Abstract
Rhodopsin, the first visual pigment identified in the animal retina, was shown to be a photosensitive membrane protein containing covalently bound retinal in the 11-cis configuration, as a chromophore. Upon photoexcitation the chromophore isomerizes in femtoseconds to all-trans, which drives the protein into the active state. Soon thereafter, another geometric isomer—9-cis retinal—was also shown to stably incorporate into the binding pocket, generating a slightly blue-shifted photosensitive protein. This pigment, coined isorhodopsin, was less photosensitive, but could also reach the active state. However, 9-cis retinal was not detected as a chromophore in any of the many animal visual pigments studied, and isorhodopsin was passed over as an exotic and little-relevant rhodopsin analog. Consequently, few in-depth studies of its photochemistry and activation mechanism have been performed. In this review, we aim to illustrate that it is unfortunate that isorhodopsin has received little attention in the visual research and literature. Elementary differences in photoexcitation of rhodopsin and isorhodopsin have already been reported. Further in-depth studies of the photochemical properties and pathways of isorhodopsin would be quite enlightening for the initial steps in vision, as well as being beneficial for biotechnological applications of retinal proteins.
1. Introduction
Rhodopsin is the first visual pigment identified, namely in the rod photoreceptor cells of the vertebrate retina [1,2]. It was shown to be a photosensitive membrane protein containing a vitamin A derivative (retinal) in the 11-cis configuration, as a chromophore, covalently bound to an internal lysine side chain via a protonated Schiff base (Figure 1) [2,3,4,5,6]. Upon absorption of a visual photon, the chromophore isomerizes in femtoseconds into the all-trans configuration, which triggers a sequel of conformational changes in the protein, driving it within milliseconds into the active state, which triggers the photoreceptor cell towards a neural response [5,7,8,9]. This response—membrane hyperpolarization—is processed by the bipolar and ganglion cells in the retina, and transmitted via the optic nerve towards the visual cortex, where it is translated within several tens of milliseconds into a visual response [10,11].
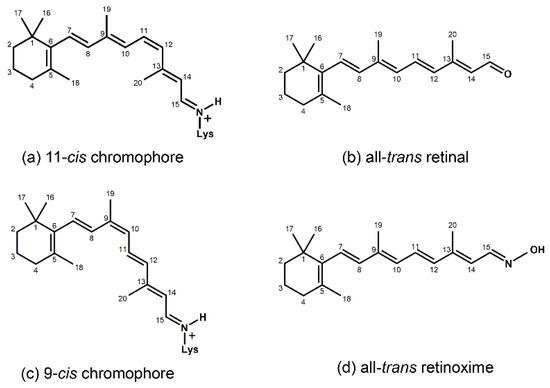
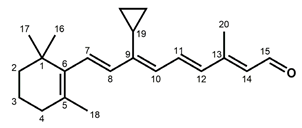
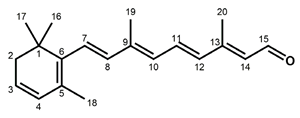
Figure 1.
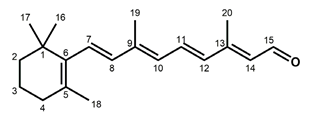
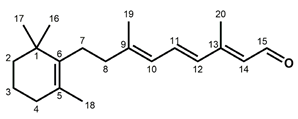
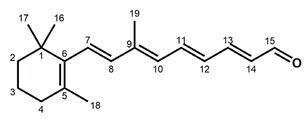
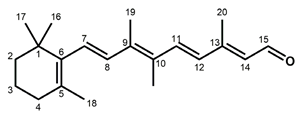
Chemical structures of the chromophore configurations in rhodopsin and isorhodopsin, and of free retinals: The retinals are covalently bound to a lysine residue as a protonated retinylidene Schiff base with the 11-cis,15-anti configuration in rhodopsin (a) and the 9-cis,15-anti configuration in isorhodopsin (c) in the “dark state” (in photophysical terminology, “ground state”). The chromophore is photoexcited into the all-trans configuration, which terminally relaxes and is eventually released as all-trans retinal (b). In the presence of hydroxylamine (HO–NH2) retinal is converted into retinoxime (d). In rhodopsin, as well as in isorhodopsin, the chromophore contains a 6-s-cis ring-polyene chain orientation and the 12-s-trans conformation. Because of the extended conjugated chain (alternating single (C–C) and double (C=C) bonds), the excitation energy is reduced, and retinal and retinoxime absorb in the UV-A region (300–420 nm). The protonated Schiff base with a lysine residue, generated upon covalent binding of the retinal with opsin (a,c), leads to further reduction in the excitation energy, red-shifting the absorbance maxima to 486 nm (isorhodopsin) and 498 nm (rhodopsin). Next to the lysine residue, the opsin binding pocket contains a docking site for the ring element and for the C19 methyl group. Binding into the pocket forces a twist and torsion in the conjugated chain. The main twist in the rhodopsin chromophore resides in the C10–C13 segment, with a torsion angle of circa 40°. The main twist in the isorhodopsin chromophore is divided over the C6–C9 and the C9–C11 segments, with a torsion angle of circa 20° over the latter. For further details and references, refer to the text.
The all-trans chromophore is eventually released as all-trans retinal by hydrolysis of the Schiff base, leaving the apoprotein opsin [7,12]. During this process, the opsin relaxes into a conformation close to that of rhodopsin, and has very little signaling activity left [13,14,15,16]. To regenerate the rhodopsin state, the all-trans retinal has to be isomerized into the 11-cis configuration which, when transported to the opsin site, will spontaneously recombine with opsin, generating rhodopsin, both in vivo and in vitro [1,5,6,17]. In the vertebrate retina, several systems for the production of 11-cis retinal are operational in the retina pigment epithelium (RPE) and cone photoreceptor cells, employing both enzymatically driven and photoactivated isomerases [18,19].
It was soon realized that the rod photoreceptor pigments had evolved for very-high-sensitivity (single-photon) black-and-white (scotopic) vision [20]. In the vertebrate realm, the absorbance maxima of the rod pigments range from 440 to 520 nm, depending on their habitat [21,22]. For color discrimination, a closely related set of pigments had evolved that cover the entire UV–deep-red region (350–640 nm), and are confined in the vertebrate cone photoreceptors [23,24,25]. In addition, it has become obvious that the visual pigments in the invertebrate realm are structured upon the same basic principle, but are slightly differently composed [26,27]. Specifically, they do not release the photogenerated all-trans chromophore, but require a second light pulse to recover the original 11-cis chromophore (bi-stable pigments) [26,28]. Thanks to the phenomenal developments in the current genome era (e.g., genome mining, recombinant DNA technology, heterologous expression), the animal rhodopsin family has expanded spectacularly over the last few decades, and the majority of these new species are bi-stable pigments involved in light-driven physiology in a wide array of tissues [27,29,30,31,32]. As far as we know now, all animal rhodopsins are involved in signaling processes, using GTP-binding proteins as second messengers, and constitute an important subfamily in the superfamily of G-protein-coupled receptors (GPCRs) [33,34]. Simultaneously, another rapidly expanding rhodopsin-like clan was discovered in unicellular organisms (the microbial rhodopsins) [35,36,37]. These pigments instead exploit an all-trans > 13-cis photoisomerization with thermal relaxation to the ground state, and mainly function as light-driven ion pumps or channels, but can also have a sensory or enzyme-modulating activity [38,39,40]. In view of their significantly different composition, photochemistry, and functionality, the microbial pigments are now classified as type-1 rhodopsins, and the animal pigments as type-2 rhodopsins.
Soon after the initial characterization of the animal rod pigment rhodopsin, it was observed that another geometric isomer—9-cis retinal—could also be stably incorporated into the binding pocket of rod opsin, generating a photosensitive protein with a slightly blue-shifted absorbance maximum (486 nm versus 498 nm for the native bovine pigment) [6,17]. This analog was termed isorhodopsin. Isorhodopsin could trigger the same photoactivation cascade in the protein, but with much lower photosensitivity. Since in that time segment 9-cis retinal had not been detected as a chromophore in any of the many animal visual pigments studied, isorhodopsin was passed over as an exotic and little-relevant rhodopsin analog. Consequently, few in-depth studies of its photochemistry and activation mechanism have been carried out. More recently, however, it was demonstrated that in certain pathologies of the retina isorhodopsin is generated in vivo, and that administration of a relatively stable 9-cis retinal derivative can restore vision in mammals that have a genetic defect in 11-cis retinal biosynthesis [41,42,43].
In this review, we aim to illustrate that it is unfortunate that isorhodopsin has received little attention in visual research and literature. So far, elementary differences between the photoexcitation of rhodopsin and isorhodopsin have already been reported. Apart from its evident medical relevance, further in-depth studies of the photochemical properties and pathways of isorhodopsin and its analog pigments, using a variety of biophysical techniques and in silico calculations, would be quite enlightening for the initial steps in vision, as well as being beneficial for biotechnological applications of retinal proteins.
2. Isorhodopsin—General Aspects
While the designation “isorhodopsin” in principle can be used to classify any retinal protein with 9-cis retinal as its chromophore, we here use it exclusively in its original context, i.e., the combination of 9-cis retinal with the bovine rod opsin. In other cases, we use the parent pigment with an “iso-” prefix.
In solution, 11-cis retinal experiences steric hindrance between the C18 methyl group and H8, and between the C20 methyl group and H10 (cf. Figure 1). As a consequence, its conformation is in equilibrium between torsional conformers around the 6–7, 10–11, and 12–13 single bonds [44]. In the binding site of the apoprotein (opsin), 11-cis retinal adopts a tight-fitting 6-s-cis,12-s-trans,15-anti conformation [45,46,47,48,49,50,51,52,53,54,55]. Nevertheless, 11-cis retinal spontaneously and rapidly incorporates into bovine opsin, generating rhodopsin [6,17]. The binding pocket contains specific anchoring space in docking sites for the ring element and the C19 methyl group, in the covalent Schiff base connection to the lysine ε-amino group, and to a lesser extent for the C20 methyl group [37,56,57,58,59,60,61,62,63,64,65,66,67,68]. In order to fit optimally, the 11-cis chromophore adopts a twist in the 10–11=12–13 segment (cf. Figure 1), while all single bonds are not significantly perturbed as compared to free 11-cis retinal [45,48,69,70,71,72,73,74,75,76,77,78,79]. The Schiff base linking the chromophore to the opsin is protonated (cf. Figure 1), and this positively charged element is stabilized by a complex counterion, which consists of a negatively charged counterion (Glu113) in combination with a hydrogen-bonded network including nearby opsin residues and bound water molecules [62,70,80,81,82,83,84,85,86,87,88,89,90,91,92]. The shift in the absorbance maximum, from circa 440 nm for a free protonated Schiff base of retinal to 498 nm in rhodopsin, strongly depends on the structure of the H-bonded network and counterion complex involving variable electrostatic interactions with the protonated Schiff base, and to a lesser extent on the properties of protein residues in the opsin binding pocket [62,81,93,94,95,96,97].
Compared to the 11-cis isomer, free 9-cis retinal is less troubled by internal steric hindrance [6,98]. Nevertheless, the rate of rhodopsin synthesis from opsin and 11-cis retinal is significantly higher than that of isorhodopsin synthesis from opsin and 9-cis retinal (5–10-fold, depending on conditions [6,17,99]). This is probably related to the circa 5 kcal/mol higher energy content of isorhodopsin relative to rhodopsin [99,100]. The ring–chain connection of the 9-cis chromophore is also 6-s-cis [101], but the induced fit of 9-cis retinal in the opsin binding pocket is suboptimal. The 9-cis chromophore is faced with strain in the ring and the C19-methyl docking sites, in the 10–11 and 14–15 single bonds, and in the 7=8 and 9=10 double bonds [48,56,57,62,69,78,102,103,104,105]. The C20 methyl group is also slightly displaced as compared to the rhodopsin chromophore [57]. In fact, the chromophore constellation in isorhodopsin has been referred to as an “induced misfit” [102]. The torsion in the 9=10 double bond of the 9-cis chromophore (circa 20°) is smaller than the torsion in the 11=12 bond of the 11-cis chromophore (circa 40°). On the other hand, the torsion in the 9=10 double bond of the 11-cis chromophore as well as in the 11=12 double bond of the 9-cis chromophore is relatively small (<5° and circa 10°, respectively) [57,79]. A very important difference between the two chromophores is that the largest torsion in the 9-cis chromophore resides in the 7=8 double bond, while in the 11-cis chromophore it resides in the 11=12 double bond.
Very informative in this context is vibrational spectroscopy (FTIR and resonance Raman). Of special relevance are the fingerprint region (1200–1300 cm−1), with a specific signature for each geometric isomer, and the hydrogen-out-of-plan (HOOP) region (800–1000 cm−1). In particular, when two HOOP vibrations are coupled in a trans-ethylenic bond, they degenerate and split up into an out-of-phase (Bg) and an in-phase (Au) component. The trans-coupled Au HOOP (HC=CH) and the cis-coupled A2 HOOP (HC=CH) absorb in the 900–990 cm−1 region [72,106,107,108,109], and their amplitude depends on the torsion in the ethylenic bond [71,72,108,110,111]. In the Raman scattering spectra, these coupled HOOPs turn up with their intensity depending on the torsion in the ethylenic bond and the loss of local symmetry. In particular, the Au HOOP in a flat structure has very low intensity in Raman spectra, but is strongly enhanced upon torsional deflection of the ethylenic bond. In infrared absorbance spectra, the coupled HOOPs are always active, but again their intensity strongly depends on the torsion in the ethylenic bond. Assignment of vibrational bands to structural elements is assisted by selective 13C or 2H labeling of the chromophore via organic synthesis, and can subsequently also validate physical computation [71,78,106,108,112,113,114]. Furthermore, solid-state NMR spectroscopy can also provide detailed structural information on the chromophore and its interaction with the binding pocket environment, using properly labeled and/or otherwise modified retinals [55,69,74,76,89,102,113,114,115,116,117,118,119,120,121,122].
While the absorbance maxima of free 11-cis and 9-cis retinal and their Schiff bases are very close [6,17], the maximum of isorhodopsin (486 ± 2 nm) is significantly blue-shifted from that of rhodopsin (498 ± 2 nm). This probably relates to the somewhat different arrangement of the complex counterion and the interaction with the binding pocket residues [102,104]. So far, a blue shift has been observed in all visual iso-pigments tested [101,123], except for the primate blue visual pigment, which apparently cannot stably bind 9-cis retinal [124,125].
3. Analog Pigments
Early on, it was realized that investigating the effects of modified retinals on pigment properties would enable significant information to be unraveled. For instance, data such as incorporation rates, spectral properties, photochemistry, G-protein activation, etc., could provide intimate insight into aspects such as binding pocket restraints, spectral tuning, and photoactivation mechanisms [45,58,63,72,112,123,126,127,128,129,130,131,132]. The majority of these analogs are based on the 11-cis isomer, but analogs of other isomers have also been studied—predominantly the 9-cis isomer (e.g., [45,101,105,113,123,126,127,131,133,134]).
It is essential to verify that the isomeric state of the analog (for instance, 9-cis or 11-cis) is retained upon incubation with opsin, since the incorporation rate is often strongly reduced, and analog pigment formation may take many hours instead of minutes (e.g., [131,135,136,137]). This is commonly performed by quantitative extraction of the chromophore as the oxime derivative (Figure 1), followed by liquid chromatographic identification [138,139]. For instance, while free 9,11-dicis and 9,13-dicis retinal are stable in organic solvents, upon incubation with opsin thermal isomerization occurs, and both isorhodopsin and the dicis-pigment are generated [131,135,136]. Furthermore, the 9,13-pigment slowly isomerizes thermally into isorhodopsin, and the 9,11-pigment is decomposed upon incubation with hydroxylamine [131,140]. For identification of these iso-pigments, several analytical data are available: the significant differences in the spectral and vibrational profiles of the 9-cis, 9,11-dicis, and 9,13-dicis retinals [141]; the different chromatographic profiles of the corresponding oxime derivatives; and the distinctly different absorbance maxima of the corresponding pigments (486, 472 and 478 nm, respectively [131,136]).
Hydroxylamine (HO–NH2) reacts with free retinals (R–C=O) under the formation of the oxime derivative (Figure 1), which is much less susceptible to thermal isomerization [17,139,142]. Hydroxylamine can also react with Schiff bases generating the oxime, but most rod pigments are stable or decompose only very slowly in the presence of hydroxylamine. This demonstrates that in the rod pigments the binding pocket is very poorly accessible to this reagent. To be able to extract the chromophore as the stable oxime, it is then essential to denature the protein chemically in the presence of a large excess of hydroxylamine [139]. On the other hand, in most cone pigments and many analog pigments the Schiff base is more accessible to hydroxylamine, and the chromophore is eventually released as the oxime derivative [17,24,123]. When this is the case for analog pigments of rhodopsin and isorhodopsin, it indicates a poor fit of the analog in the binding pocket, rendering the Schiff base region accessible to small hydrophilic reagents.
An alternative way to identify the isomeric state of the chromophore, allowing in situ observation, is to employ vibrational spectroscopy. The fingerprint region of the vibrational spectra, reflecting single C–C bond stretching vibrations, is very characteristic of the isomeric configuration of retinals and retinylidene chromophores. This is usually sufficient for identification [78,106,141,143]. In this review, we focus on the HOOP vibrations (HC=CH) and (HC=CH), since they provide information on the structure of the chromophore, as explained above. In rhodopsin and isorhodopsin, this pertains to the 7=8 and 11=12 HOOPs (Figure 1). In the 7,8-dihydro analogs, the 7=8 double bonds are reduced to a single bond, and only the 11=12 HOOP remains. Table 1 shows the vibrational assignment for these HOOPs in rhodopsin, isorhodopsin, and selected analog pigments, based on resonance Raman and FTIR spectroscopy. When a vibration cannot be observed or is weak, it indicates that the corresponding ethylenic bond is largely free from or low in torsion. Here, we only present rhodopsin and isorhodopsin, but the 11-cis and 9-cis pigments of the red cone pigment iodopsin and of squid and octopus rhodopsin exhibit HOOP vibrational frequencies that are very close to the rod pigments [47,144,145,146,147,148]. The data in Table 1 show several interesting features. In agreement with other studies, the 11-cis chromophore in the native rhodopsin only shows strain in the 11=12 double bond. In contrast, the 9-cis chromophore in isorhodopsin exhibits less strain in the 11=12 double bond, but clear indication of strain in the 7=8 double bond. This probably reflects the poor fit of the ring and the C19 methyl group in their docking site [57,102,104]. In the 7,8-dihydro pigments, the 7-8 bond is now single and less restrained, which increases the flexibility around the ring–chain connection. This does not release the strain in the 11=12 double bond, and therefore does not make much difference for the 11-cis chromophore, but the 7,8-dihydro 9-cis chromophore probably fits better than the unmodified 9-cis chromophore (see Section 4). As expected, elimination of the C20 methyl group, resulting in the 13-desmethyl pigment, reduces the strain in the 11=12 double bond and, thus, the intensity of the 11=12 coupled HOOP in the 11-cis chromophore. In contrast, the addition of a methyl group at C10, resulting in the 10-methyl pigment, perturbs the fit of the 11-cis chromophore and increases the strain in the 11=12 double bond.

Table 1.
HOOP frequencies of selected 11-cis and 9-cis pigments.
Particularly remarkable is the strong and opposite effect on the intensity of the HOOP vibrations upon replacing the C19 methyl group with the more voluminous cyclopropyl group (Table 1). In the case of the 11-cis chromophore, this clearly enhances the 7=8 HOOP, reflecting more strain in the 7=8 double bond, and weakens the 11=12 HOOP, reflecting less strain in the 11=12 double bond. This reflects a much poorer fit in the binding pocket, as compared to the unmodified 11-cis retinal. In contrast, in the case of the 9-cis chromophore, the strain in the 7=8 double bond is strongly reduced, while the strain in the 11=12 bond is increased. This more closely resembles the HOOP profile of the unmodified 11-cis chromophore, and suggests that the 9-cis cyclopropyl analog creates a better induced fit in the binding pocket than its parent 9-cis retinal. This is discussed further in Section 4.
In this context, as well as in view of its photochemical properties, it is also noteworthy to look at the 3,4-dehydro analog (Table 1). In fact, the 11-cis isomer of this analog is also found in vertebrate visual pigments—in particular, those of freshwater and coastal vertebrates [123,156]. The 3,4-dehydro-retinal is also denoted as retinal A2, to differentiate it from the abundant retinal (Figure 1)—also denoted as retinal A1. The more extended conjugated chain in retinal A2 further decreases the excitation energy, thereby red-shifting the absorbance band by about 20 nm. As a chromophore, retinal A2 red-shifts the absorbance maximum by 20–40 nm in rod pigments and up to 70 nm in cone pigments, as compared to retinal A1 [17,22,101,157]. The extra double bond in the ring (Table 1) also induces more stiffness in the ring dynamics. The HOOP frequencies of the 11-cis 3,4-dehydro-chromophore have not been reported but, as discussed below, it probably fits equally well as the 11-cis A1 chromophore. However, the intensity of the 7=8 HOOP vibration in the 9-cis 3,4-dehydro-chromophore is enhanced relative to the 9-cis A1 chromophore (Table 1), suggesting an increase in the strain and in the “misfit” in the binding pocket.
4. Photochemistry
The photochemistry of iso-pigments has mainly been studied in isorhodopsin. Among the animal rhodopsins, the photochemical cascade of bovine rod rhodopsin has been investigated in most detail, with a variety of techniques (ultrarapid optical and Raman spectroscopy of the kinetics of selected transitions, crystallography and vibrational and NMR spectroscopy of stabilized photo-intermediates, advanced in silico computational procedures, etc.) (e.g., [79,82,102,158,159,160,161,162,163,164,165,166] and references therein). A global scheme is presented in Figure 2, and we first discuss this in some detail. The photocascade is triggered by isomerization of the photoexcited chromophore towards the all-trans configuration, which results in formation of the first photo-intermediate bathorhodopsin (Batho) within femtoseconds. This intermediate is red-shifted from the ground-state rhodopsin [167,168]. The efficiency of this isomerization reaction is astonishing, occurring within femtoseconds with a quantum yield of 0.65 (i.e., two out of three photons are productive) [169,170,171,172]. Rapid kinetic studies indicate that in free 11-cis retinal, as well as its protonated Schiff base, photoisomerization is more selective towards the all-trans isomer, and occurs within picoseconds, with quantum yields between 0.2 and 0.3 [173,174,175]. Hence, in the rhodopsin context, this conversion is surprisingly efficient and selective in 11-cis to all-trans. This is facilitated by the constraints of the binding site and the twist in the C10-C13 segment of the chromophore.
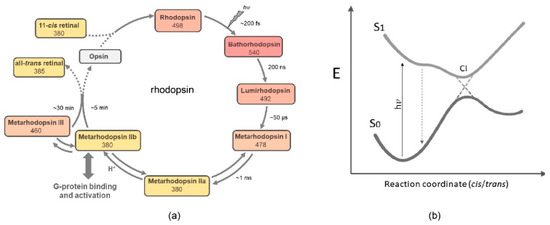
Figure 2.
Global presentation of the predominant photochemical pathway in rhodopsin: (a) The 11-cis,15-anti chromophore configuration in rhodopsin is photoisomerized into all-trans. It remains all-trans in all photo-intermediates, except for Meta III, which contains a 15-syn protonated Schiff base [58]. The early photo-intermediates also contain a protonated Schiff base, and relax thermally to deprotonated Meta II states. In many visual pigments, Batho is in equilibrium with a blue-shifted intermediate (BSI) [176,177]. (b) Simplified schematic of a conical intersection where the excited chromophore at the S1 potential energy surface can cross over to the S0 potential energy surface of the photoproduct. The S1 surface can contain thermal transitions, as suggested for isorhodopsin (see text). For rhodopsin, very early interstate mixing between the S1 and S2 states has been proposed, which would lead to excited state population splitting [178].
A very effective combination of a selectively deuterated chromophore at the 11 and/or 12 position with femtosecond spectroscopy and advanced quantum chemical computation resolved many issues in the photoisomerization process of bovine rhodopsin [179]. The global picture arises that, after photoexcitation of the chromophore into the Franck–Condon state, it rapidly relaxes along a barrierless trajectory on the potential surface, to a minimal-energy conical intersection (Figure 2). Here, productive resonance of the electronic wave packet at the excited-state potential surface, with torsional and HOOP vibrational modes in the twisted C10–C13 segment of the 11-cis chromophore, can prime very effective crossover to a ground-state energy surface, generating a hot all-transoid state (photorhodopsin) after tens of fs [171,178,180]. This relaxes thermally within about 200 fs into the photoproduct Batho, which contains a still highly twisted all-trans chromophore, but is stable below 130 K [78,108,167]. Recent advanced QM/MM simulations suggest excited-state population splitting resulting in several waves of productive crossover to the photoproduct’s ground state in the first 150 fs, promoted by vibronic coupling, vibrational phase matching, and electrostatic interactions in the binding site [178].
At room temperature, the circa 35 kcal of excitation energy stored in Batho [181,182,183] drives further relaxation via several intermediates until the metarhodopsin IIa-IIb equilibrium is reached within ms. This relaxation process subtly rearranges chromophores, protein residues, and H-bonded hydrated networks up to the metarhodopsin stage [12,184,185,186,187,188], where the Schiff base transfers its proton, the counterion and another Glu residue at the intracellular side of the protein become protonated, and an interhelical activity switch reshuffles helical segments to open up binding residues for the G protein [12,189,190,191,192,193,194,195]. The chromophore is subsequently slowly released via hydrolysis of the Schiff base to generate the nearly inactive apoprotein opsin [7,13,15,196]. In vivo, however, the active state is rapidly inactivated through phosphorylation by rhodopsin kinase, followed by arrestin binding, which blocks further activation of the G protein [197]. As documented in the Introduction, in order to retrieve the rhodopsin pigment, the opsin end-state of the photocascade has to be refurbished with 11-cis retinal, so as to spontaneously regenerate rhodopsin in accord with the circa 10 kcal lower energy level of the holoprotein [100].
Soon after its discovery, it was already established that, as from bathorhodopsin, excitation of isorhodopsin yields the same photocascade as rhodopsin, albeit with significantly lower photosensitivity (i.e., quantum yield of 0.25 [99,154,172,198]). As a result, isorhodopsin also signals the G protein, albeit with reduced kinetics at low light intensity [43,199]. Overall, the isorhodopsin photocascade significantly differs in several aspects from that of rhodopsin, namely, the pigment (re)generation rate, the photoisomerization mechanism, the kinetics and quantum yield, and the temperature dependence. Pigment regeneration is discussed in Section 2. The other aspects are probably interrelated, and are discussed here in context.
It was established that the chromophore in Batho is still bound via a protonated Schiff base, but has an all-trans configuration and a highly twisted conformation in the single bonds, whereby the C19 and C20 methyl groups and the ring are barely displaced [57,82,167,184,200,201]. Very informative in this context are the vibrational spectra—in particular, the fingerprint region and the HOOP and torsional region [78,107,108,153,202]. The general consensus is that upon photoexcitation rhodopsin and isorhodopsin generate the same first photo-intermediate—bathorhodopsin (Figure 2). Indeed “both” Bathos exhibit very similar spectral and vibrational properties [48,70,72,85,107,108,144,146,150,152,153,167,176,203,204,205]. However, it has been reported that the photo-intermediates Batho and Lumi may subtly differ between rhodopsin and isorhodopsin with respect to their absorbance maxima, molar absorbance, vibrational features, and the position of the C20 methyl group [57,104,205,206,207].
The exceptionally high quantum yield of rhodopsin—which is hardly affected by wavelength or temperature [169,170,208]—in combination with its ultrafast photoisomerization [179,180,202,209,210,211,212,213], is interpreted as reflecting a vibronically coherent photochemical process. This would involve productive vibronic coupling of nuclear vibrational modes of the isomerizing 10-11=12-13 segment and the excited electronic wave packet, resulting in a barrierless S1 potential energy surface trajectory to a minimal-energy conical intersection, with very efficient crossover to the S0 surface of the trans photoproduct via a bicycle pedal motion [79,153,166,170,178,179,180,207,210,214,215,216,217,218,219,220]. The important contribution of the coupled 11=12 HOOP vibrational phase is perfectly illustrated by analysis of deuterated analogs, where the 11,12-dideutero derivative (DC=CD) even slightly increases the quantum yield (Table 2), while the mono-deuterated analogs substantially decrease the quantum yield [179], similar to other 11- or 12-mono-substituted analogs [149,221]. Deuteration does not affect the rate of Batho formation (Table 2). The twist in the 10-11=12-13 segment of the polyene chain in the ground state would substantially prime the entire process. However, the chromophore is tightly constrained in the binding pocket, especially with respect to the ring and C19 methyl group, and at the femtosecond-to-picosecond timescale other motions in the binding pocket are restricted to the hydrated hydrogen-bonded network and very small displacements of the protein residue side chains [222,223,224]. Hence, 11-cis ⇒ trans isomerization would require a kind of concerted motion in the 10–14 segment, for which volume-conserving “bicycle-pedal”, “hula-twist”, and “wring-a-wet-towel” trajectories have been proposed, and slight displacement of the C20 methyl group is required [79,111,225]. A bicycle pedal motion now seems to be generally accepted [178,207,217].
However, for isorhodopsin, this situation is markedly different. First of all, the photoisomerization rate is reduced significantly, since Batho formation only levels off at about 600 fs, compared to the 200 fs for rhodopsin [158,209,213,226]. In addition, the quantum yield of isorhodopsin is not only significantly reduced (Table 2), but is also strongly temperature- and wavelength-dependent. At 77 K, the quantum yield is reduced to 0.16 at wavelengths ≤ 460 nm, and decreases further to 0.09 at 540 nm [99,169]. This has been interpreted as a small activation barrier along the S1 potential energy surface. However, it should be realized that compared to the 11-cis chromophore, the strain in the 9-cis chromophore is distributed quite differently. Most of the strain in the 9-cis chromophore is located in the 7=8 trans double bond, of which the skeletal vibrational modes and A2 HOOP are counterproductive for isomerization of the 9=10 cis bond—also because of the fixed position of the C19 methyl group. Furthermore, the strain in the 9=10-11=12-13 segment is relatively modest (cf. Section 2). Hence, we also take the view, that the photoisomerization trajectory of isorhodopsin must deviate from that of rhodopsin [57,79,207]. In this context, it is relevant to inspect Table 2.
First of all, Table 2 presents some exceptions to the common feature that 9-cis pigments are blue-shifted relative to the corresponding 11-cis pigments. For instance, when the 5=6 or 7=8 double bonds are reduced to single bonds, the corresponding 9-cis and 11-cis pigments have very similar spectral properties ([227] and Table 2), suggesting that the increase in flexibility around the ring segment improves the induced fit of the 9-cis chromophore. Nevertheless, it has been reported that the Batho ↔ BSI equilibrium behaves differently in the 9-cis 5,6-dihydropigment as compared to the corresponding 11-cis pigment [227]. More spectacular is the effect of a cyclopropyl substituent at C9. Here, the 11-cis pigment is blue-shifted, while the 9-cis pigment is significantly red-shifted, and even passes by the 11-cis pigment (Table 2).

Table 2.
Optical and photochemical parameters of selected 11-cis and 9-cis pigments #.
Table 2.
Optical and photochemical parameters of selected 11-cis and 9-cis pigments #.
| Retinal * | Isomer | λmax Pigment (nm ± 0.02) | Photochemistry | References | |
|---|---|---|---|---|---|
| Time for Batho to Max | Quantum Yield | ||||
| Native (A1) | 11-cis 9-cis | 498 486 | 200 fs 600 fs | 0.65 ± 0.02 0.25 ± 0.04 | [72,99,158,169,170,172,208,209,213,226] |
| 7,8-Dihydro A1 | 11-cis 9-cis | 426 428 | - - | 0.68 ± 0.06 0.39 ± 0.04 | [105,127,134,228,229] |
| 9-Cyclopropyl A1 | 11-cis 9-cis | 492 504 | - - | 0.08 ± 0.04 0.39 ± 0.04 | [105,154] |
| 10-Methyl A1 | 11-cis 9-cis | 506 498 | - - | 0.55 ± 0.07 <0.2 | [130,198,230] |
| 11,12-D2 A1 | 11-cis 9-cis | 498 - | 200 fs - | 0.69 ± 0.02 - | [179] |
| 12-D A1 | 11-cis 9-cis | 498 - | 200 fs - | 0.48 ± 0.03 - | [179] |
| 13-Desmethyl A1 | 11-cis 9-cis | 496 486 | 400 fs - | 0.46 ± 0.04 - | [72,137,221,231,232,233,234] |
| 14-Fluoro A1 | 11-cis 9-cis | 528 510 | - - | 0.55 ± 0.10 0.40 ± 0.06 | [149,198,235,236] |
| 3,4-Dehydro A1 (A2) | 11-cis 9-cis | 518 500 | - - | 0.63 ± 0.04 0.10 ± 0.02 | [155,208,237,238] |
# All data taken at room temperature. -: “data not available”. * For full chemical structures, see Table 1.
The effects on the quantum yield of the photoisomerization are also instructive. The substantial decrease in the quantum yield of the mono-deuterated 11-cis pigment (12-D) is similar to that of the 11-D analog [179], as explained above. Where tested, the quantum yield of the 11-cis pigments is higher in most cases than that of the corresponding 9-cis pigments. A significant increase in the quantum yield of analog 11-cis pigments has not yet been observed, but for 9-cis pigments this is clearly the case for the 7,8-dihydro, 9-cyclopropyl, and 14-F analogs. Again, the 9-cyclopropyl pigments are exceptional, as the 11-cis quantum yield drops precipitously while the 9-cis quantum yield increases substantially (Table 2).
How can all of these data fit into a plausible photoisomerization trajectory for isorhodopsin? The astonishing decrease in the quantum yield of the 11-cis 9-cyclopropyl-pigment evokes the deduction that an increase in the twist in the 7=8 double bond with a decrease in the twist in the 11=12 double bond (cf. Table 1) is counterproductive for photoisomerization. While this thesis does need further experimental and computational underpinning, it is supported by the 7,8-dihydro data, showing that with a flexible 7–8 single bond presenting solitary C–H vibrations the quantum yield of the iso-pigment is substantially enhanced. It is further supported by our observation that in 9-cyclopropyl-isorhodopsin a decrease in the 7=8 strain with an increase in the 11=12 strain (cf. Table 1) is accompanied by an increase in the quantum yield (Table 2). Unfortunately, we do not have the HOOP profiles of the 14-F analogs, but further studies on these pigments are also strongly recommended.
We can therefore conclude that in isorhodopsin the 11=12 trans double bond is intimately involved in the photoisomerization of the 9=10 cis double bond. In fact, QM/MM trajectory simulations of excited isorhodopsin in combination with ultrarapid spectroscopy predicted up to 23% production of a 9,11-dicis photoproduct [79,207]. The more recent article [207] derives two distinct trajectories on the S1 potential energy surface, leading to two distinct conical intersections: a faster unproductive trajectory, producing isorhodopsin and 9,11-dicis rhodopsin, and—due to sterical interaction with the protein—a slower trajectory producing the all-trans photoproduct via a forward pedal rotation. However, 9,11-dicis rhodopsin has never been observed as a photoproduct of isorhodopsin. Moreover, we would expect stronger sterical interaction with the protein in the case of the 9-cyclopropyl derivative, thereby reducing the quantum yield. In the 7,8-dihydro- and 9-cyclopropyl-isorhodopsin, an absent or reduced twist in the 7=8 trans double bond, in combination with a larger twist in the 11=12 trans double bond, results in an increase in the quantum yield. Hence, we can infer that the vibrational phase of the 11=12 trans double bond plays an important role in the photoisomerization of the 9-cis chromophore in isorhodopsin. Our thesis is that on the S1 potential surface of excited isorhodopsin, a (major?) trajectory leads to a conical intersection and crossover under trans ⇒ cis isomerization of the 11=12 double bond, resulting in a hot ground-state 9,11-dicis-like chromophore. In the overheated binding pocket with strong vibrational activity, this can thermally return to the original state (isorhodopsin), or can transition via a bicycle pedal mechanism to a highly twisted all-transoid Batho-like photoproduct. This would slow down the entire process, and may also explain the increase in stimulated emission [178,207]. It is also consistent with the very slow incorporation of 9,11-dicis retinal into the opsin at room temperature, the higher energy level of the 9,11-dicis pigment, and its susceptibility to attack by hydroxylamine [79,131,140], indicative of a very unstable induced fit in the 9,11-dicis pigment. It is also conceivable that cryotemperatures and lower-energy photons, which reduce the vibrational excitement, promote return to the isorhodopsin ground state rather than isomerization to all-trans, resulting in a reduction in the isomerization quantum yield. In conclusion, we propose that in the excited binding pocket of isorhodopsin a hot 9,11-dicisoid photoproduct is generated, and a thermal trajectory leads to significant all-trans formation. Both trajectories are enhanced by the torsion and vibrational activity of the 11=12 bond.
In this context, it is noteworthy that the enhanced quantum yields of the isorhodopsin analogs all level out around 0.4 (Table 2)—still substantially below that of rhodopsin. This may be related to the intrinsically slower torsional velocity and less straightforward dynamics in the 9-cis isomer [79,207,226,239].
Finally, we note again that 3,4-dehydroretinal shows interesting behavior (Table 1 and Table 2). The quantum yield of 3,4-dehydrorhodopsin is hardly reduced. Although HOOP frequencies are not available, this is consistent with the concept that the isomerization trajectory of rhodopsin is concentrated in a fast and productive bicycle pedal motion in the C9-C13 reaction coordinate, and is scarcely influenced by the dynamics of the ring [57,178,207,210]. In contrast, the quantum yield of 3,4-dehydro-isorhodopsin is significantly reduced relative to isorhodopsin itself (Table 2). Again, this is consistent with the stronger 7=8 HOOP intensity since, as discussed above, the twist in the 7=8 double bond is counterproductive in the iso-pigments. This would further impede the photoisomerization trajectory in 3,4-dehydro-isorhodopsin towards the all-trans photoproduct via the 9,11-dicisoid photoproduct, reducing the quantum yield. In further support, similar to isorhodopsin, the quantum yield of 3,4-dehydro-isorhodopsin is temperature-dependent, dropping to 0.05 at 80 K [155].
5. Physiological and Medical Relevance
Iso-pigments not only can be generated with animal opsins, but also have been observed in the microbial opsin family, that without exception employs all-trans retinal for its chromophore, which is photoisomerized to the 13-cis configuration, and eventually thermally reverts to the all-trans ground state [36,38]. For instance, the microbial prototype bacteriorhodopsin (BR) generates a 9-cis photoproduct upon sustained illumination, and this phenomenon may also occur in other microbial rhodopsins [36,240,241]. The putative microbial proton pump MR (middle rhodopsin) can even slowly incorporate 9-cis retinal, generating the iso-pigment [242]. Nevertheless, most microbial opsins do not generate iso-pigments, in contrast to most animal opsins.
Initially, visual pigments were isolated and laboriously purified from natural sources [17,22,123,243,244]. For solubilization, mild natural surfactants such as bile acids and digitonin were used [1], since cone pigments in particular were not very stable when dissolved with the commercially available surface-active agents [17,244,245]. Later on, mild synthetic detergents such as CHAPSO, OG, and DDM were developed [246,247,248], and now dominate the retinal protein field. This was also of great help when the recombinant DNA era arrived in the 1980s and proficient heterologous hosts for the production of the apoprotein (opsin) in cell culture were developed [249,250,251,252,253]. In order to generate the holoproteins, 11-cis retinal had to be supplied to the cell culture, but this compound has several disadvantages, including low thermal stability in biological environments [17,142], not being commercially available, and elaborate purification after chemical synthesis or after photoproduction from all-trans retinal [17,254,255]. Conversely, 9-cis retinal has better thermal stability [6] and is commercially available; hence, it seems to be a more suitable alternative.
This would not cause any problem for bi-stable rhodopsins, such as invertebrate visual pigments and melanopsins, since after photoexcitation they do not release all-trans retinal, but halt in a stable metarhodopsin state that can be photoisomerized to the corresponding 11-cis pigment [26,27,256]. In fact, the recent first crystal structure of an arthropod rhodopsin (jumping spider) was achieved with the iso-pigment [257]. When investigating the slower pathways in a rhodopsin photocycle, iso-pigments would also be a proper alternative, except for mutants or analog pigments, where the photocascades may deviate [113,133,154]. When using iso-pigments in vitro or in cell culture, one must take into account the much slower incorporation rate of 9-cis retinal into the opsin, the lower thermodynamic stability, and the lower photosensitivity of iso-pigments [258,259,260,261,262]. In particular, when using 9-cis retinal in physiological studies—for instance, when scanning for chaperones stabilizing rhodopsin mutants, investigating rhodopsin oligomerization in vivo, or studying organoid differentiation—, it is essential to verify essential data with 11-cis retinal [263,264,265,266,267,268,269].
Unexpectedly, 9-cis retinal has come to play an important role in treating certain pathologies of the vertebrate retina. Genetic defects in enzymes involved in the pathways responsible for furnishing the rod photoreceptor with 11-cis retinal—such as the isomerohydrolase RPE65, 11-cis retinol dehydrogenases, or lecithin retinol acyltransferase (LRAT)—can lead to night blindness, retinitis pigmentosa, or retinal degeneration [270,271,272]. Subcutaneous injection of 11-cis retinal in mutant mice did not properly relieve the symptoms [273]. When it was observed that mutant mice slowly produced isorhodopsin when kept in darkness, which enhanced the rod photoreceptor response [43,199], the option presented itself to test application of the more stable alternative 9-cis retinal. Indeed, early intervention in mutant mice with intravenously or intraperitoneally administered 9-cis retinal attenuated retinal loss of function and degeneration in a dose-dependent manner [274,275,276]. For oral delivery, 9-cis retinal was not very suitable, but 9-cis retinyl acetate—the acetate ester of the corresponding alcohol—proved to be sufficiently stable, and was rapidly absorbed and intracellularly hydrolyzed to 9-cis retinol, which did reach the retina [42]. Following intracellular enzymatic oxidation to 9-cis retinal, isorhodopsin was generated, which in mutant mice again attenuated retinal pathology [42,277,278]. An elaborate study in human patients also showed beneficial effects of oral delivery of 9-cis retinyl acetate, yielding sustained improvement in visual field and visual acuity of up to 70% of the patients [41,279]. Further improvement in pharmacological treatment was achieved by using a slow-release chitosan-9-cis-retinal conjugate [280]. This prodrug was tested in mice and dogs, and shown to be slowly absorbed from the gastrointestinal tract, resulting in sustained plasma levels of 9-cis-retinol and dose-dependent recovery of visual function lasting for several weeks [280].
Other physiological effects of retinols, such as production and function of the corresponding retinoic acids, are beyond the scope of this paper, and we refer to a relevant recent review [281].
6. Overview and Prospects
We expect to have provided compelling evidence that 9-cis retinal, while photophysiologically much less abundant and photophysicochemically much less exposed than 11-cis retinal, presents quite an attractive profile for detailed further studies.
For medical applications, the use of 9-cis retinal or derivatives will probably be restricted to retinal defects in 11-cis retinal production, since its potential in other retinal pathologies seems to be limited [282,283]. Testing analogs for this purpose is a complex option, since the physiological stability and pigment regeneration rate of the analog, along with the spectral properties and photoisomerization quantum yield of the corresponding iso-pigments, should not be problematic. For instance, 9-cis 7,8-dihydroretinal would improve the quantum yield, but would significantly blue-shift the photosensitivity of the rod photoreceptor, potentially disturbing the visual sensation. On the other hand, a locked analog, preventing photoisomerization, has been shown to have a protective effect against light-induced retinal degeneration [284], and to selectively block rod opsin consumption of chromophores while largely sparing cone opsins, prolonging cone-dependent vision [280].
For studies at a physiological, cellular, or in vitro level, 9-cis 7,8-dihydroretinal would actually be an interesting alternative to 9-cis retinal itself, since the spectral shift would then not be very disturbing, and the induced fit and the photoisomerization quantum yield would be closer to those with 11-cis retinal. In fact, it should be worthwhile to test whether the combination of the 7,8-dihydro element with a 14-F substitution would red-shift the corresponding iso-pigment closer to rhodopsin without reducing the quantum yield (Table 2).
However, most interesting are the photochemical mechanism and potential of the iso-pigment family. We have presented that, while rhodopsin is almost evolutionarily “finalized”—exhibiting ultrafast, nearly unidirectional, and very efficient photo-activation—isorhodopsin is much less effective. Nevertheless, the fascinating feature of isorhodopsin is that its mechanism and performance can be adapted and/or improved by retinal analogs. From that point of view, isorhodopsins offer great promise, which definitely needs to be further explored.
We have postulated an adapted photoisomerization trajectory for isorhodopsin, which should be verified by similar experimental and theoretical approaches as used for rhodopsin. Femtosecond spectroscopy, supported by advanced QM/MM computation, in combination with di- and mono-deuteration of the 7=8 double bond and/or of the 11=12 double bond, should yield principal information on their contribution to the rate and quantum yield of Batho formation by isorhodopsin. We postulate that mono-deuteration of the 7=8 double bond would reduce its counterproductive effect, thereby enhancing the quantum yield, while mono-deuteration of the 11=12 double bond would slow down generation of the 9,11-dicis product, thereby probably reducing the quantum yield and the rate of Batho formation. Similar studies with 3,4-dehydro, 7,8-dihydro-, and 9-cyclopropyl-isorhodopsin could reveal whether the same mechanistic principle is indeed active in these pigments, which should also make their photoproduct quantum yield temperature-dependent. In this context, analysis of 8–18, 11–13, or 12–14 ring-fused isorhodopsin analogs would also be very informative [101,285]. For instance, in the 8,18 methano-analog, the 7=8 double bond is mono-substituted and nearly flat, and we would postulate an increase in the quantum yield for the corresponding iso-pigment. The photoisomerization trajectory we propose for isorhodopsin would be perturbed or prohibited in the 11–13 and 12–14 ring-fused isorhodopsin analogs, and it would be very relevant to investigate whether alternative photoisomerization trajectories can be selected.
From another perspective, it would be very valuable if femtosecond structural studies became available for animal (iso)rhodopsins, as has recently been accomplished for microbial rhodopsins using femtosecond XFEL crystallography [222,286]. For animal pigments, it would probably be advisable to use cryo-EM in combination with the more easily prepared and more stable nanodisc environment [287], in an ultrarapid mode when possible. Eventually, in silico ab initio structural model building exploiting the newly available algorithms [288,289,290] will allow rapid testing of the chromophoric potential of theoretical promising retinal analogs for (iso)rhodopsins [291]. Atomic insight into the photoisomerization trajectories of isorhodopsins will be of utmost importance, not only from a photochemical point of view, but also for the design of retinal proteins with selected properties—both for bioengineering purposes and in optogenetics [292,293].
Author Contributions
Conceptualization: J.L. and W.J.d.G.; writing—original draft preparation, W.J.d.G.; writing—review and editing, W.J.d.G. and J.L. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Acknowledgments
The authors express their gratitude to Srividya Ganapathy (University of California at San Diego, School of Medicine) for her help with preparation of the figures.
Conflicts of Interest
The authors declare no conflict of interest.
Abbreviations
| BR | Bacteriorhodopsin |
| CHAPSO | 3-[(3-Cholamidopropyl)dimethylammonio]-2-hydroxy-1-propanesulfonate |
| Cryo-EM | Cryo-electron microscopy |
| DDM | Dodecylmaltoside |
| DFT | Density functional theory |
| FTIR | Fourier-transform infrared |
| Glu | Glutamate residue in the protein |
| HOOP | Hydrogen out-of-plane vibration |
| LRAT | Lecithin retinol acyltransferase |
| NMR | Nuclear magnetic resonance |
| OG | Octyl glucoside |
| QM/MM | Quantum-mechanical/molecular-mechanical |
| XFEL | X-ray free-electron laser |
References
- Tansley, K. The regeneration of visual purple: Its relation to dark adaptation andnight blindness. J. Physiol. 1931, 71, 442–458. [Google Scholar] [CrossRef] [PubMed]
- Wald, G. Carotenoids and the visual cycle. J. Gen. Physiol. 1935, 19, 351–371. [Google Scholar] [CrossRef] [PubMed]
- Honig, B.; Ebrey, T.G. Structure and spectra of chromophore of visual pigments. Annu. Rev. Biophys. Bioeng. 1974, 3, 151–177. [Google Scholar] [CrossRef] [PubMed]
- Morton, R.A.; Goodwin, T.W. Preparation of Retinene in Vitro. Nature 1944, 153, 405–406. [Google Scholar] [CrossRef]
- Morton, R.A.; Pitt, G.A.J. Visual pigments. Fortschr. Chem. Organ. Nat. 1957, 14, 244–316. [Google Scholar]
- Hubbard, R.; Wald, G. Cis-trans isomers of vitamin A and retinene in the rhodopsin system. J. Gen. Physiol. 1952, 36, 269–315. [Google Scholar] [CrossRef]
- Wald, G. The biochemistry of vision. Annu. Rev. Biochem. 1953, 22, 497–526. [Google Scholar] [CrossRef]
- Baylor, D.A. Photoreceptor signals and vision. Invest. Ophthalmol. Vis. Sci. 1987, 28, 34–49. [Google Scholar]
- Bloomfield, S.A.; Dacheux, R.F. Rod vision: Pathways and processing in the mammalian retina. Prog. Retin. Eye Res. 2001, 20, 351–384. [Google Scholar] [CrossRef]
- Porciatti, V.; Pizzorusso, T.; Maffei, L. The visual physiology of the wild type mouse determined with pattern VEPs. Vis. Res. 1999, 39, 3071–3081. [Google Scholar] [CrossRef]
- McConnell, S.K. The generation of neuronal diversity in the central nervous system. Annu. Rev. Neurosci. 1991, 14, 269–300. [Google Scholar] [CrossRef] [PubMed]
- Hofmann, K.P. Late photoproducts and signaling states of bovine rhodopsin. In Molecular Mechanisms in Visual Transduction; Stavenga, D.G., DeGrip, W.J., Pugh, E.N., Jr., Eds.; Elsevier Science Pub.: Amsterdam, The Netherlands, 2000; Volume 3, pp. 91–142. [Google Scholar]
- Rothschild, K.J.; Gillespie, J.; DeGrip, W.J. Evidence for rhodopsin refolding during the decay of meta II. Biophys. J. 1987, 51, 345–350. [Google Scholar] [CrossRef]
- Park, J.H.; Scheerer, P.; Hofmann, K.P.; Choe, H.-W.; Ernst, O.P. Crystal structure of the ligand-free G-protein-coupled receptor opsin. Nature 2008, 454, 183–187. [Google Scholar] [CrossRef] [PubMed]
- Jäger, S.; Palczewski, K.; Hofmann, K.P. Opsin/all-trans-retinal complex activates transducin by different mechanisms than photolyzed rhodopsin. Biochemistry 1996, 35, 2901–2908. [Google Scholar] [CrossRef]
- Sato, S.; Jastrzebska, B.; Engel, A.; Palczewski, K.; Kefalov, V.J. Apo-Opsin Exists in Equilibrium Between a Predominant Inactive and a Rare Highly Active State. J. Neurosci. 2019, 39, 212–223. [Google Scholar] [CrossRef]
- Hubbard, R.; Brown, P.K.; Bownds, M.D. Methodology of vitamin A and visual pigments. Methods Enzym. 1971, 18C, 615–653. [Google Scholar]
- Kiser, P.D.; Golczak, M.; Maeda, A.; Palczewski, K. Key enzymes of the retinoid (visual) cycle in vertebrate retina. Biochim. Biophys. Acta-Mol. Cell Biol. Lipids 2012, 1821, 137–151. [Google Scholar] [CrossRef]
- Choi, E.H.; Daruwalla, A.; Suh, S.; Leinonen, H.; Palczewski, K. Retinoids in the visual cycle: Role of the retinal G protein-coupled receptor. J. Lipid Res. 2021, 62, 100040. [Google Scholar] [CrossRef]
- Hecht, S.; Shlaer, S.; Pirenne, M.H. Energy, quanta, and vision. J. Gen. Physiol. 1942, 25, 819–840. [Google Scholar] [CrossRef]
- Musilova, Z.; Cortesi, F.; Matschiner, M.; Davies, W.I.L.; Patel, J.S.; Stieb, S.M.; de Busserolles, F.; Malmstrom, M.; Torresen, O.K.; Brown, C.J.; et al. Vision using multiple distinct rod opsins in deep-sea fishes. Science 2019, 364, 588–592. [Google Scholar] [CrossRef]
- Dartnall, H.J.A. The Identity and Distribution of Visual Pigments in the Animal Kingdom. In The Visual Process, 1st ed.; Davson, H., Ed.; Academic Press: New York, NY, USA, 1962; Volume 2, pp. 367–426. [Google Scholar]
- Crescitelli, F. The natural history of visual pigments: 1990. Prog. Retin. Res. 1991, 11, 1–32. [Google Scholar] [CrossRef]
- Imamoto, Y.; Shichida, Y. Cone visual pigments. Biochim. Biophys. Acta-Bioenerg. 2014, 1837, 664–673. [Google Scholar] [CrossRef] [PubMed]
- Yokoyama, S.; Yokoyama, R. Comparative Molecular Biology of Visual Pigments. In Molecular Mechanisms in Visual Transduction; Stavenga, D.G., DeGrip, W.J., Pugh, E.N., Jr., Eds.; Elsevier Science Pub.: Amsterdam, The Netherlands, 2000; Volume 3, pp. 257–296. [Google Scholar]
- Gärtner, W. Invertebrate visual pigments. In Molecular Mechanisms in Visual Transduction; Stavenga, D.G., DeGrip, W.J., Pugh, E.N., Jr., Eds.; Elsevier Science Pub.: Amsterdam, The Netherlands, 2000; Volume 3, pp. 298–388. [Google Scholar]
- Tsukamoto, H.; Terakita, A. Diversity and functional properties of bistable pigments. Photochem. Photobiol. Sci. 2010, 9, 1435–1443. [Google Scholar] [CrossRef]
- Stavenga, D.G. Insect retinal pigments: Spectral characteristics and physiological functions. Prog. Retin. Eye Res. 1995, 15, 231–259. [Google Scholar] [CrossRef]
- Bertolucci, C.; Foà, A. Extraocular photoreception and circadian entrainment in nonmammalian vertebrates. Chronobiol. Int. 2004, 21, 501–519. [Google Scholar] [CrossRef]
- Peirson, S.N.; Halford, S.; Foster, R.G. The evolution of irradiance detection: Melanopsin and the non-visual opsins. Philos. Trans. R. Soc. B—Biol. Sci. 2009, 364, 2849–2865. [Google Scholar]
- Koyanagi, M.; Terakita, A. Diversity of animal opsin-based pigments and their optogenetic potential. Biochim. Biophys. Acta-Bioenerg. 2014, 1837, 710–716. [Google Scholar] [CrossRef]
- Davies, W.I.L.; Tamai, T.K.; Zheng, L.; Fu, J.K.; Rihel, J.; Foster, R.G.; Whitmore, D.; Hankins, M.W. An extended family of novel vertebrate photopigments is widely expressed and displays a diversity of function. Genome Res. 2015, 25, 1666–1679. [Google Scholar] [CrossRef]
- Palczewski, K. G protein-coupled receptor rhodopsin. Annu. Rev. Biochem. 2006, 75, 743–767. [Google Scholar] [CrossRef]
- Kühn, H. Interactions between photoexcited rhodopsin and light-activated enzymes in rods. Prog. Retin. Res. 1984, 3, 123–156. [Google Scholar] [CrossRef]
- Oesterhelt, D.; Stoeckenius, W. Rhodopsin-like Protein from the Purple Membrane of Halobacterium halobium. Nat. New Biol. 1971, 233, 149–152. [Google Scholar] [CrossRef] [PubMed]
- Oesterhelt, D. The structure and mechanism of the family of retinal proteins from halophilic archaea. Curr. Opin. Struct. Biol. 1998, 8, 489–500. [Google Scholar] [CrossRef]
- Nagata, T.; Inoue, K. Rhodopsins at a glance. J. Cell Sci. 2022, 134, jcs258989. [Google Scholar] [CrossRef] [PubMed]
- Rozenberg, A.; Inoue, K.; Kandori, H.; Béjà, O. Microbial Rhodopsins: The Last Two Decades. Annu. Rev. Microbiol. 2021, 75, 427–447. [Google Scholar] [CrossRef] [PubMed]
- Mukherjee, S.; Hegemann, P.; Broser, M. Enzymerhodopsins: Novel photoregulated catalysts for optogenetics. Curr. Opin. Struct. Biol. 2019, 57, 118–126. [Google Scholar] [CrossRef]
- Govorunova, E.G.; Sineshchekov, O.A.; Li, H.; Spudich, J.L. Microbial Rhodopsins: Diversity, Mechanisms, and Optogenetic Applications. Annu. Rev. Biochem. 2017, 86, 845–872. [Google Scholar] [CrossRef]
- Koenekoop, R.K.; Sui, R.F.; Sallum, J.; van den Born, L.I.; Ajlan, R.; Khan, A.; den Hollander, A.I.; Cremers, F.P.M.; Mendola, J.D.; Bittner, A.K.; et al. Oral 9-cis retinoid for childhood blindness due to Leber congenital amaurosis caused by RPE65 or LRAT mutations: An open-label phase 1b trial. Lancet 2014, 384, 1513–1520. [Google Scholar] [CrossRef]
- Maeda, T.; Maeda, A.; Casadesus, G.; Palczewski, K.; Margaron, P. Evaluation of 9-cis-retinyl acetate therapy in RPE65−/− mice. Invest. Ophthalmol. Vis. Sci. 2009, 50, 4368–4378. [Google Scholar] [CrossRef]
- Fan, J.; Rohrer, B.; Moiseyev, G.; Ma, J.-X.; Crouch, R.K. Isorhodopsin rather than rhodopsin mediates rod function in RPE65 knock-out mice. Proc. Natl. Acad. Sci. USA 2003, 100, 13662–13667. [Google Scholar] [CrossRef]
- Honig, B.; Karplus, M. Implications of Torsional Potential of Retinal Isomers for Visual Excitation. Nature 1971, 229, 558–560. [Google Scholar] [CrossRef]
- Nakanishi, K.; Crouch, R.K. Application of artificial pigments to structure determination and study of photoinduced transformations of retinal proteins. Isr. J. Chem. 1995, 35, 253–272. [Google Scholar] [CrossRef]
- Ebrey, T.G.; Govindjee, R.; Honig, B.; Pollock, E.; Chan, W.; Crouch, R.K.; Yudd, A.; Nakanishi, K. Properties of several sterically modified retinal analogs and their photosensitive pigments. Biochemistry 1975, 14, 3933–3941. [Google Scholar] [CrossRef]
- Huang, L.; Deng, H.; Weng, G.; Koutalos, Y.; Ebrey, T.G.; Groesbeek, M.; Lugtenburg, J.; Tsuda, M.; Callender, R.H. A resonance raman study of the C=N configurations of octopus rhodopsin, bathorhodopsin, and isorhodopsin. Biochemistry 1996, 35, 8504–8510. [Google Scholar] [CrossRef] [PubMed]
- Bagley, K.A.; Balogh-Nair, V.; Croteau, A.A.; Dollinger, G.; Ebrey, T.G.; Eisenstein, L.; Hong, M.K.; Nakanishi, K.; Vittitow, J. Fourier-transform infrared difference spectroscopy of rhodopsin and its photoproducts at low temperature. Biochemistry 1985, 24, 6055–6071. [Google Scholar] [CrossRef]
- Deng, H.; Huang, L.; Groesbeek, M.; Lugtenburg, J.; Callender, R.H. Vibrational analysis of a retinal protonated schiff-base analog. J. Phys. Chem. 1994, 98, 4776–4779. [Google Scholar] [CrossRef]
- Kandori, H.; Matuoka, S.; Shichida, Y.; Yoshizawa, T.; Ito, M.; Tsukida, K.; Balogh-Nair, V.; Nakanishi, K. Mechanism of isomerization of rhodopsin studied by use of 11-cis-locked rhodopsin analogs excited with a picosecond laser pulse. Biochemistry 1989, 28, 6460–6467. [Google Scholar] [CrossRef]
- Ito, M.; Hiroshima, T.; Tsukida, K.; Shichida, Y.; Yoshizawa, T. A novel rhodopsin analog possessing the conformationally 6-s-cis-fixed retinylidene chromophore. J. Chem. Soc. Chem. Commun. 1985, 1443–1444. [Google Scholar] [CrossRef]
- Ito, M.; Mantani, Y.; Tsukida, K.; Shichida, Y.; Ioshida, S.; Fukada, Y.; Yoshizawa, T. A novel rhodopsin analog with bicyclic retinal involving the 8–18 bonded structure in the chromophore. J. Nutr. Sci. Vitam. 1988, 34, 641–646. [Google Scholar] [CrossRef]
- Ito, M.; Katsuta, Y.; Imamoto, Y.; Shichida, Y.; Yoshizawa, T. Conformational analysis of the rhodopsin chromophore using bicyclic retinal analogues. Photochem. Photobiol. 1992, 56, 915–919. [Google Scholar] [CrossRef]
- Fukada, Y.; Shichida, Y.; Yoshizawa, T.; Ito, M.; Kodama, A.; Tsukida, K. Studies on structure and function of rhodopsin by use of cyclopentatrienylidene 11-cis-locked-rhodopsin. Biochemistry 1984, 23, 5826–5832. [Google Scholar] [CrossRef]
- Mollevanger, L.C.P.J.; Kentgens, A.P.M.; Pardoen, J.A.; Courtin, J.M.L.; Veeman, W.S.; Lugtenburg, J.; DeGrip, W.J. High-resolution solid-state 13C-NMR study of carbons C-5 and C-12 of the chromophore of bovine rhodopsin: Evidence for a 6-S-cis conformation with negative-charge perturbation near C-12. Eur. J. Biochem. 1987, 163, 9–14. [Google Scholar] [CrossRef] [PubMed]
- Jäger, F.; Jäger, S.; Kräutle, O.; Friedman, N.; Sheves, M.; Hofmann, K.P.; Siebert, F. Interactions of the β-ionone ring with the protein in the visual pigment rhodopsin control the activation mechanism. An FTIR and fluorescence study on artificial vertebrate rhodopsins. Biochemistry 1994, 33, 7389–7397. [Google Scholar] [CrossRef] [PubMed]
- Nakamichi, H.; Buss, V.; Okada, T. Photoisomerization mechanism of rhodopsin and 9-cis-rhodopsin revealed by X-ray crystallography. Biophys. J. 2007, 92, L106–L108. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Vogel, R.; Lüdeke, S.; Siebert, F.; Sakmar, T.P.; Hirshfeld, A.; Sheves, M. Agonists and partial agonists of rhodopsin: Retinal polyene methylation affects receptor activation. Biochemistry 2006, 45, 1640–1652. [Google Scholar] [CrossRef]
- Ganter, U.M.; Schmid, E.D.; Perez-Sala, D.; Rando, R.R.; Siebert, F. Removal of the 9-methyl group of retinal inhibits signal transduction in the visual process. A Fourier-transform infrared and biochemical investigation. Biochemistry 1989, 28, 5954–5962. [Google Scholar] [CrossRef]
- Khorana, H.G. Rhodopsin, photoreceptor of the rod cell—An emerging pattern for structure and function. J. Biol. Chem. 1992, 267, 1–4. [Google Scholar] [CrossRef]
- DeGrip, W.J.; Bonting, S.L.; Daemen, F.J.M. The binding site of retinaldehyde in cattle rhodopsin. Biochim. Biophys. Acta 1973, 303, 189–193. [Google Scholar] [CrossRef]
- Sekharan, S.; Sugihara, M.; Weingart, O.; Okada, T.; Buss, V. Protein assistance in the photoisomerization of rhodopsin and 9-cis-rhodopsin-Insights from experiment and theory. J. Am. Chem. Soc. 2007, 129, 1052–1054. [Google Scholar] [CrossRef]
- Vogel, R.; Fan, G.-B.; Sheves, M.; Siebert, F. The molecular origin of the inhibition of transducin activation in rhodopsin lacking the 9-methyl group of the retinal chromophore: A UV-Vis and FTIR spectroscopic study. Biochemistry 2000, 39, 8895–8908. [Google Scholar] [CrossRef]
- Ganter, U.M.; Gärtner, W.; Siebert, F. The influence of the 13-methyl group of the retinal on the photoreaction of rhodopsin revealed by FTIR difference spectroscopy. Eur. Biophys. J. 1990, 18, 295–299. [Google Scholar] [CrossRef]
- Ebrey, T.G.; Tsuda, M.; Sassenrath, G.; West, J.L.; Waddell, W.H. Light activation of bovine rod phosphodiesterase by non-physiological visual pigments. FEBS Lett. 1980, 116, 217–219. [Google Scholar] [CrossRef]
- Okada, T.; Sugihara, M.; Bondar, A.-N.; Elstner, M.; Entel, P.; Buss, V. The retinal conformation and its environment in rhodopsin in light of a new 2.2 Å crystal structure. J. Mol. Biol. 2004, 342, 571–583. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Edwards, P.C.; Burghammer, M.; Villa, C.; Schertler, G.F.X. Structure of bovine rhodopsin in a trigonal crystal form. J. Mol. Biol. 2004, 343, 1409–1438. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.-J.; Bovee-Geurts, P.H.M.; Lugtenburg, J.; DeGrip, W.J. Constraints of the 9-methyl group binding pocket of the rhodopsin chromophore probed by 9-halogeno substitution. Biochemistry 2004, 43, 14802–14810. [Google Scholar] [CrossRef]
- Smith, S.O.; Palings, I.; Miley, M.E.; Courtin, J.M.L.; de Groot, H.J.M.; Lugtenburg, J.; Mathies, R.A.; Griffin, R.G. Solid-state NMR studies of the mechanism of the opsin shift in the visual pigment rhodopsin. Biochemistry 1990, 29, 8158–8164. [Google Scholar] [CrossRef]
- Rothschild, K.J.; Cantore, W.A.; Marrero, H. Fourier transform infrared difference spectra of intermediates in rhodopsin bleaching. Science 1983, 219, 1333–1335. [Google Scholar] [CrossRef]
- Eyring, G.; Curry, B.; Broek, A.; Lugtenburg, J.; Mathies, R.A. Assignment and interpretation of hydrogen out-of-plane vibrations in the resonance Raman spectra of rhodopsin and bathorhodopsin. Biochemistry 1982, 21, 384–393. [Google Scholar] [CrossRef]
- Kochendoerfer, G.G.; Verdegem, P.J.E.; Van der Hoef, I.; Lugtenburg, J.; Mathies, R.A. Retinal analog study of the role of steric interactions in the excited state isomerization dynamics of rhodopsin. Biochemistry 1996, 35, 16230–16240. [Google Scholar] [CrossRef]
- Verdegem, P.J.E.; Bovee-Geurts, P.H.M.; DeGrip, W.J.; Lugtenburg, J.; de Groot, H.J.M. Retinylidene ligand structure in bovine rhodopsin, metarhodopsin I, and 10-methylrhodopsin from internuclear distance measurements using 13C-labeling and 1-D rotational resonance MAS NMR. Biochemistry 1999, 38, 11316–11324. [Google Scholar] [CrossRef]
- Feng, X.; Verdegem, P.J.E.; Lee, Y.K.; Sandström, D.; Edén, M.; Bovee-Geurts, P.H.M.; DeGrip, W.J.; Lugtenburg, J.; de Groot, H.J.M.; Levitt, M.H. Direct determination of a molecular torsional angle in the membrane protein rhodopsin by solid-state NMR. J. Am. Chem. Soc. 1997, 119, 6853–6857. [Google Scholar] [CrossRef]
- Gröbner, G.; Choi, G.; Burnett, I.J.; Glaubitz, C.; Verdegem, P.J.E.; Lugtenburg, J.; Watts, A. Photoreceptor rhodopsin: Structural and conformational study of its chromophore 11-cis retinal in oriented membranes by deuterium solid state NMR. FEBS Lett. 1998, 422, 201–204. [Google Scholar] [CrossRef]
- Carravetta, M.; Zhao, X.; Johannessen, O.G.; Lai, W.C.; Verhoeven, M.A.; Bovee-Geurts, P.H.M.; Verdegem, P.J.E.; Kiihne, S.R.; Luthman, H.; de Groot, H.J.M.; et al. Protein-induced bonding perturbation of the rhodopsin chromophore detected by double-quantum solid-state NMR. J. Am. Chem. Soc. 2004, 126, 3948–3953. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Brown, M.F.; Heyn, M.P.; Job, C.; Kim, S.; Moltke, S.; Nakanishi, K.; Nevzorov, A.A.; Struts, A.V.; Salgado, G.F.J.; Wallat, I. Solid-State 2H NMR spectroscopy of retinal proteins in aligned membranes. Biochim. Biophys. Acta-Biomembr. 2007, 1768, 2979–3000. [Google Scholar] [CrossRef] [PubMed]
- Palings, I.; Pardoen, J.A.; VanDenBerg, E.M.M.; Winkel, C.; Lugtenburg, J.; Mathies, R.A. Assignment of fingerprint vibrations in the resonance Raman spectra of rhodopsin, isorhodopsin, and bathorhodopsin: Implications for chromophore structure and environment. Biochemistry 1987, 26, 2544–2556. [Google Scholar] [CrossRef] [PubMed]
- Chung, W.C.; Nanbu, S.; Ishida, T. QM/MM trajectory surface hopping approach to photoisomerization of rhodopsin and isorhodopsin: The origin of faster and more efficient isomerization for rhodopsin. J. Phys. Chem. B 2012, 116, 8009–8023. [Google Scholar] [CrossRef]
- Sugihara, M.; Buss, V.; Entel, P.; Hafner, J. The nature of the complex counterion of the chromophore in rhodopsin. J. Phys. Chem. B 2004, 108, 3673–3680. [Google Scholar] [CrossRef]
- Sekharan, S.; Sugihara, M.; Buss, V. Origin of spectral tuning in rhodopsin—It is not the binding pocket. Angew. Chem. Int. Ed. 2007, 46, 269–271. [Google Scholar] [CrossRef]
- Ernst, O.P.; Lodowski, D.T.; Elstner, M.; Hegemann, P.; Brown, L.S.; Kandori, H. Microbial and animal rhodopsins: Structures, functions, and molecular mechanisms. Chem. Rev. 2014, 114, 126–163. [Google Scholar] [CrossRef]
- Lesca, E.; Panneels, V.; Schertler, G.F.X. The role of water molecules in phototransduction of retinal proteins and G protein-coupled receptors. Faraday Discuss. 2018, 207, 27–37. [Google Scholar] [CrossRef]
- Nikolaev, D.M.; Shtyrov, A.A.; Mereshchenko, A.S.; Panov, M.S.; Tveryanovich, Y.S.; Ryazantsev, M.N. An assessment of water placement algorithms in quantum mechanics/molecular mechanics modeling: The case of rhodopsins’ first spectral absorption band maxima. Phys. Chem. Chem. Phys. 2020, 22, 18114–18123. [Google Scholar] [CrossRef]
- Siebert, F.; Mäntele, W.; Gerwert, K. Fourier-transform infrared spectroscopy applied to rhodopsin. The problem of the protonation state of the retinylidene Schiff base re-investigated. Eur. J. Biochem. 1983, 136, 119–127. [Google Scholar] [CrossRef] [PubMed]
- Verhoeven, M.A.; Creemers, A.F.L.; Bovee-Geurts, P.H.M.; DeGrip, W.J.; Lugtenburg, J.; de Groot, H.J.M. Ultra-high-field MAS NMR assay of a multispin labeled ligand bound to its G-protein receptor target in the natural membrane environment: Electronic structure of the retinylidene chromophore in rhodopsin. Biochemistry 2001, 40, 3282–3288. [Google Scholar] [CrossRef] [PubMed]
- Okada, T.; Fujiyoshi, Y.; Silow, M.; Navarro, J.; Landau, E.M.; Shichida, Y. Functional role of internal water molecules in rhodopsin revealed by X-ray crystallography. Proc. Natl. Acad. Sci. USA 2002, 99, 5982–5987. [Google Scholar] [CrossRef] [PubMed]
- Sakmar, T.P.; Franke, R.R.; Khorana, H.G. Glutamic acid-113 serves as the retinylidene Schiff base counterion in bovine rhodopsin. Proc. Natl. Acad. Sci. USA 1989, 86, 8309–8313. [Google Scholar] [CrossRef]
- Creemers, A.F.L.; Klaassen, C.H.W.; Bovee-Geurts, P.H.M.; Kelle, R.; Kragl, U.; Raap, J.; DeGrip, W.J.; Lugtenburg, J.; de Groot, H.J.M. 15N Solid state NMR evidence for a compex Schiff base counterion in the visual G-protein coupled receptor rhodopsin. Biochemistry 1999, 38, 7195–7199. [Google Scholar] [CrossRef]
- Deng, H.; Huang, L.W.; Callender, R.H.; Ebrey, T.G. Evidence for a bound water molecule next to the retinal Schiff base in bacteriorhodopsin and rhodopsin: A resonance raman study of the Schiff base hydrogen/deuterium exchange. Biophys. J. 1994, 66, 1129–1136. [Google Scholar] [CrossRef][Green Version]
- Nagata, T.; Terakita, A.; Kandori, H.; Kojima, D.; Shichida, Y.; Maeda, A. Water and peptide backbone structure in the active center of bovine rhodopsin. Biochemistry 1997, 36, 6164–6170. [Google Scholar] [CrossRef]
- Nagata, T.; Terakita, A.; Kandori, H.; Shichida, Y.; Maeda, A. The hydrogen-bonding network of water molecules and the peptide backbone in the region connecting Asp83, Gly120, and Glu113 in bovine rhodopsin. Biochemistry 1998, 37, 17216–17222. [Google Scholar] [CrossRef]
- Fujimoto, K.J. Electronic Couplings and Electrostatic Interactions Behind the Light Absorption of Retinal Proteins. Front. Mol. Biosci. 2021, 8, 752700. [Google Scholar] [CrossRef]
- Shtyrov, A.A.; Nikolaev, D.M.; Mironov, V.N.; Vasin, A.V.; Panov, M.S.; Tveryanovich, Y.S.; Ryazantsev, M.N. Simple Models to Study Spectral Properties of Microbial and Animal Rhodopsins: Evaluation of the Electrostatic Effect of Charged and Polar Residues on the First Absorption Band Maxima. Int. J. Mol. Sci. 2021, 22, 3029. [Google Scholar] [CrossRef]
- Honig, B.; Dinur, U.; Nakanishi, K.; Balogh-Nair, V.; Gawinowicz, M.A.; Arnaboldi, M.; Motto, M.G. An external point-charge model for wavelength regulation in visual pigments. J. Am. Chem. Soc. 1979, 101, 7084–7086. [Google Scholar] [CrossRef]
- Demoulin, B.; Maiuri, M.; Berbasova, T.; Geiger, J.H.; Borhan, B.; Garavelli, M.; Cerullo, G.; Rivalta, I. Control of Protonated Schiff Base Excited State Decay within Visual Protein Mimics: A Unified Model for Retinal Chromophores. Chem. Eur. J. 2021, 27, 16389–16400. [Google Scholar] [CrossRef] [PubMed]
- Agathangelou, D.; Roy, P.P.; Del Carmen Marín, M.; Ferré, N.; Olivucci, M.; Buckup, T.; Léonard, J.; Haacke, S. Sub-picosecond C=C bond photo-isomerization: Evidence for the role of excited state mixing. Compt. Rendus. Phys. 2021, 22, 1–28. [Google Scholar] [CrossRef]
- Curry, B.; Palings, I.; Broek, A.D.; Pardoen, J.A.; Lugtenburg, J.; Mathies, R.A. Vibrational Analysis of the Retinal Isomers. In Advances in Infrared and Raman Spectroscopy; Clark, R.J.H., Hester, R.E., Eds.; Wiley, Heyden: Chichester, UK, 1985; Volume 12, pp. 115–178. [Google Scholar]
- Birge, R.R.; Einterz, C.M.; Knapp, H.M.; Murray, L.P. The nature of the primary photochemical events in rhodopsin and isorhodopsin. Biophys. J. 1988, 53, 367–385. [Google Scholar] [CrossRef][Green Version]
- Cooper, A. Energetics of rhodopsin and isorhodopsin. FEBS Lett. 1979, 100, 382–384. [Google Scholar] [CrossRef]
- Makino, C.L.; Kraft, T.W.; Mathies, R.A.; Lugtenburg, J.; Miley, M.E.; van der Steen, R.; Baylor, D.A. Effects of modified chromophores on the spectral sensitivity of salamander, squirrel and macaque cones. J. Physiol. 1990, 424, 545–560. [Google Scholar] [CrossRef]
- Creemers, A.F.L.; Bovee-Geurts, P.H.M.; DeGrip, W.J.; Lugtenburg, J.; de Groot, H.J.M. Solid-state NMR analysis of ligand-receptor interactions reveals an induced misfit in the binding site of isorhodopsin. Biochemistry 2004, 43, 16011–16018. [Google Scholar] [CrossRef]
- Mirzadegan, T.; Liu, R.S.H. Probing the visual pigment rhodopsin and its analogs by molecular modeling analysis and computer graphics. Prog. Retin. Res. 1991, 11, 57–74. [Google Scholar] [CrossRef]
- Nakamichi, H.; Okada, T. X-ray crystallographic analysis of 9-cis-rhodopsin, a model analogue visual pigment. Photochem. Photobiol. 2007, 83, 232–235. [Google Scholar] [CrossRef]
- Bovee-Geurts, P.H.M.; Lugtenburg, J.; DeGrip, W.J. Coupled HOOP signature correlates with quantum yield of isorhodopsin and analog pigments. Biochim. Biophys. Acta—Bioenerg. 2017, 1858, 118–125. [Google Scholar] [CrossRef]
- Mathies, R.A.; Smith, S.O.; Palings, I. Determination of retinal chromophore structure in rhodopsins. In Resonance Raman Spectra of Polyenes and Aromatics; Spiro, T.G., Ed.; John Wiley & Sons: New York, NY, USA, 1987; Volume 2, pp. 59–108. [Google Scholar]
- Palings, I.; Van den Berg, E.M.M.; Lugtenburg, J.; Mathies, R.A. Complete assignment of the hydrogen out-of-plane wagging vibrations of bathorhodopsin: Chromophore structure and energy storage in the primary photoproduct of vision. Biochemistry 1989, 28, 1498–1507. [Google Scholar] [CrossRef] [PubMed]
- Eyring, G.; Curry, B.; Mathies, R.A.; Fransen, R.; Palings, I.; Lugtenburg, J. Interpretation of the resonance Raman spectrum of bathorhodopsin based on visual pigment analogues. Biochemistry 1980, 19, 2410–2418. [Google Scholar] [CrossRef] [PubMed]
- Mathies, R.A.; Freedman, T.B.; Stryer, L. Resonance Raman studies of conformation of retinal in rhodopsin and isorhodopsin. J. Mol. Biol. 1977, 109, 367–372. [Google Scholar] [CrossRef]
- Fahmy, K.; Siebert, F.; Grossjean, M.F.; Tavan, P. Photoisomerization in bacteriorhodopsin studied by FTIR, linear dichroism and photoselection experiments combined with quantum chemical theoretical-analysis. J. Mol. Struct. 1989, 214, 257–288. [Google Scholar] [CrossRef]
- Warshel, A.; Barboy, N. Energy storage and reaction pathways in the first step of the vision process. J. Am. Chem. Soc. 1982, 104, 1469–1476. [Google Scholar] [CrossRef]
- Dawadi, P.B.S.; Lugtenburg, J. Synthesis and use of stable isotope enriched retinals in the field of vitamin A. Molecules 2010, 15, 1825–1872. [Google Scholar] [CrossRef] [PubMed]
- Liu, R.S.H.; Asato, A.E. The binding site of opsin based on analog studies with isomeric, fluorinated, alkylated, and other modified retinals. In Chemistry and Biology of Synthetic Retinoids; Dawson, M.I., Okamura, W.H., Eds.; CRC Press, Inc.: Boca Raton, FL, USA, 1990; pp. 52–75. [Google Scholar]
- Lugtenburg, J.; Mathies, R.A.; Griffin, R.G.; Herzfeld, J. Structure and function of rhodopsins from solid state NMR and resonance Raman spectroscopy of isotopic retinal derivatives. Trends. Biochem. Sci. 1988, 13, 388–393. [Google Scholar] [CrossRef]
- DeGrip, W.J.; DeLange, F.; Klaassen, C.H.W.; Verdegem, P.J.E.; Wallace-Williams, S.E.; Creemers, A.F.L.; Bergo, V.B.; Bovee-Geurts, P.H.M.; Raap, J.; Rothschild, K.J.; et al. Photoactivation of rhodopsin: Interplay between protein and chromophore. In Rhodopsins and Phototransduction; Goode, J.A., Ed.; John Wiley & Sons, Ltd.: Chichester, UK, 1999; Volume 224, pp. 102–118. [Google Scholar]
- Creemers, A.F.L.; Kiihne, S.R.; Bovee-Geurts, P.H.M.; DeGrip, W.J.; Lugtenburg, J.; de Groot, H.J.M. 1H and 13C MAS NMR evidence for pronounced ligand-protein interactions involving the ionone ring of the retinylidene chromophore in rhodopsin. Proc. Natl. Acad. Sci. USA 2002, 99, 9101–9106. [Google Scholar] [CrossRef]
- Kiihne, S.R.; Creemers, A.F.L.; DeGrip, W.J.; Bovee-Geurts, P.H.M.; Lugtenburg, J.; de Groot, H.J.M. Selective interface detection: Mapping binding site contacts in membrane proteins by NMR spectroscopy. J. Am. Chem. Soc. 2005, 127, 5734–5735. [Google Scholar] [CrossRef]
- Brinkmann, A.; Sternberg, U.; Bovee-Geurts, P.H.M.; Fernández Fernández, I.; Lugtenburg, J.; Kentgens, A.P.M.; DeGrip, W.J. Insight into the chromophore of rhodopsin and its Meta-II photointermediate by 19F solid-state NMR and chemical shift tensor calculations. Phys. Chem. Chem. Phys. 2018, 20, 30174–30188. [Google Scholar] [CrossRef]
- Carravetta, M.; Edén, M.; Johannessen, O.G.; Luthman, H.; Verdegem, P.J.E.; Lugtenburg, J.; Sebald, A.; Levitt, M.H. Estimation of carbon-carbon bond lengths and medium-range internuclear distances by solid-state nuclear magnetic resonance. J. Am. Chem. Soc. 2001, 123, 10628–10638. [Google Scholar] [CrossRef] [PubMed]
- Peersen, O.B.; Smith, S.O. Rotational resonance NMR of biological membranes. Concepts Magn. Reson. 1993, 5, 303–317. [Google Scholar] [CrossRef]
- Spooner, P.J.R.; Sharples, J.M.; Verhoeven, M.A.; Lugtenburg, J.; Glaubitz, C.; Watts, A. Relative orientation between the β-ionone ring and the polyene chain for the chromophore of rhodopsin in native membranes. Biochemistry 2002, 41, 7549–7555. [Google Scholar] [CrossRef] [PubMed]
- Spooner, P.J.R.; Sharples, J.M.; Goodall, S.C.; Bovee-Geurts, P.H.M.; Verhoeven, M.A.; Lugtenburg, J.; Pistorius, A.M.A.; DeGrip, W.J.; Watts, A. The ring of the rhodopsin chromophore in a hydrophobic activation switch within the binding pocket. J. Mol. Biol. 2004, 343, 719–730. [Google Scholar] [CrossRef]
- Yoshizawa, T. Photophysiological functions of visual pigments. Adv. Biophys. 1984, 17, 5–67. [Google Scholar] [CrossRef]
- Nonaka, Y.; Hanai, S.; Katayama, K.; Imai, H.; Kandori, H. Unique Retinal Binding Pocket of Primate Blue-Sensitive Visual Pigment. Biochemistry 2020, 59, 2602–2607. [Google Scholar] [CrossRef]
- Fasick, J.I.; Lee, N.; Oprian, D.D. Spectral tuning in the human blue cone pigment. Biochemistry 1999, 38, 11593–11596. [Google Scholar] [CrossRef]
- Kropf, A.; Whittenberger, B.P.; Goff, S.P.; Waggoner, A.S. The spectral properties of some visual pigment analogs. Exp. Eye Res. 1973, 17, 591–606. [Google Scholar] [CrossRef]
- Derguini, F.; Nakanishi, K. Synthetic rhodopsin analogs. Photobiochem. Photobiophys. 1986, 13, 259–283. [Google Scholar]
- Álvarez, R.; Vaz, B.; Gronemeyer, H.; de Lera, A.R. Functions, therapeutic applications, and synthesis of retinoids and carotenoids. Chem. Rev. 2014, 114, 1–125. [Google Scholar] [CrossRef]
- Kahremany, S.; Sander, C.L.; Tochtrop, G.P.; Kubas, A.; Palczewski, K. Z-isomerization of retinoids through combination of monochromatic photoisomerization and metal catalysis. Org. Biomol. Chem. 2019, 17, 8125–8139. [Google Scholar] [CrossRef] [PubMed]
- DeLange, F.; Bovee-Geurts, P.H.M.; VanOostrum, J.; Portier, M.D.; Verdegem, P.J.E.; Lugtenburg, J.; DeGrip, W.J. An additional methyl group at the 10-position of retinal dramatically slows down the kinetics of the rhodopsin photocascade. Biochemistry 1998, 37, 1411–1420. [Google Scholar] [PubMed]
- Trehan, A.; Liu, R.S.H.; Shichida, Y.; Imamoto, Y.; Nakamura, K.; Yoshizawa, T. On retention of chromophore configuration of rhodopsin isomers derived from three dicis retinal isomers. Bioorg. Chem. 1990, 18, 30–40. [Google Scholar]
- DeGrip, W.J.; VanOostrum, J.; Bovee-Geurts, P.H.M.; van der Steen, R.; VanAmsterdam, L.J.P.; Groesbeek, M.; Lugtenburg, J. 10,20-Methanorhodopsins: (7E,9E,13E)-10,20-methanorhodopsin and (7E,9Z,13Z)-10,20-methanorhodopsin-11-cis-Locked rhodopsin analog pigments with unusual thermal and photo-stability. Eur. J. Biochem. 1990, 191, 211–220. [Google Scholar]
- Wang, Y.-J.; Bovee-Geurts, P.H.M.; Lugtenburg, J.; DeGrip, W.J. Alpha-retinals as rhodopsin chromophores—Preference for the 9-Z configuration and partial agonist activity. Photochem. Photobiol. 2008, 84, 889–894. [Google Scholar] [CrossRef] [PubMed]
- DeGrip, W.J.; Bovee-Geurts, P.H.M.; Van der Hoef, I.; Lugtenburg, J. 7,8-Dihydro-retinals outperform the native retinals in conferring photosensitivity to visual opsin. J. Am. Chem. Soc. 2007, 129, 13265–13269. [Google Scholar] [CrossRef]
- Zhu, Y.; Liu, R.S.H. Divergent pathways in photobleaching of 7,9-dicis-rhodopsin and 9,11-dicis-12-fluororhodopsin: One-photon-two-bond and one-photon- one-bond isomerization. Biochemistry 1993, 32, 10233–10238. [Google Scholar] [CrossRef]
- Shichida, Y.; Nakamura, K.; Yoshizawa, T.; Trehan, A.; Denny, M.; Liu, R.S.H. 9,13-dicis-Rhodopsin and Its One-Photon-One-Double-Bond Isomerization. Biochemistry 1988, 27, 6495–6499. [Google Scholar] [CrossRef]
- Liu, R.S.H.; Mirzadegan, T. The shape of a three-dimensional binding-site of rhodopsin based on molecular modeling analyses of isomeric and other visual pigment analogs. Bioorganic studies of visual pigments. 11. J. Am. Chem. Soc. 1988, 110, 8617–8623. [Google Scholar] [CrossRef]
- Groenendijk, G.W.T.; DeGrip, W.J.; Daemen, F.J.M. Identification and characterization of syn-isomers and anti-isomers of retinaloximes. Anal. Biochem. 1979, 99, 304–310. [Google Scholar] [CrossRef]
- Groenendijk, G.W.T.; DeGrip, W.J.; Daemen, F.J.M. Quantitative determination of retinals with complete retention of their geometric configuration. Biochim. Biophys. Acta 1980, 617, 430–438. [Google Scholar] [CrossRef]
- Kini, A.; Matsumoto, H.; Liu, R.S.H. 9,11-Di-cis-retinal and 9,11-Di-cis-rhodopsin. Bioorg. Chem. 1980, 9, 406–410. [Google Scholar] [CrossRef]
- Zhu, Y.; Ganapathy, S.; Trehan, A.; Asato, A.E.; Liu, R.S.H. FT-IR spectra of all sixteen isomers of retinal, their isolation, and other spectroscopic properties. Tetrahedron 1992, 48, 10061–10074. [Google Scholar] [CrossRef]
- Groenendijk, G.W.T.; Jacobs, C.W.M.; Bonting, S.L.; Daemen, F.J.M. Dark isomerization of retinals in the presence of phosphatidylethanolamine. Eur. J. Biochem. 1980, 106, 119–128. [Google Scholar] [CrossRef]
- Braiman, M.S.; Rothschild, K.J. Fourier transform infrared techniques for probing membrane protein structure. Annu. Rev. Biophys. Biophys. Chem. 1988, 17, 541–570. [Google Scholar] [CrossRef]
- Kochendoerfer, G.G.; Lin, S.W.; Sakmar, T.P.; Mathies, R.A. How color visual pigments are tuned. Trends. Biochem. Sci. 1999, 24, 300–305. [Google Scholar] [CrossRef]
- Hirano, T.; Fujioka, N.; Imai, H.; Kandori, H.; Wada, A.; Ito, M.; Shichida, Y. Assignment of the vibrational modes of the chromophores of iodopsin and bathoiodopsin: Low-temperature Fourier transform infrared spectroscopy of 13C and 2H-labeled iodopsins. Biochemistry 2006, 45, 1285–1294. [Google Scholar] [CrossRef]
- Lin, S.W.; Imamoto, Y.; Fukada, Y.; Shichida, Y.; Yoshizawa, T.; Mathies, R.A. What makes red visual pigments red? A resonance Raman microprobe study of retinal chromophore structure in iodopsin. Biochemistry 1994, 33, 2151–2160. [Google Scholar] [CrossRef]
- Pande, C.; Pande, A.; Yue, K.T.; Callender, R.H.; Ebrey, T.G.; Tsuda, M. Resonance Raman spectroscopy of octopus rhodopsin and its photoproducts. Biochemistry 1987, 26, 4941–4947. [Google Scholar] [CrossRef]
- Bagley, K.A.; Eisenstein, L.; Ebrey, T.G.; Tsuda, M. A comparative study of the infrared difference spectra for octopus and bovine rhodopsins and their bathorhodopsin photointermediates. Biochemistry 1989, 28, 3366–3373. [Google Scholar] [CrossRef]
- Bovee-Geurts, P.H.M.; Fernández Fernández, I.; Liu, R.S.H.; Mathies, R.A.; Lugtenburg, J.; DeGrip, W.J. Fluoro derivatives of retinal illuminate the decisive role of the C12-H element in photoisomerization and rhodopsin activation. J. Am. Chem. Soc. 2009, 131, 17933–17942. [Google Scholar] [CrossRef] [PubMed]
- Oseroff, A.R.; Callender, R.H. Resonance Raman spectroscopy of rhodopsin in retinal disk membranes. Biochemistry 1974, 13, 4243–4248. [Google Scholar] [CrossRef] [PubMed]
- Popp, A.; Ujj, L.; Atkinson, G.H. Vibrational spectra of room-temperature rhodopsin: Concentration dependence in picosecond resonance coherent anti-stokes Raman scattering. Biophys. Chem. 1995, 56, 129–135. [Google Scholar] [CrossRef]
- Jäger, F.; Ujj, L.; Atkinson, G.H. Vibrational spectrum of bathorhodopsin in the room-temperature rhodopsin photoreaction. J. Am. Chem. Soc. 1997, 119, 12610–12618. [Google Scholar] [CrossRef]
- Lin, S.W.; Groesbeek, M.; Van der Hoef, I.; Verdegem, P.J.E.; Lugtenburg, J.; Mathies, R.A. Vibrational assignment of torsional normal modes of rhodopsin: Probing excited-state isomerization dynamics along the reactive C11=C12 torsion coordinate. J. Phys. Chem. B 1998, 102, 2787–2806. [Google Scholar] [CrossRef]
- DeGrip, W.J.; Bovee-Geurts, P.H.M.; Wang, Y.-J.; Verhoeven, M.A.; Lugtenburg, J. Cyclopropyl and isopropyl derivatives of 11-cis and 9-cis retinals at C-9 and C-13: Subtle steric differences with major effects on ligand efficacy in rhodopsin. J. Nat. Prod. 2011, 74, 383–390. [Google Scholar] [CrossRef]
- Barry, B.; Mathies, R.A.; Pardoen, J.A.; Lugtenburg, J. Raman microscope and quantum yield studies on the primary photochemistry of A2-visual pigments. Biophys. J. 1987, 52, 603–610. [Google Scholar] [CrossRef]
- Bridges, C.D.B. The rhodopsin-porphyropsin visual system. In Photochemistry of Vision; Dartnall, H.J.A., Ed.; Springer: Berlin/Heidelberg, Germany, 1972; Volume VII/1, pp. 417–480. [Google Scholar]
- Imai, H.; Hirano, T.; Terakita, A.; Shichida, Y.; Muthyala, R.S.; Chen, R.-L.; Colmenares, L.U.; Liu, R.S.H. Probing for the threshold energy for visual transduction: Red-shifted visual pigment analogs from 3-methoxy-3-dehydroretinal and related compounds. Photochem. Photobiol. 1999, 70, 111–115. [Google Scholar] [CrossRef]
- Polli, D.; Rivalta, I.; Nenov, A.; Weingart, O.; Garavelli, M.; Cerullo, G. Tracking the primary photoconversion events in rhodopsins by ultrafast optical spectroscopy. Photochem. Photobiol. Sci. 2015, 14, 213–228. [Google Scholar] [CrossRef]
- Panneels, V.; Wu, W.T.; Tsai, C.-J.; Nogly, P.; Rheinberger, J.; Jaeger, K.; Cicchetti, G.; Gati, C.; Kick, L.M.; Sala, L.; et al. Time-resolved structural studies with serial crystallography: A new light on retinal proteins. Struct. Dyn. 2015, 2, 041718. [Google Scholar] [CrossRef]
- Yoshizawa, T.; Kandori, H. Primary photochemical events in the rhodopsin molecule. Prog. Retin. Res. 1991, 11, 33–55. [Google Scholar] [CrossRef]
- Farrens, D.L. What site-directed labeling studies tell us about the mechanism of rhodopsin activation and G-protein binding. Photochem. Photobiol. Sci. 2010, 9, 1466–1474. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Smith, S.O. Structure and activation of the visual pigment rhodopsin. Annu. Rev. Biophys. 2010, 39, 309–328. [Google Scholar] [CrossRef] [PubMed]
- Brown, L.S.; Ladizhansky, V. Membrane proteins in their native habitat as seen by solid-state NMR spectroscopy. Protein Sci. 2015, 24, 1333–1346. [Google Scholar] [CrossRef] [PubMed]
- Mathies, R.A.; Lugtenburg, J. The primary photoreaction of rhodopsin. In Molecular Mechanisms in Visual Transduction; Stavenga, D.G., DeGrip, W.J., Pugh, E.N., Jr., Eds.; Elsevier Science Pub.: Amsterdam, The Netherlands, 2000; Volume 3, pp. 55–90. [Google Scholar]
- DeGrip, W.J.; Rothschild, K.J. Structure and mechanism of vertebrate visual pigments. In Molecular Mechanisms in Visual Transduction; Stavenga, D.G., DeGrip, W.J., Pugh, E.N., Jr., Eds.; Elsevier Science Pub.: Amsterdam, The Netherlands, 2000; Volume 3, pp. 1–54. [Google Scholar]
- Weingart, O.; Schapiro, I.; Buss, V. Photochemistry of visual pigment chromophore models by ab initio molecular dynamics. J. Phys. Chem. B 2007, 111, 3782–3788. [Google Scholar] [CrossRef] [PubMed]
- Yoshizawa, T.; Wald, G. Pre-lumirhodopsin and the bleaching of visual pigments. Nature 1963, 197, 1279–1286. [Google Scholar] [CrossRef] [PubMed]
- Yoshizawa, T. The behaviour of visual pigments at low temperatures. In Photochemistry of Vision; Dartnall, H.J.A., Ed.; Springer: Berlin/Heidelberg, Germany, 1972; Volume VII/I, pp. 146–179. [Google Scholar]
- Hurley, J.B.; Ebrey, T.G.; Honig, B.; Ottolenghi, M. Temperature and wavelength effects on photochemistry of rhodopsin, isorhodopsin, bacteriorhodopsin and their photoproducts. Nature 1977, 270, 540–542. [Google Scholar] [CrossRef]
- Kim, J.E.; Tauber, M.J.; Mathies, R.A. Wavelength dependent cis-trans isomerization in vision. Biochemistry 2001, 40, 13774–13778. [Google Scholar] [CrossRef]
- Kim, J.E.; Tauber, M.J.; Mathies, R.A. Analysis of the mode-specific excited-state energy distribution and wavelength-dependent photoreaction quantum yield in rhodopsin. Biophys. J. 2003, 84, 2492–2501. [Google Scholar] [CrossRef]
- Hubbard, R.; Kropf, A. The action of light on rhodopsin. Proc. Natl. Acad. Sci. USA 1958, 44, 130–139. [Google Scholar] [CrossRef]
- Kropf, A.; Hubbard, R. The photoisomerization of retinal. Photochem. Photobiol. 1970, 12, 249–260. [Google Scholar] [CrossRef] [PubMed]
- Becker, R.S.; Freedman, K. A comprehensive investigation of the mechanism and photophysics of isomerization of a protonated and unprotonated Schiff-base of 11-cis-retinal. J. Am. Chem. Soc. 1985, 107, 1477–1485. [Google Scholar] [CrossRef]
- Becker, R.S.; Freedman, K.; Hutchinson, J.A.; Noe, L.J. Kinetic study of the photoisomerization of a protonated Schiff-base of 11-cis-retinal over the picosecond-to-second time regimes. J. Am. Chem. Soc. 1985, 107, 3942–3944. [Google Scholar] [CrossRef]
- Lewis, J.W.; Hug, S.J.; Wallace-Williams, S.E.; Kliger, D.S. Direct evidence for an equilibrium between early photolysis intermediates of rhodopsin. J. Am. Chem. Soc. 1990, 112, 6711–6712. [Google Scholar] [CrossRef]
- Birge, R.R. Nature of the primary photochemical events in rhodopsin and bacteriorhodopsin. Biochim. Biophys. Acta 1990, 1016, 293–327. [Google Scholar] [CrossRef]
- Yang, X.; Manathunga, M.; Gozem, S.; Léonard, J.; Andruniów, T.; Olivucci, M. Quantum-classical simulations of rhodopsin reveal excited-state population splitting and its effects on quantum efficiency. Nat. Chem. 2022, 14, 441–449. [Google Scholar] [CrossRef]
- Schnedermann, C.; Yang, X.; Liebel, M.; Spillane, K.M.; Lugtenburg, J.; Fernández, I.; Valentini, A.; Schapiro, I.; Olivucci, M.; Kukura, P.; et al. Evidence for a vibrational phase-dependent isotope effect on the photochemistry of vision. Nat. Chem. 2018, 10, 449–455. [Google Scholar] [CrossRef]
- Johnson, P.J.M.; Halpin, A.; Morizumi, T.; Prokhorenko, V.I.; Ernst, O.P.; Miller, R.J.D. Local vibrational coherences drive the primary photochemistry of vision. Nat. Chem. 2015, 7, 980–986. [Google Scholar] [CrossRef]
- Schick, G.A.; Cooper, T.M.; Holloway, R.A.; Murray, L.P.; Birge, R.R. Energy-storage in the primary photochemical events of rhodopsin and isorhodopsin. Biochemistry 1987, 26, 2556–2562. [Google Scholar] [CrossRef]
- Cooper, A. Energy uptake in the first step of visual excitation. Nature 1979, 282, 531–533. [Google Scholar] [CrossRef]
- Birge, R.R.; Vought, B.W. Energetics of rhodopsin photobleaching: Photocalorimetric studies of energy storage in early and later intermediates. Methods Enzym. 2000, 315, 143–163. [Google Scholar]
- Nakamichi, H.; Okada, T. Local peptide movement in the photoreaction intermediate of rhodopsin. Proc. Natl. Acad. Sci. USA 2006, 103, 12729–12734. [Google Scholar] [CrossRef] [PubMed]
- Lewis, J.W.; Jäger, S.; Kliger, D.S. Absorbance changes by aromatic amino acid side chains in early rhodopsin photointermediates. Photochem. Photobiol. 1997, 66, 741–746. [Google Scholar] [CrossRef] [PubMed]
- Doukas, A.G.; Aton, B.; Callender, R.H.; Ebrey, T.G. Resonance Raman studies of bovine metarhodopsin I and metarhodopsin II. Biochemistry 1978, 17, 2430–2435. [Google Scholar] [CrossRef]
- Furutani, Y.; Kandori, H.; Shichida, Y. Structural changes in lumirhodopsin and metarhodopsin I studied by their photoreactions at 77 K. Biochemistry 2003, 42, 8494–8500. [Google Scholar] [CrossRef]
- Schertler, G.F.X. Structure of rhodopsin and the metarhodopsin I photointermediate. Curr. Opin. Struct. Biol. 2005, 15, 408–415. [Google Scholar] [CrossRef]
- Vogel, R.; Sakmar, T.P.; Sheves, M.; Siebert, F. Coupling of protonation switches during rhodopsin activation. Photochem. Photobiol. 2007, 83, 286–292. [Google Scholar] [CrossRef]
- Pope, A.L.; Sanchez-Reyes, O.B.; South, K.; Zaitseva, E.; Ziliox, M.; Vogel, R.; Reeves, P.J.; Smith, S.O. A Conserved Proline Hinge Mediates Helix Dynamics and Activation of Rhodopsin. Structure 2020, 28, 1004–1013. [Google Scholar] [CrossRef]
- Vogel, R.; Mahalingam, M.; Lüdeke, S.; Huber, T.; Siebert, F.; Sakmar, T.P. Functional role of the “Ionic Lock”—An interhelical hydrogen-bond network in family a heptahelical receptors. J. Mol. Biol. 2008, 380, 648–655. [Google Scholar] [CrossRef]
- Jäger, F.; Fahmy, K.; Sakmar, T.P.; Siebert, F. Identification of glutamic acid 113 as the Schiff base proton acceptor in the metarhodopsin II photointermediate of rhodopsin. Biochemistry 1994, 33, 10878–10882. [Google Scholar] [CrossRef]
- Maeda, A. Internal water molecules as mobile polar groups for light-induced proton translocation in bacteriorhodopsin and rhodopsin as studied by difference FTIR spectroscopy. Biochemistry 2001, 66, 1256–1268. [Google Scholar] [PubMed]
- Furutani, Y.; Shichida, Y.; Kandori, H. Structural changes of water molecules during the photoactivation processes in bovine rhodopsin. Biochemistry 2003, 42, 9619–9625. [Google Scholar] [CrossRef] [PubMed]
- Choe, H.-W.; Kim, Y.J.; Park, J.H.; Morizumi, T.; Pai, E.F.; Krauß, N.; Hofmann, K.P.; Scheerer, P.; Ernst, O.P. Crystal structure of metarhodopsin II. Nature 2011, 471, 651–655. [Google Scholar] [CrossRef] [PubMed]
- Jastrzebska, B.; Palczewski, K.; Golczak, M. Role of bulk water in hydrolysis of the rhodopsin chromophore. J. Biol. Chem. 2011, 286, 18930–18937. [Google Scholar] [CrossRef] [PubMed]
- Ranganathan, R.; Stevens, C.F. Arrestin binding determines the rate of inactivation of the G protein-coupled receptor rhodopsin in vivo. Cell 1995, 81, 841–848. [Google Scholar] [CrossRef]
- Liu, R.S.H.; Crescitelli, F.; Denny, M.; Matsumoto, H.; Asato, A.E. Photosensitivity of 10-substituted visual pigment analogues: Detection of a specific secondary opsin-retinal interaction. Biochemistry 1986, 25, 7026–7030. [Google Scholar] [CrossRef]
- Fan, J.; Woodruff, M.L.; Cilluffo, M.C.; Crouch, R.K.; Fain, G.L. Opsin activation of transduction in the rods of dark-reared Rpe65 knockout mice. J. Physiol. 2005, 568, 83–95. [Google Scholar] [CrossRef]
- Concistrè, M.; Gansmüller, A.; McLean, N.; Johannessen, O.G.; Marín-Montesinos, I.; Bovee-Geurts, P.H.M.; Verdegem, P.J.E.; Lugtenburg, J.; Brown, R.C.D.; DeGrip, W.J.; et al. Double-quantum 13C nuclear magnetic resonance of bathorhodopsin, the first photointermediate in mammalian vision. J. Am. Chem. Soc. 2008, 130, 10490–10491. [Google Scholar] [CrossRef]
- Nakamichi, H.; Okada, T. Crystallographic analysis of primary visual photochemistry. Angew. Chem. Int. Ed. 2006, 45, 4270–4273. [Google Scholar] [CrossRef]
- Kukura, P.; McCamant, D.W.; Yoon, S.; Wandschneider, D.B.; Mathies, R.A. Structural observation of the primary isomerization in vision with femtosecond-stimulated Raman. Science 2005, 310, 1006–1009. [Google Scholar] [CrossRef]
- DeLange, F.; Bovee-Geurts, P.H.M.; Pistorius, A.M.A.; Rothschild, K.J.; DeGrip, W.J. Probing intramolecular orientations in rhodopsin and metarhodopsin II by polarized infrared difference spectroscopy. Biochemistry 1999, 38, 13200–13209. [Google Scholar] [CrossRef] [PubMed]
- Popp, A.; Ujj, L.; Atkinson, G.H. Bathorhodopsin structure in the room-temperature rhodopsin photosequence: Picosecond time-resolved coherent anti-Stokes Raman scattering. Proc. Natl. Acad. Sci. USA 1996, 93, 372–376. [Google Scholar] [CrossRef] [PubMed]
- Mao, B.; Ebrey, T.G.; Crouch, R. Bathoproducts of rhodopsin, isorhodopsin-i, and isorhodopsin-ii. Biophys. J. 1980, 29, 247–256. [Google Scholar] [CrossRef]
- Lewis, J.W.; Liang, J.; Ebrey, T.G.; Sheves, M.; Livnah, N.; Kuwata, O.; Jäger, S.; Kliger, D.S. Early photolysis intermediates of gecko and bovine artificial visual pigments. Biochemistry 1997, 36, 14593–14600. [Google Scholar] [CrossRef]
- Polli, D.; Weingart, O.; Brida, D.; Poli, E.; Maiuri, M.; Spillane, K.M.; Bottoni, A.; Kukura, P.; Mathies, R.A.; Cerullo, G.; et al. Wavepacket splitting and two-pathway deactivation in the photoexcited visual pigment isorhodopsin. Angew. Chem. Int. Ed. 2014, 53, 2504–2507. [Google Scholar] [CrossRef]
- Dartnall, H.J.A. The photosensitivities of visual pigments in presence of hydroxylamine. Vis. Res. 1968, 8, 339–358. [Google Scholar] [CrossRef]
- Schoenlein, R.W.; Peteanu, L.A.; Mathies, R.A.; Shank, C.V. The first step in vision: Femtosecond isomerization of rhodopsin. Science 1991, 254, 412–415. [Google Scholar] [CrossRef]
- Wang, Q.; Schoenlein, R.W.; Peteanu, L.A.; Mathies, R.A.; Shank, C.V. Vibrationally coherent photochemistry in the femtosecond primary event of vision. Science 1994, 266, 422–424. [Google Scholar] [CrossRef]
- McCamant, D.W.; Kukura, P.; Mathies, R.A. Femtosecond stimulated Raman study of excited-state evolution in bacteriorhodopsin. J. Phys. Chem. B 2005, 109, 10449–10457. [Google Scholar] [CrossRef]
- Kim, J.E.; McCamant, D.W.; Zhu, L.Y.; Mathies, R.A. Resonance Raman structural evidence that the cis-to-trans isomerization in rhodopsin occurs in femtoseconds. J. Phys. Chem. B 2001, 105, 1240–1249. [Google Scholar] [CrossRef]
- Peteanu, L.A.; Schoenlein, R.W.; Wang, Q.; Mathies, R.A.; Shank, C.V. The first step in vision occurs in femtoseconds: Complete blue and red spectral studies. Proc. Natl. Acad. Sci. USA 1993, 90, 11762–11766. [Google Scholar] [CrossRef] [PubMed]
- Weingart, O.; Altoè, P.; Stenta, M.; Bottoni, A.; Orlandi, G.; Garavelli, M. Product formation in rhodopsin by fast hydrogen motions. Phys. Chem. Chem. Phys. 2011, 13, 3645–3648. [Google Scholar] [CrossRef] [PubMed]
- Schapiro, I.; Ryazantsev, M.N.; Frutos, L.M.; Ferré, N.; Lindh, R.; Olivucci, M. The ultrafast photoisomerizations of rhodopsin and bathorhodopsin are modulated by bond length alternation and HOOP driven electronic effects. J. Am. Chem. Soc. 2011, 133, 3354–3364. [Google Scholar] [CrossRef] [PubMed]
- Strambi, A.; Coto, P.B.; Frutos, L.M.; Ferré, N.; Olivucci, M. Relationship between the excited state relaxation paths of rhodopsin and isorhodopsin. J. Am. Chem. Soc. 2008, 130, 3382–3388. [Google Scholar] [CrossRef] [PubMed]
- Polli, D.; Altoè, P.; Weingart, O.; Spillane, K.M.; Manzoni, C.; Brida, D.; Tomasello, G.; Orlandi, G.; Kukura, P.; Mathies, R.A.; et al. Conical intersection dynamics of the primary photoisomerization event in vision. Nature 2010, 467, 440–443. [Google Scholar] [CrossRef]
- Weingart, O. The twisted C11=C12 bond of the rhodopsin chromophore—A photochemical hot spot. J. Am. Chem. Soc. 2007, 129, 10618–10619. [Google Scholar] [CrossRef]
- Gozem, S.; Luk, H.L.; Schapiro, I.; Olivucci, M. Theory and Simulation of the Ultrafast Double-Bond lsomerization of Biological Chromophores. Chem. Rev. 2017, 117, 13502–13565. [Google Scholar] [CrossRef]
- Weingart, O.; Garavelli, M. Modelling vibrational coherence in the primary rhodopsin photoproduct. J. Chem. Phys. 2012, 137, 22A523. [Google Scholar] [CrossRef]
- Verhoeven, M.A.; Bovee-Geurts, P.H.M.; de Groot, H.J.M.; Lugtenburg, J.; DeGrip, W.J. Methyl substituents at the 11- or 12-position of retinal profoundly and differentially affect photochemistry and signalling activity of rhodopsin. J. Mol. Biol. 2006, 363, 98–113. [Google Scholar] [CrossRef]
- Nogly, P.; Weinert, T.; James, D.; Carbajo, S.; Ozerov, D.; Furrer, A.; Gashi, D.; Borin, V.; Skopintsev, P.; Jaeger, K.; et al. Retinal isomerization in bacteriorhodopsin captured by a femtosecond X-ray laser. Science 2018, 361, 145–151. [Google Scholar] [CrossRef]
- Skopintsev, P.; Ehrenberg, D.; Weinert, T.; James, D.; Kar, R.K.; Johnson, P.J.M.; Ozerov, D.; Furrer, A.; Martiel, I.; Dworkowski, F.; et al. Femtosecond-to-millisecond structural changes in a light-driven sodium pump. Nature 2020, 583, 314–322. [Google Scholar] [CrossRef] [PubMed]
- Oda, K.; Nomura, T.; Nakane, T.; Yamashita, K.; Inoue, K.; Ito, S.; Vierock, J.; Hirata, K.; Maturana, A.D.; Katayama, K.; et al. Time-resolved serial femtosecond crystallography reveals early structural changes in channelrhodopsin. eLife 2021, 10, 62389. [Google Scholar] [CrossRef] [PubMed]
- Liu, R.S.H.; Asato, A.E. Photochemistry of polyenes 22. The primary process of vision and the structure of bathorhodopsin: A mechanism for photoisomerization of polyenes. Proc. Natl. Acad. Sci. USA 1985, 82, 259–263. [Google Scholar] [CrossRef]
- Schoenlein, R.W.; Peteanu, L.A.; Wang, Q.; Mathies, R.A.; Shank, C.V. Femtosecond Dynamics of Cis-Trans Isomerization in a Visual Pigment Analog: Isorhodopsin. J. Phys. Chem. 1993, 97, 12087–12092. [Google Scholar] [CrossRef]
- Albeck, A.; Friedman, N.; Ottolenghi, M.; Sheves, M.; Einterz, C.M.; Hug, S.J.; Lewis, J.W.; Kliger, D.S. Photolysis intermediates of the artificial visual pigment cis-5,6-dihydro-isorhodopsin. Biophys. J. 1989, 55, 233–241. [Google Scholar] [CrossRef]
- Muto, O.; Tokunaga, F.; Yoshizawa, T.; Kamat, V.; Blatchly, H.A.; Balogh-Nair, V.; Nakanishi, K. Photochemical reaction of 7,8-dihydrorhodopsin at low temperatures. Biochim. Biophys. Acta-Lipids Lipid Metab. 1984, 766, 597–602. [Google Scholar] [CrossRef]
- Koutalos, Y.; Ebrey, T.G.; Tsuda, M.; Odashima, K.; Lien, T.; Park, M.H.; Shimizu, N.; Derguini, F.; Nakanishi, K.; Gilson, H.R.; et al. Regeneration of bovine and octopus opsins in situ with natural and artificial retinals. Biochemistry 1989, 28, 2732–2739. [Google Scholar] [CrossRef]
- Asato, A.E.; Denny, M.; Matsumoto, H.; Mirzadegan, T.; Ripka, W.C.; Crescitelli, F.; Liu, R.S.H. Study of the shape of the binding site of bovine opsin using 10-substituted retinal isomers. Biochemistry 1986, 25, 7021–7026. [Google Scholar] [CrossRef]
- Wang, Q.; Kochendoerfer, G.G.; Schoenlein, R.W.; Verdegem, P.J.E.; Lugtenburg, J.; Mathies, R.A.; Shank, C.V. Femtosecond spectroscopy of a 13-demethylrhodopsin visual pigment analogue: The role of non-bonded interactions in the isomerization process. J. Phys. Chem. B 1996, 100, 17388–17394. [Google Scholar] [CrossRef]
- Shichida, Y.; Kropf, A.; Yoshizawa, T. Photochemical reactions of 13-demethyl visual pigment analogues at low temperatures. Biochemistry 1981, 20, 1962–1968. [Google Scholar] [CrossRef]
- Nelson, R.; Deriel, J.K.; Kropf, A. 13-Desmethyl rhodopsin and 13-desmethyl isorhodopsin—Visual pigment analogues. Proc. Natl. Acad. Sci. USA 1970, 66, 531–538. [Google Scholar] [CrossRef] [PubMed]
- Blatz, P.E.; Lin, M.; Balasubramaniyan, P.; Balasubramaniyan, V.; Dewhurst, P.B. A new series of synthetic visual pigments from cattle opsin and homologs of retinal. J. Am. Chem. Soc. 1969, 91, 5930–5931. [Google Scholar] [CrossRef] [PubMed]
- Colmenares, L.U.; Asato, A.E.; Denny, M.; Mead, D.; Zingoni, J.P.; Liu, R.S.H. NMR studies of fluorinated visual pigment analogs. Biochem. Biophys. Res. Commun. 1991, 179, 1337–1343. [Google Scholar] [CrossRef]
- Asato, A.E.; Matsumoto, H.; Denny, M.; Liu, R.S.H. Fluorinated rhodopsin analogs from 10-fluororetinal and 14-fluororetinal. J. Am. Chem. Soc. 1978, 100, 5957–5960. [Google Scholar] [CrossRef]
- de Grip, W.J. (Leiden University, Leiden, The Netherlands); Bovee-Geurts, P.H.M. (Radboudumc, Nijmegen, The Netherlands); Wada, A. (Kobe Pharmaceutical University, Kobe, Japan); Lugtenburg, J. (Leiden University, Leiden, The Netherlands). Photosensitivity and Spectral Properties of 11-cis and 9-cis 3,4-Dehydrorhodopsin. Personal Communication. 2022. [Google Scholar]
- Azuma, M.; Azuma, K.; Kito, Y. Circular dichroism of visual pigment analogues containing 3-dehydroretinal and 5,6-epoxy-3-dehydroretinal as the chromophore. Biochim. Biophys. Acta 1973, 295, 520–527. [Google Scholar] [CrossRef]
- Chung, W.C.; Nanbu, S.; Ishida, T. Nonadiabatic ab initio dynamics of a model protonated Schiff base of 9-cis retinal. J. Phys. Chem. A 2010, 114, 8190–8201. [Google Scholar] [CrossRef]
- Popp, A.; Wolperdinger, M.; Hampp, N.; Bräuchle, C.; Oesterhelt, D. Photochemical conversion of the O-intermediate to 9-cis-retinal- containing products in bacteriorhodopsin films. Biophys. J. 1993, 65, 1449–1459. [Google Scholar] [CrossRef]
- Mei, G.X.; Cavini, C.M.; Mamaeva, N.; Wang, P.; DeGrip, W.J.; Rothschild, K.J. Optical switching between long-lived states of opsin transmembrane voltage sensors. Photochem. Photobiol. 2021, 97, 1001–1015. [Google Scholar] [CrossRef]
- Mori, A.; Yagasaki, J.; Homma, M.; Reissig, L.; Sudo, Y. Investigation of the chromophore binding cavity in the 11-cis acceptable microbial rhodopsin MR. Chem. Phys. 2013, 419, 23–29. [Google Scholar] [CrossRef]
- DeGrip, W.J.; Daemen, F.J.M.; Bonting, S.L. Isolation and purification of bovine rhodopsin. Methods Enzym. 1980, 67, 301–320. [Google Scholar]
- Yoshizawa, T.; Wald, G. Photochemistry of iodopsin. Nature 1967, 214, 566–571. [Google Scholar] [CrossRef] [PubMed]
- Okano, T.; Fukada, Y.; Artamonov, I.D.; Yoshizawa, T. Purification of cone visual pigments from chicken retina. Biochemistry 1989, 28, 8848–8856. [Google Scholar] [CrossRef] [PubMed]
- Kropf, A. A New Detergent for the Study of Visual Pigments. Vis. Res. 1982, 22, 495–497. [Google Scholar] [CrossRef]
- Stubbs, G.W.; Smith, H.G.; Litman, B.J. Alkyl glucosides as effective solubilizing agents for bovine rhodopsin—A comparison with several commonly used detergents. Biochim. Biophys. Acta 1976, 426, 46–56. [Google Scholar] [CrossRef]
- DeGrip, W.J.; Bovee-Geurts, P.H.M. Synthesis and properties of alkylglucosides with mild detergent action: Improved synthesis and purification of β-1-octyl, -nonyl- and -decyl-glucose. Synthesis of β-1-undecylglucose and β-1-dodecylmaltose. Chem. Phys. Lipids 1979, 23, 321–335. [Google Scholar] [CrossRef]
- Oprian, D.D.; Molday, R.S.; Kaufman, R.J.; Khorana, H.G. Expression of a synthetic bovine rhodopsin gene in monkey kidney cells. Proc. Natl. Acad. Sci. USA 1987, 84, 8874–8878. [Google Scholar] [CrossRef]
- Nathans, J.; Weitz, C.J.; Agarwal, N.; Nir, I.; Papermaster, D.S. Production of bovine rhodopsin by mammalian cell lines expressing cloned c-DNA: Spectrophotometry and subcellular localization. Vis. Res. 1989, 29, 907–914. [Google Scholar] [CrossRef]
- Reeves, P.J.; Callewaert, N.; Contreras, R.; Khorana, H.G. Structure and function in rhodopsin: High-level expression of rhodopsin with restricted and homogeneous N-glycosylation by a tetracycline-inducible N-acetylglucosaminyltransferase I-negative HEK293S stable mammalian cell line. Proc. Natl. Acad. Sci. USA 2002, 99, 13419–13424. [Google Scholar] [CrossRef]
- Janssen, J.J.M.; VanDeVen, W.J.M.; VanGroningen-Luyben, W.A.H.M.; Roosien, J.; Vlak, J.M.; DeGrip, W.J. Synthesis of functional bovine opsin in insect cells under control of the baculovirus polyhedrin promotor. Mol. Biol. Rep. 1988, 13, 65–71. [Google Scholar] [CrossRef]
- Klaassen, C.H.W.; Bovee-Geurts, P.H.M.; DeCaluwé, G.L.J.; DeGrip, W.J. Large-scale production and purification of functional recombinant bovine rhodopsin using the baculovirus expression system. Biochem. J. 1999, 342, 293–300. [Google Scholar] [CrossRef]
- Groenendijk, G.W.T.; Jansen, P.A.A.; Daemen, F.J.M.; Bonting, S.L. Analysis of geometrically isomeric vitamin A compounds. Methods Enzym. 1980, 67, 203–220. [Google Scholar]
- Ganapathy, S.; Liu, R.S.H. Photoisomerization of sixteen isomers of retinal. Initial product distribution in direct and sensitized irradiation. Photochem. Photobiol. 1992, 56, 959–964. [Google Scholar] [CrossRef]
- Davies, W.I.L.; Hankins, M.W.; Foster, R.G. Vertebrate ancient opsin and melanopsin: Divergent irradiance detectors. Photochem. Photobiol. Sci. 2010, 9, 1444–1457. [Google Scholar] [CrossRef] [PubMed]
- Varma, N.; Mutt, E.; Mühle, J.; Panneels, V.; Terakita, A.; Deupi, X.; Nogly, P.; Schertler, G.F.X.; Lesca, E. Crystal structure of jumping spider rhodopsin-1 as a light sensitive GPCR. Proc. Natl. Acad. Sci. USA 2019, 116, 14547–14556. [Google Scholar] [CrossRef] [PubMed]
- Bailes, H.J.; Zhuang, L.-Y.; Lucas, R.J. Reproducible and sustained regulation of Gαs signalling using a metazoan opsin as an optogenetic tool. PLoS ONE 2012, 7, e30774. [Google Scholar] [CrossRef]
- Church, J.R.; Haugaard Olsen, J.M.; Schapiro, I. The Impact of Retinal Configuration on the Protein-Chromophore Interactions in Bistable Jumping Spider Rhodopsin-1. Molecules 2022, 27, 71. [Google Scholar] [CrossRef]
- Davies, W.I.L.; Tay, B.-H.; Zheng, L.; Danks, J.A.; Brenner, S.; Foster, R.G.; Collin, S.P.; Hankins, M.W.; Venkatesh, B.; Hunt, D.M. Evolution and functional characterisation of melanopsins in a deep-sea chimaera (Elephant shark, Callorhinchus milii). PLoS ONE 2012, 7, e51276. [Google Scholar] [CrossRef]
- Herrera-Hernández, M.G.; Ramon, E.; Lupala, C.S.; Tena-Campos, M.; Pérez, J.J.; Garriga, P. Flavonoid allosteric modulation of mutated visual rhodopsin associated with retinitis pigmentosa. Sci. Rep. 2017, 7, 11167. [Google Scholar] [CrossRef]
- Gragg, M.; Park, P.S.-H. Misfolded rhodopsin mutants display variable aggregation properties. Biochim. Biophys. Acta-Mol. Basis Dis. 2018, 1864, 2938–2948. [Google Scholar] [CrossRef]
- Pasqualetto, G.; Pileggi, E.; Schepelmann, M.; Varricchio, C.; Rozanowska, M.; Brancale, A.; Bassetto, M. Ligand-based rational design, synthesis and evaluation of novel potential chemical chaperones for opsin. Eur. J. Med. Chem. 2021, 226, 113841. [Google Scholar] [CrossRef]
- Zhang, T.; Cao, L.-H.; Kumar, S.; Enemchukwu, N.O.; Zhang, N.; Lambert, A.; Zhao, X.C.; Jones, A.; Wang, S.X.; Dennis, E.M.; et al. Dimerization of visual pigments in vivo. Proc. Natl. Acad. Sci. USA 2016, 113, 9093–9098. [Google Scholar] [CrossRef] [PubMed]
- Behnen, P.; Felline, A.; Comitato, A.; Di Salvo, M.T.; Raimondi, F.; Gulati, S.; Kahremany, S.; Palczewski, K.; Marigo, V.; Fanelli, F. A Small Chaperone Improves Folding and Routing of Rhodopsin Mutants Linked to Inherited Blindness. iScience 2018, 4, 1–19. [Google Scholar] [CrossRef] [PubMed]
- Cao, P.X.; Sun, W.; Kramp, K.; Zheng, M.; Salom, D.; Jastrzebska, B.; Jin, H.; Palczewski, K.; Feng, Z.Y. Light-sensitive coupling of rhodopsin and melanopsin to Gi/o and Gq signal transduction in Caenorhabditis elegans. FASEB J. 2012, 26, 480–491. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.Y.; Chen, Y.; Jastrzebska, B.; Golczak, M.; Gulati, S.; Tang, H.; Seibel, W.; Li, X.Y.; Jin, H.; Han, Y.; et al. A novel small molecule chaperone of rod opsin and its potential therapy for retinal degeneration. Nat. Commun. 2018, 9, 1976. [Google Scholar] [CrossRef]
- Kaya, K.D.; Chen, H.Y.; Brooks, M.J.; Kelley, R.A.; Shimada, H.; Nagashima, K.; de Val, N.; Drinnan, C.T.; Gieser, L.; Kruczek, K.; et al. Transcriptome-based molecular staging of human stem cell-derived retinal organoids uncovers accelerated photoreceptor differentiation by 9-cis retinal. Mol. Vis. 2019, 25, 663–678. [Google Scholar]
- Ohgane, K.; Dodo, K.; Hashimoto, Y. Structural development study of a novel pharmacological chaperone for folding-defective rhodopsin mutants responsible for retinitis pigmentosa. Yakugaku Zasshi 2011, 131, 325–334. [Google Scholar] [CrossRef]
- Kiser, P.D.; Palczewski, K. Retinoids and Retinal Diseases. Annu. Rev. Vis. Sci. 2016, 2, 197–234. [Google Scholar] [CrossRef]
- Sullivan, L.S.; Daiger, S.P. Inherited retinal degeneration: Exceptional genetic and clinical heterogeneity. Mol. Med. Today 1996, 2, 380–386. [Google Scholar] [CrossRef]
- Rattner, A.; Sun, H.; Nathans, J. Molecular genetics of human retinal disease. Annu. Rev. Genet. 1999, 33, 89–131. [Google Scholar] [CrossRef]
- Fan, J.; Crouch, R.K.; Kono, M. Light prevents exogenous 11-cis retinal from maintaining cone photoreceptors in chromophore-deficient mice. Invest. Ophthalmol. Vis. Sci. 2011, 52, 2412–2416. [Google Scholar] [CrossRef][Green Version]
- VanHooser, J.P.; Alemán, T.S.; He, Y.-G.; Cideciyan, A.V.; Kuksa, V.; Pittler, S.J.; Stone, E.M.; Jacobson, S.G.; Palczewski, K. Rapid restoration of visual pigment and function with oral retinoid in a mouse model of childhood blindness. Proc. Natl. Acad. Sci. USA 2000, 97, 8623–8628. [Google Scholar] [CrossRef] [PubMed]
- VanHooser, J.P.; Liang, Y.; Maeda, T.; Kuksa, V.; Jang, G.-F.; He, Y.-G.; Rieke, F.; Fong, H.K.W.; Detwiler, P.B.; Palczewski, K. Recovery of visual functions in a mouse model of Leber congenital amaurosis. J. Biol. Chem. 2002, 277, 19173–19182. [Google Scholar] [CrossRef] [PubMed]
- Maeda, A.; Maeda, T.; Palczewski, K. Improvement in rod and cone function in mouse model of Fundus albipunctatus after pharmacologic treatment with 9-cis-retinal. Invest. Ophthalmol. Vis. Sci. 2006, 47, 4540–4546. [Google Scholar] [CrossRef] [PubMed]
- Maeda, T.; Dong, Z.Q.; Jin, H.; Sawada, O.; Gao, S.Q.; Utkhede, D.; Monk, W.; Palczewska, G.; Palczewski, K. QLT091001, a 9-cis-retinal analog, is well-tolerated by retinas of mice with impaired visual cycles. Invest. Ophthalmol. Vis. Sci. 2013, 54, 455–466. [Google Scholar] [CrossRef] [PubMed]
- Maeda, T.; Maeda, A.; Leahy, P.; Saperstein, D.A.; Palczewski, K. Effects of long-term administration of 9-cis-retinyl acetate on visual function in mice. Invest. Ophthalmol. Vis. Sci. 2009, 50, 322–333. [Google Scholar] [CrossRef]
- Scholl, H.P.N.; Moore, A.T.; Koenekoop, R.K.; Wen, Y.Q.; Fishman, G.A.; van den Born, L.I.; Bittner, A.; Bowles, K.; Fletcher, E.C.; Collison, F.T.; et al. Safety and Proof-of-Concept Study of Oral QLT091001 in Retinitis Pigmentosa Due to Inherited Deficiencies of Retinal Pigment Epithelial 65 Protein (RPE65) or Lecithin: Retinol Acyltransferase (LRAT). PLoS ONE 2015, 10, e0143846. [Google Scholar] [CrossRef]
- Gao, S.Q.; Kahremany, S.; Zhang, J.Y.; Jastrzebska, B.; Querubin, J.; Petersen-Jones, S.M.; Palczewski, K. Retinal-chitosan Conjugates Effectively Deliver Active Chromophores to Retinal Photoreceptor Cells in Blind Mice and Dogs. Mol. Pharm. 2018, 93, 438–452. [Google Scholar] [CrossRef]
- Dowling, J.E. Vitamin A: Its many roles—From vision and synaptic plasticity to infant mortality. J. Comp. Physiol. A 2020, 206, 389–399. [Google Scholar] [CrossRef]
- Sokolov, M.; Yadav, R.P.; Brooks, C.; Artemyev, N.O. Chaperones and retinal disorders. In Molecular Chaperones in Human Disorders; Donev, R., Ed.; Elsevier Inc.: Amsterdam, The Netherlands, 2019; pp. 85–117. [Google Scholar]
- Veritti, D.; Sarao, V.; Samassa, F.; Danese, C.; Lowenstein, A.; Schmidt-Erfurth, U.; Lanzetta, P. State-of-the art pharmacotherapy for non-neovascular age-related macular degeneration. Expert Opin. Pharm. 2020, 21, 773–784. [Google Scholar] [CrossRef]
- Gao, S.Q.; Parmar, T.; Palczewska, G.; Dong, Z.Q.; Golczak, M.; Palczewski, K.; Jastrzebska, B. Protective Effect of a Locked Retinal Chromophore Analog against Light-Induced Retinal Degeneration. Mol. Pharm. 2018, 94, 1132–1144. [Google Scholar] [CrossRef]
- Hirano, T.; Lim, I.T.; Kim, D.M.; Zheng, X.-G.; Yoshihara, K.; Oyama, Y.; Imai, H.; Shichida, Y.; Ishiguro, M. Constraints of opsin structure on the ligand-binding site: Studies with ring-fused retinals. Photochem. Photobiol. 2002, 76, 606–615. [Google Scholar] [CrossRef]
- Yun, J.-H.; Li, X.X.; Park, J.-H.; Wang, Y.; Ohki, M.; Jin, Z.Y.; Lee, W.; Park, S.-Y.; Hu, H.; Li, C.F.; et al. Non-cryogenic structure of a chloride pump provides crucial clues to temperature-dependent channel transport efficiency. J. Biol. Chem. 2019, 294, 794–804. [Google Scholar] [CrossRef] [PubMed]
- Zhang, M.; Gui, M.; Wang, Z.-F.; Gorgulla, C.; Yu, J.J.; Wu, H.; Sun, Z.-Y.J.; Klenk, C.; Merklinger, L.; Morstein, L.; et al. Cryo-EM structure of an activated GPCR-G protein complex in lipid nanodiscs. Nat. Struct. Mol. Biol. 2021, 28, 258–267. [Google Scholar] [CrossRef] [PubMed]
- Humphreys, I.R.; Pei, J.M.; Baek, M.; Krishnakumar, A.; Anishchenko, I.; Ovchinnikov, S.; Zhang, J.; Ness, T.J.; Banjade, S.; Bagde, S.R.; et al. Computed structures of core eukaryotic protein complexes. Science 2021, 374, 1340. [Google Scholar] [CrossRef] [PubMed]
- Jumper, J.; Evans, R.; Pritzel, A.; Green, T.; Figurnov, M.; Ronneberger, O.; Tunyasuvunakool, K.; Bates, R.; Zidek, A.; Potapenko, A.; et al. Highly accurate protein structure prediction with AlphaFold. Nature 2021, 596, 583–589. [Google Scholar] [CrossRef] [PubMed]
- Pedraza-González, L.; Barneschi, L.; Padula, D.; De Vico, L.; Olivucci, M. Evolution of the Automatic Rhodopsin Modeling (ARM) Protocol. Top Curr. Chem. 2022, 380, 21. [Google Scholar] [CrossRef] [PubMed]
- El-Tahawy, M.M.T.; Conti, I.; Bonfanti, M.; Nenov, A.; Garavelli, M. Tailoring Spectral and Photochemical Properties of Bioinspired Retinal Mimics by in Silico Engineering. Angew. Chem. Int. Ed. 2020, 59, 20619–20627. [Google Scholar] [CrossRef]
- Tan, P.; He, L.; Huang, Y.; Zhou, Y. Optophysiology: Illuminating cell physiology with optogenetics. Physiol. Rev. 2022, 102, 1263–1325. [Google Scholar] [CrossRef]
- Gilhooley, M.J.; Lindner, M.; Palumaa, T.; Hughes, S.; Peirson, S.N.; Hankins, M.W. A systematic comparison of optogenetic approaches to visual restoration. Mol. Ther. Methods Clin. Dev. 2022, 25, 111–123. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).