Chemical Modification of Novel Glycosidases from Lactobacillus plantarum Using Hyaluronic Acid: Effects on High Specificity against 6-Phosphate Glucopyranoside
Abstract
1. Introduction
2. Materials and Methods
2.1. General Description
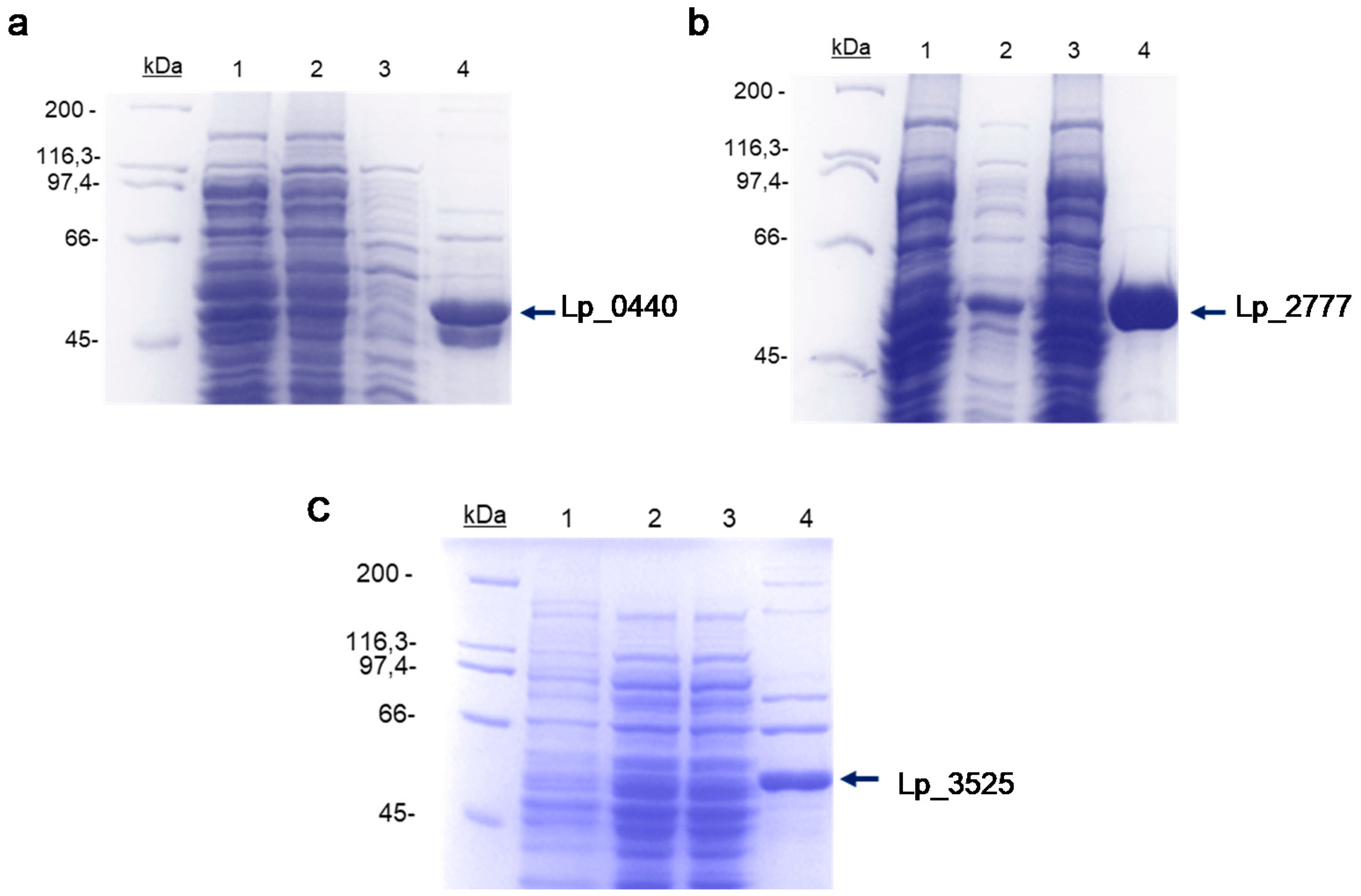
2.2. Expression of Lp_ 0440, Lp_2777, and Lp_3525 Genes in Escherichia coli and Purification of the Recombinant Proteins
2.3. Physical Modification of the New LpGs
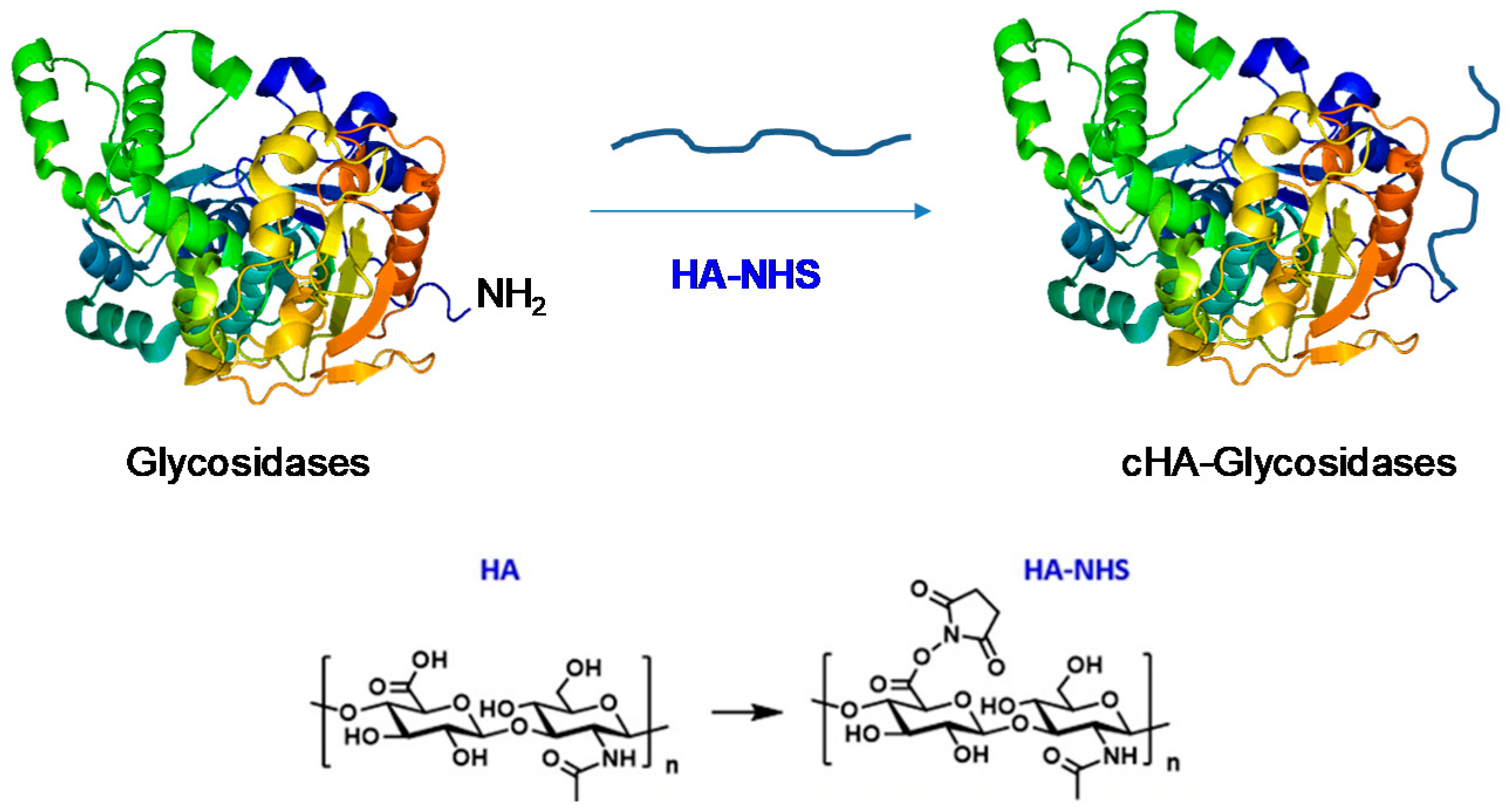
2.4. Covalent Modification of Lp Glycosidases with Hyaluronic Acid (HA)
2.5. Native Electrophoresis Assay
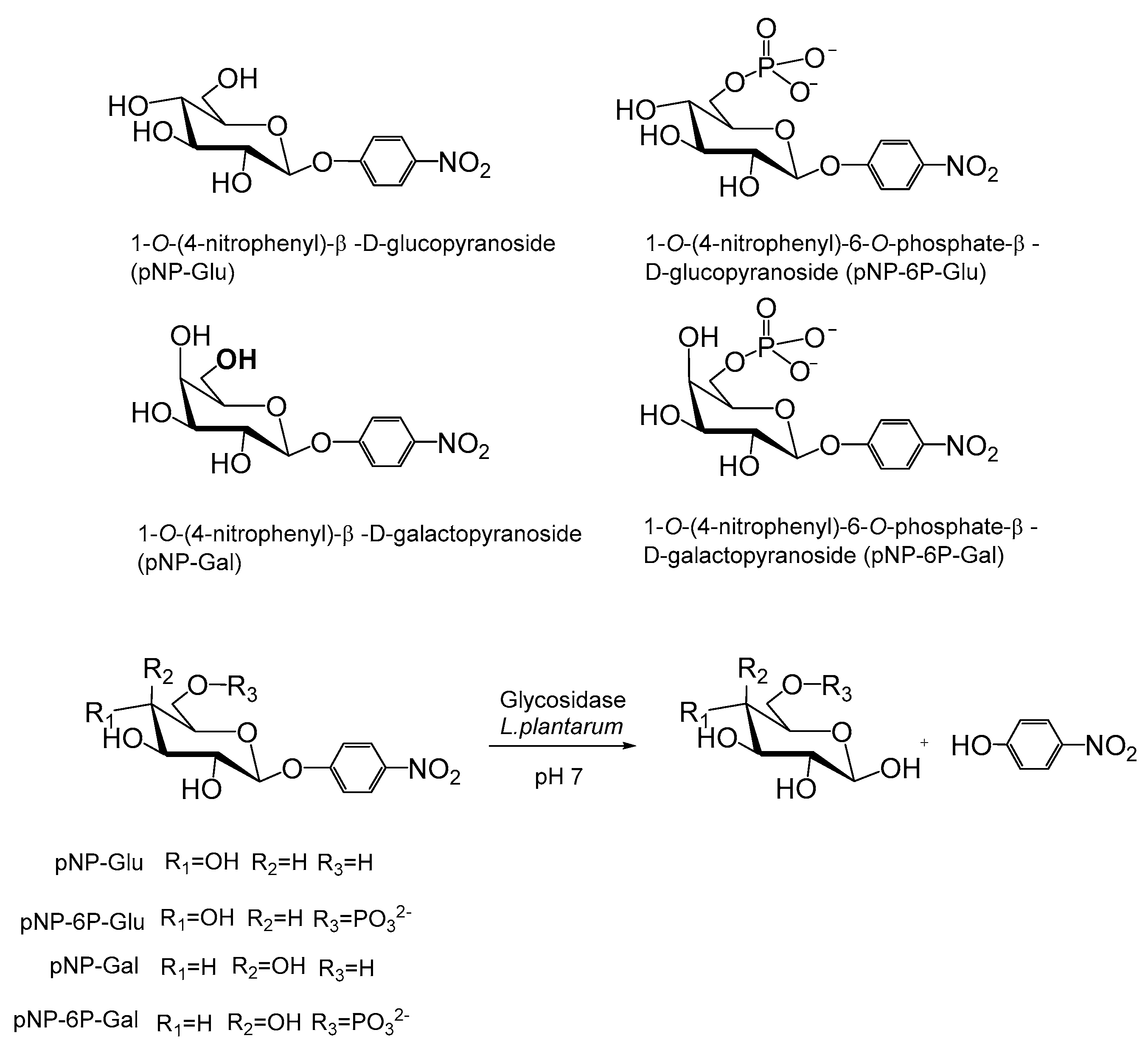
2.6. Synthesis of 4-Nitropheny,6-Phosphate-β-d-Glucopyranoside (pNP-6P-Glu)
2.7. Enzymatic Activity Assay of Glycosidases in the Hydrolysis of p-Nitrophenyl-Glycopyranosides
3. Results and Discussion
3.1. Production and Purification of the Different Glycosidases
3.2. Modification of Different LpGs by Hyaluronic Acid (HA)
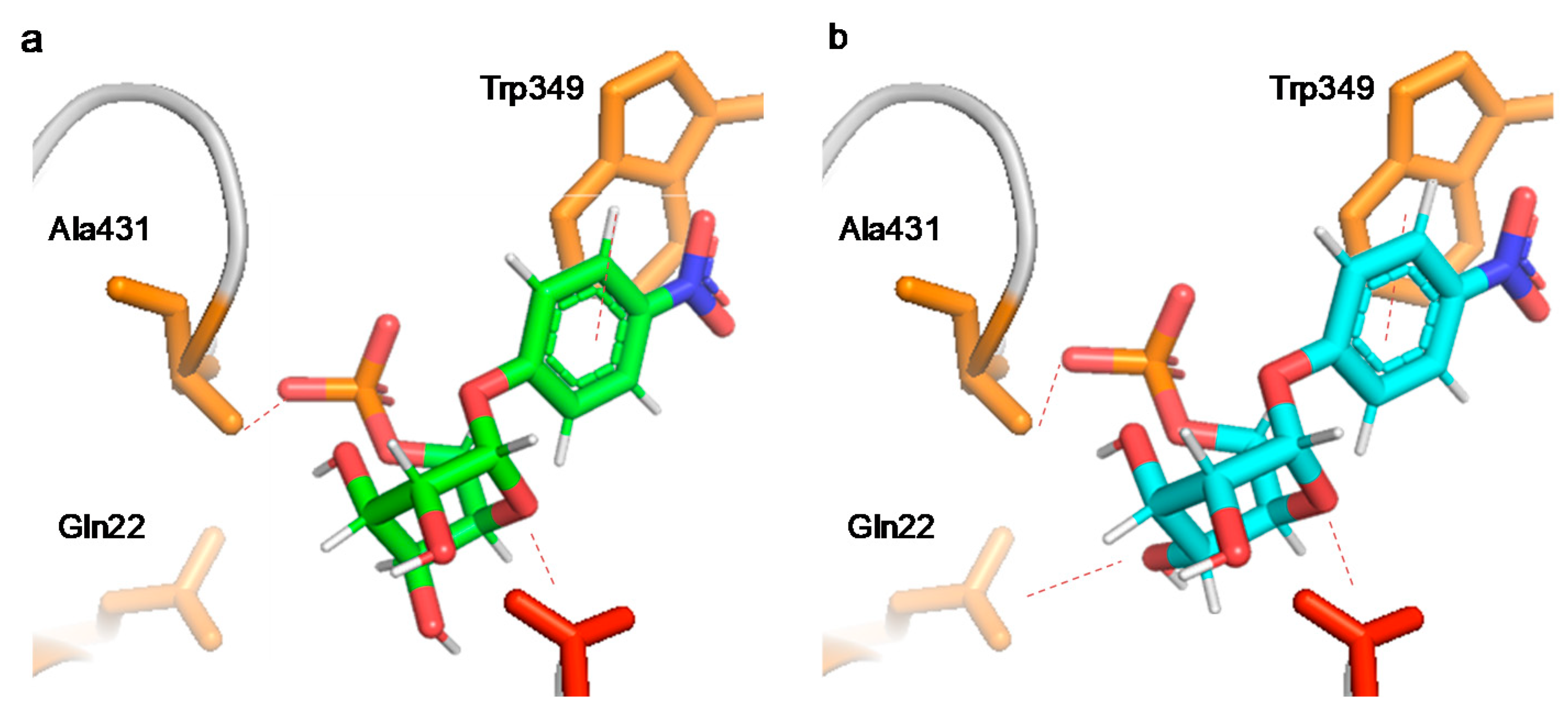
3.3. Glycosidase Activity, Specificity, and Regioselectivity of the Different Modified Glycosidases
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Spicer, C.D.; Davis, B.G. Selective chemical protein modification. Nat. Commun. 2014, 5, 4740. [Google Scholar] [CrossRef]
- MacDonald, J.I.; Munch, H.K.; Moore, T.; Francis, M.B. One-step site-specific modification of native proteins with 2-pyridinecarboxyaldehydes. Nat. Methods 2015, 11, 326–331. [Google Scholar] [CrossRef] [PubMed]
- Chan, A.O.Y.; Ho, C.M.; Chong, H.C.; Leung, Y.C.; Huang, J.S.; Wong, M.K.; Che, C.M. Modification of N-terminal α-amino groups of peptides and proteins using ketenes. J. Am. Chem. Soc. 2012, 134, 2589–2598. [Google Scholar] [CrossRef] [PubMed]
- Lai, Z.W.; Petrera, A.; Schilling, O. Protein amino-terminal modifications and proteomic approaches for N-terminal profiling. Chem. Boil. 2015, 24, 71–79. [Google Scholar] [CrossRef] [PubMed]
- Filice, M.; Romero, O.; Guisán, J.M.; Palomo, J.M. trans,trans-2,4-Hexadiene incorporation on enzymes for site-specific immobilization and fluorescent labeling. Org. Biomol. Chem. 2011, 9, 5535–5540. [Google Scholar] [CrossRef] [PubMed]
- Wu, B.; Wijma, H.J.; Song, L.; Rozeboom, H.J.; Poloni, C.; Tian, Y.; Arif, M.I.; Nuijens, T.; Quaedflieg, P.J.L.M.; Szymanski, W.; et al. Versatile peptide C-terminal functionalization via a computationally engineered peptide amidase. ACS Catal. 2016, 6, 5405–5414. [Google Scholar] [CrossRef]
- Díaz-Rodríguez, A.; Davis, B.G. Chemical modification in the creation of novel biocatalysts. Chem. Boil. 2011, 15, 211–219. [Google Scholar] [CrossRef]
- Romero, O.; Filice, M.; de las Rivas, B.; Carrasco-Lopez, C.; Klett, J.; Morreale, A.; Hermoso, J.A.; Guisan, J.M.; Abian, O.; Palomo, J.M. Semisynthetic peptide-lipase conjugates for improved biotransformations. Chem. Commun. 2012, 48, 9053–9055. [Google Scholar] [CrossRef]
- Gutarra, M.L.E.; Romero, O.; Abian, O.; Torres, F.A.G.; Freire, D.M.G.; Castro, A.M.; Guisan, J.M.; Palomo, J.M.; Freire, D.M.G. Enzyme surface glycosylation in the solid phase: Improved activity and selectivity of Candida antarctica lipase B. Chem. Cat. Chem. 2011, 3, 1902–1910. [Google Scholar] [CrossRef]
- Callaghan, C.; Redmond, M.; Filice, M.; Alnoch, R.C.; Mateo, C.; Palomo, J.M. Biocatalytic process optimization for the production of high-added-value 6-O-hydroxy and 3-O-hydroxy glycosyl building blocks. Chem. Cat. Chem. 2017, 9, 2536–2543. [Google Scholar] [CrossRef]
- Perugino, G.; Trincone, A.; Rossi, M.; Moracci, M. Oligosaccharide synthesis by glycosynthases. Trends Biotechnol. 2004, 22, 31–37. [Google Scholar] [CrossRef]
- Ajisaka, K.; Yamamoto, Y. Control of the regioselectivity in the enzymatic syntheses of oligosaccharides using glycosidases. Trends Glycosci. Glycotechnol. 2002, 14, 1–11. (In Japanese) [Google Scholar] [CrossRef]
- García, C.; Hoyos, P.; Hernáiz, M.J. Enzymatic synthesis of carbohydrates and glycoconjugates using lipases and glycosidases in green solvents. Biocatal. Biotrans. 2018, 36, 131–140. [Google Scholar] [CrossRef]
- Harit, V.K.; Ramesh, N.G. Amino-functionalized iminocyclitols: Synthetic glycomimetics of medicinal interest. RSC Adv. 2016, 6, 109528–109607. [Google Scholar] [CrossRef]
- Seeberger, P.H.; Werz, D.B. Synthesis and medical applications of oligosaccharides. Nat. Cell Boil. 2007, 446, 1046–1051. [Google Scholar] [CrossRef]
- Murrey, H.E.; Hsieh-Wilson, L.C. The chemical neurobiology of carbohydrates. Chem. Rev. 2008, 108, 1708–1731. [Google Scholar] [CrossRef]
- Campbell, C.T.; Yarema, K.J. Large-scale approaches for glycobiology. Genome Boil. 2005, 6, 236. [Google Scholar] [CrossRef]
- Filice, M.; Ubiali, D.; Fernandez-Lafuente, R.; Fernandez-Lorente, G.; Guisán, J.M.; Palomo, J.M.; Terreni, M. A chemo-biocatalytic approach in the synthesis of β-O-naphtylmethyl-N-peracetylated lactosamine. J. Mol. Catal. B Enzym. 2008, 52, 106–112. [Google Scholar] [CrossRef]
- Chen, L.; Huang, G. The antioxidant activity of derivatized cushaw polysaccharides. Int. J. Boil. Macromol. 2019, 128, 1–4. [Google Scholar] [CrossRef]
- Pinheiro, V.B.; Taylor, A.I.; Cozens, C.; Abramov, M.; Renders, M.; Zhang, S.; Chaput, J.C.; Wengel, J.; Peak-Chew, S.-Y.; McLaughlin, S.H.; et al. Synthetic genetic polymers capable of heredity and evolution. Science 2012, 336, 341–344. [Google Scholar] [CrossRef]
- Herdewijn, P. Nucleic acids with a six-membered ‘carbohydrate’ mimic in the backbone. Chem. Biodivers. 2010, 7, 1–59. [Google Scholar] [CrossRef]
- Wright, T.A.; Page, R.C.; Konkolewicz, D. Polymer conjugation of proteins as a synthetic post-translational modification to impact their stability and activity. Polym. Chem. 2019, 10, 434–454. [Google Scholar] [CrossRef]
- Romero, O.; Rivero, C.W.; Guisan, J.M.; Palomo, J.M.; Leite, L. Novel enzyme-polymer conjugates for biotechnological applications. PeerJ 2013, 1, e27. [Google Scholar] [CrossRef]
- Montagner, I.M.; Merlo, A.; Carpanese, D.; Pietà, A.D.; Mero, A.; Grigoletto, A.; Loregian, A.; Renier, D.; Campisi, M.; Zanovello, P.; et al. A site-selective hyaluronan-interferonα2a conjugate for the treatment of ovarian cancer. J. Control. Release 2016, 236, 79–89. [Google Scholar] [CrossRef]
- Huang, G.; Huang, H. Application of hyaluronic acid as carriers in drug delivery. Drug Deliv. 2018, 25, 766–772. [Google Scholar] [CrossRef]
- Curiel, J.A.; Rivas, B.D.L.; Mancheño, J.M.; Muñoz, R. The pURI family of expression vectors: A versatile set of ligation independent cloning plasmids for producing recombinant His-fusion proteins. Protein Expr. Purif. 2011, 76, 44–53. [Google Scholar] [CrossRef]
- Chevallet, M.; Luche, S.; Rabilloud, T. Silver staining of proteins in polyacrylamide gels. Nat. Protoc. 2006, 1, 1852–1858. [Google Scholar] [CrossRef]
- Marasco, R.; Muscariello, L.; Varcamonti, M.; De Felice, M.; Sacco, M. Expression of the bglH gene of Lactobacillus plantarum is controlled by carbon catabolite repression. J. Bacteriol. 1998, 180, 3400–3404. [Google Scholar]
- Michalska, K.; Tan, K.; Li, H.; Hatzos-Skintges, C.; Bearden, J.; Babnigg, G.; Joachimiak, A. GH1-family 6-P-β-glucosidases from human microbiome lactic acid bacteria. Acta Crystallogr. Sect. D Boil. Crystallogr. 2013, 69, 451–463. [Google Scholar] [CrossRef]
- De las Rivas, B.; Curiel, J.A.; Mancheño, J.M.; Muñoz, R. Expression vectors for enzyme restriction- and ligation-independent cloning for producing recombinant His-fusion proteins. Biotechnol. Prog. 2007, 23, 680–686. [Google Scholar] [CrossRef]
- Mero, A.; Pasqualin, M.; Campisi, M.; Renier, D.; Pasut, G. Conjugation of hyaluronan to proteins. Carbohydr. Polym. 2013, 92, 2163–2170. [Google Scholar] [CrossRef]
- Yang, J.-A.; Kim, E.-S.; Kwon, J.H.; Kim, H.; Shin, J.H.; Yun, S.H.; Choi, K.Y.; Hahn, S.K. Transdermal delivery of hyaluronic acid–Human growth hormone conjugate. Biomaterials 2012, 33, 5947–5954. [Google Scholar] [CrossRef]
- Mero, A.; Campisi, M. Hyaluronic acid bioconjugates for the delivery of bioactive molecules. Polymers 2014, 6, 346–369. [Google Scholar] [CrossRef]
- Martínez-Sanz, E.; Ossipov, D.A.; Hilborn, J.; Larsson, S.; Jonsson, K.B.; Varghese, O.P. Bone reservoir: Injectable hyaluronic acid hydrogel for minimal invasive bone augmentation. J. Control. Release 2011, 152, 232–240. [Google Scholar] [CrossRef]
- Yang, J.-A.; Park, K.; Jung, H.; Kim, H.; Hong, S.W.; Yoon, S.K.; Hahn, S.K. Target specific hyaluronic acid–Interferon alpha conjugate for the treatment of hepatitis C virus infection. Biomaterials 2011, 32, 8722–8729. [Google Scholar] [CrossRef]
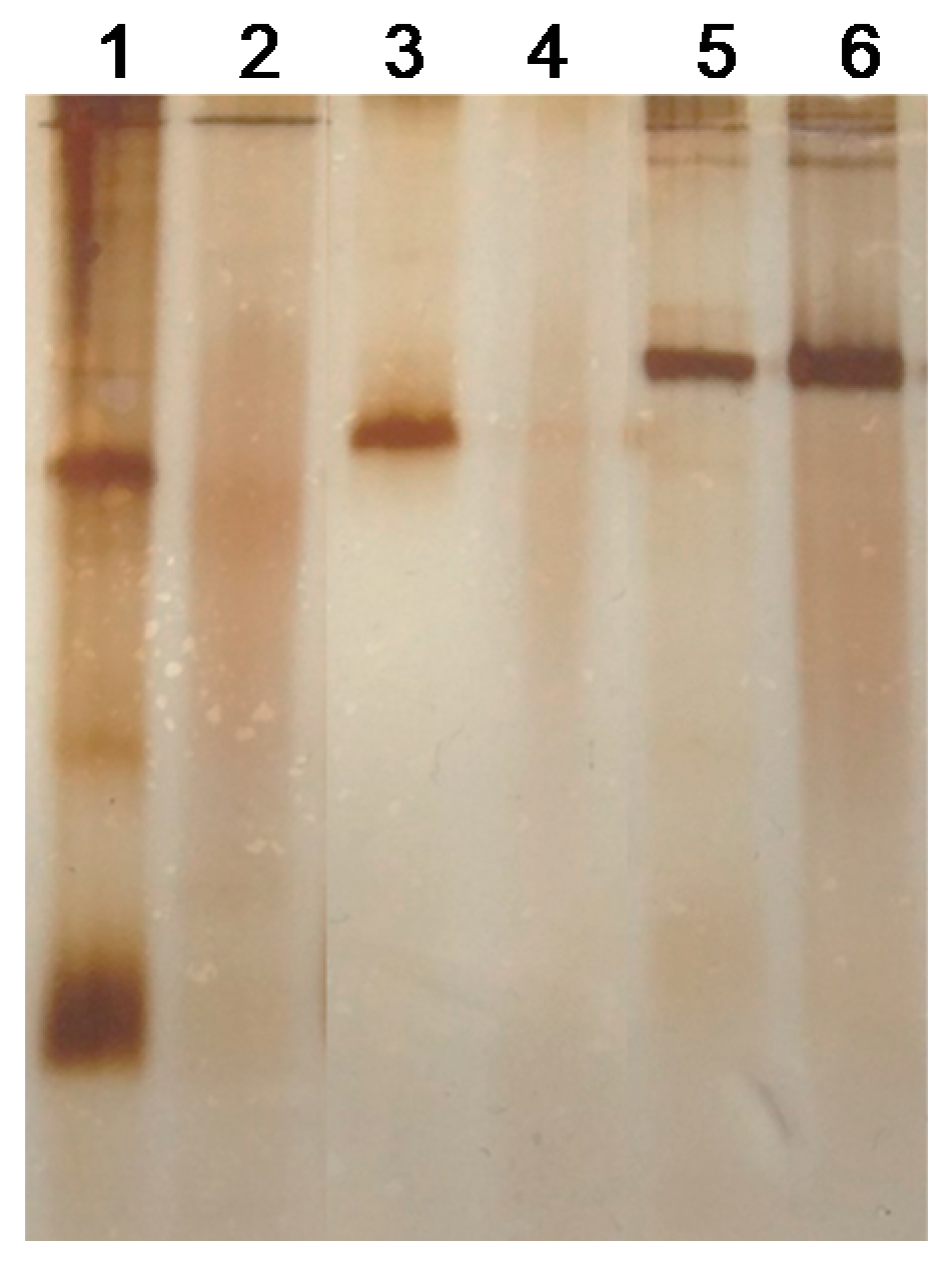
| Entry | Biocatalyst | Activity (∆Abs/min) a | |||
|---|---|---|---|---|---|
| pNP-Glu b | pNP-6P-Glu c | pNP-Gal b | pNP-6P-Gal b | ||
| 1 | Lp_0440 | 39.20 ± 1.20 | 39.1 ± 1.10 | 15.6 | 0 |
| 2 | HA-Lp_0440 | 5.80 ± 0.20 | 3.30 ± 0.10 | 5.1 | 0 |
| 3 | cHA-Lp_0440 | 13.10 ± 0.39 | 1.90 ± 0.10 | 7 | 0 |
| 4 | Lp_2777 | 0 | 270 ± 8 | 0 | 1.4 ± 0.07 |
| 5 | HA-Lp_2777 | 0 | 0 | 0 | 0 |
| 6 | cHA-Lp_2777 | 0 | 80 ± 2 | 0 | 0.5 ± 0.04 |
| 7 | Lp_3525 | 0 | 335 ± 10 | 0 | 12.4 ± 0.38 |
| 8 | HA-Lp_3525 | 0 | 20 ± 0.60 | 0 | 0 |
| 9 | cHA-Lp_3525 | 0 | 252 ± 7.5 | 0 | 0 |
| Entry | Biocatalyst | Activity (∆Abs/min) a | |||
|---|---|---|---|---|---|
| pNP-Glu b | pNP-6P-Glu c | pNP-Gal b | pNP-6P-Gal b | ||
| 1 | Lp_0440 | 0 | 0 | 17 ± 0.50 | 0 |
| 2 | HA-Lp_0440 | 0 | 0 | 1.6 ± 0.13 | 0 |
| 3 | cHA-Lp_0440 | 0 | 0 | 11.4 ± 0.34 | 0 |
| 4 | Lp_2777 | 0 | 69 ± 2 | 0 | 3.2 ± 0.01 |
| 5 | HA-Lp_2777 | 0 | 0 | 0 | 0.7 ± 0.07 |
| 6 | cHA-Lp_2777 | 0 | 8.7 ± 0.26 | 0 | 0.8 ± 0.08 |
| 7 | Lp_3525 | 0 | 300 ± 9 | 0 | 5.5 ± 0.17 |
| 8 | HA-Lp_3525 | 0 | 0 | 0 | 0 |
| 9 | cHA-Lp_3525 | 0 | 0.3 ± 0.03 | 0 | 3 ± 0.15 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Perez-Rizquez, C.; Lopez-Tejedor, D.; Plaza-Vinuesa, L.; de las Rivas, B.; Muñoz, R.; Cumella, J.; Palomo, J.M. Chemical Modification of Novel Glycosidases from Lactobacillus plantarum Using Hyaluronic Acid: Effects on High Specificity against 6-Phosphate Glucopyranoside. Coatings 2019, 9, 311. https://doi.org/10.3390/coatings9050311
Perez-Rizquez C, Lopez-Tejedor D, Plaza-Vinuesa L, de las Rivas B, Muñoz R, Cumella J, Palomo JM. Chemical Modification of Novel Glycosidases from Lactobacillus plantarum Using Hyaluronic Acid: Effects on High Specificity against 6-Phosphate Glucopyranoside. Coatings. 2019; 9(5):311. https://doi.org/10.3390/coatings9050311
Chicago/Turabian StylePerez-Rizquez, Carlos, David Lopez-Tejedor, Laura Plaza-Vinuesa, Blanca de las Rivas, Rosario Muñoz, Jose Cumella, and Jose M. Palomo. 2019. "Chemical Modification of Novel Glycosidases from Lactobacillus plantarum Using Hyaluronic Acid: Effects on High Specificity against 6-Phosphate Glucopyranoside" Coatings 9, no. 5: 311. https://doi.org/10.3390/coatings9050311
APA StylePerez-Rizquez, C., Lopez-Tejedor, D., Plaza-Vinuesa, L., de las Rivas, B., Muñoz, R., Cumella, J., & Palomo, J. M. (2019). Chemical Modification of Novel Glycosidases from Lactobacillus plantarum Using Hyaluronic Acid: Effects on High Specificity against 6-Phosphate Glucopyranoside. Coatings, 9(5), 311. https://doi.org/10.3390/coatings9050311






