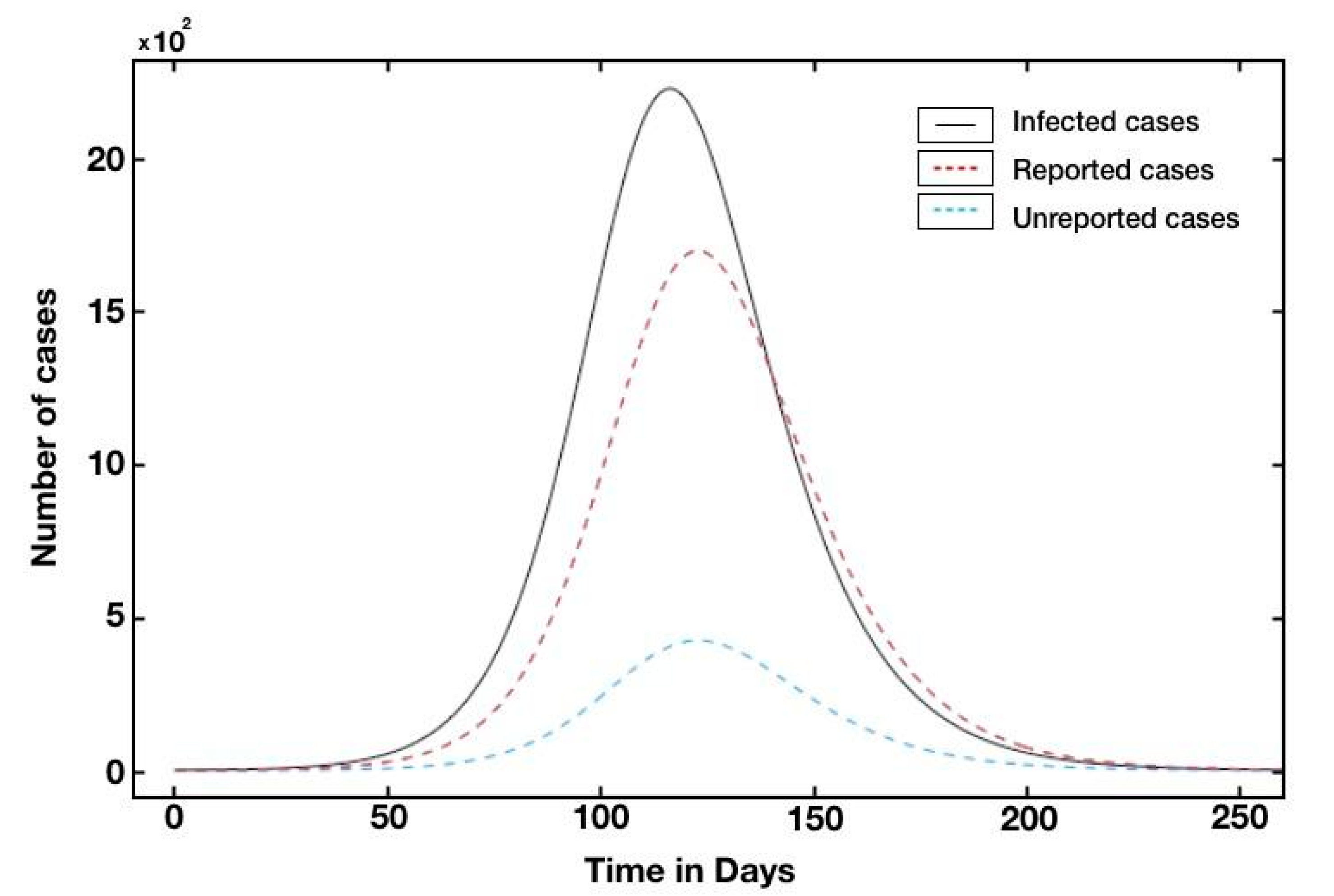
| Countries | R0 | Reference |
|---|---|---|
| Iran | 2.30 | [12] |
| South Korea | 2.60 | [13] |
| Singapore | 1.54 | [14] |
| Japan | 2.20 | [19] |
| Israel | 1.26 | [15] |
| Algeria | 2.55 | [16] |
| USA | 4.02 | [17] |
| Brazil | 2.81 | [18] |
| China | 6.6 | [20] |
| Symbol | Interpretation |
|---|---|
| Time at which the epidemic started | |
| Number of susceptible at time | |
| Number of asymptomatic infectious at time | |
| Number of unreported symptomatic infectious at time | |
| 1/ | Average time during which asymptomatic are asymptomatic |
| f | Fraction of asymptomatic that become reported symptomatic |
| Rate at which asymptomatic become reported symptomatic | |
| Natural death rate | |
| Number of births per unit time | |
| Fraction of infectives recovering with immunity against reinfection | |
| Rate at which asymptomatic become unreported symptomatic | |
| 1/ | Average time symptomatic infectious have symptoms |
| Cumulative | 2nd | 3rd | 4rd | 5th | 6th | 7th | 8th |
|---|---|---|---|---|---|---|---|
| Reported cases | 14,872 | 15,551 | 16,191 | 17,142 | 17,972 | 18,020 | 18,321 |
| Predicted cases | 15,138 | 16,524 | 17,027 | 19,658 | 19,427 | 19,701 | 19,890 |
| τ | Λ | µ | ||||
|---|---|---|---|---|---|---|
| 215.08 | 0.08 | 145.48 | −4.88 | 7.1 × 10−8 | 9.40 | 1.20% |
| Country | t0 | τ | I0 | U0 | R0 |
|---|---|---|---|---|---|
| Qatar | −5 | 7.10 × 10−8 | 10.1 | 1.3 | 2.42 |
| Saudi Arabia | −1 | 0.58 × 10−8 | 26.4 | 3.3 | 2.45 |
| UAE | −1 | 7.60 × 10−8 | 12.4 | 1.8 | 2.19 |
| Bahrain | −20 | 10.4 × 10−8 | 09.8 | 1.5 | 2.19 |
| Kuwait | −22 | 3.74 × 10−8 | 11.0 | 1.4 | 2.37 |
| Oman | −7 | 3.39 × 10−8 | 14.9 | 2.2 | 2.20 |
| France | −6 | 0.38 × 10−8 | 09.2 | 0.9 | 2.84 |
| Italy | −7 | 0.42 × 10−8 | 17.1 | 7.3 | 2.80 |
| New York | −4 | 6.48 × 10−8 | 03.1 | 1.0 | 3.62 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sallahi, N.; Park, H.; El Mellouhi, F.; Rachdi, M.; Ouassou, I.; Belhaouari, S.; Arredouani, A.; Bensmail, H. Using Unstated Cases to Correct for COVID-19 Pandemic Outbreak and Its Impact on Easing the Intervention for Qatar. Biology 2021, 10, 463. https://doi.org/10.3390/biology10060463
Sallahi N, Park H, El Mellouhi F, Rachdi M, Ouassou I, Belhaouari S, Arredouani A, Bensmail H. Using Unstated Cases to Correct for COVID-19 Pandemic Outbreak and Its Impact on Easing the Intervention for Qatar. Biology. 2021; 10(6):463. https://doi.org/10.3390/biology10060463
Chicago/Turabian StyleSallahi, Narjiss, Heesoo Park, Fedwa El Mellouhi, Mustapha Rachdi, Idir Ouassou, Samir Belhaouari, Abdelilah Arredouani, and Halima Bensmail. 2021. "Using Unstated Cases to Correct for COVID-19 Pandemic Outbreak and Its Impact on Easing the Intervention for Qatar" Biology 10, no. 6: 463. https://doi.org/10.3390/biology10060463
APA StyleSallahi, N., Park, H., El Mellouhi, F., Rachdi, M., Ouassou, I., Belhaouari, S., Arredouani, A., & Bensmail, H. (2021). Using Unstated Cases to Correct for COVID-19 Pandemic Outbreak and Its Impact on Easing the Intervention for Qatar. Biology, 10(6), 463. https://doi.org/10.3390/biology10060463








