Research on the Formation Mechanisms of Red Stains on Outdoor Marble Cultural Relics at Beijing Confucian Temple and the Imperial College
Abstract
1. Introduction
2. Materials and Methods
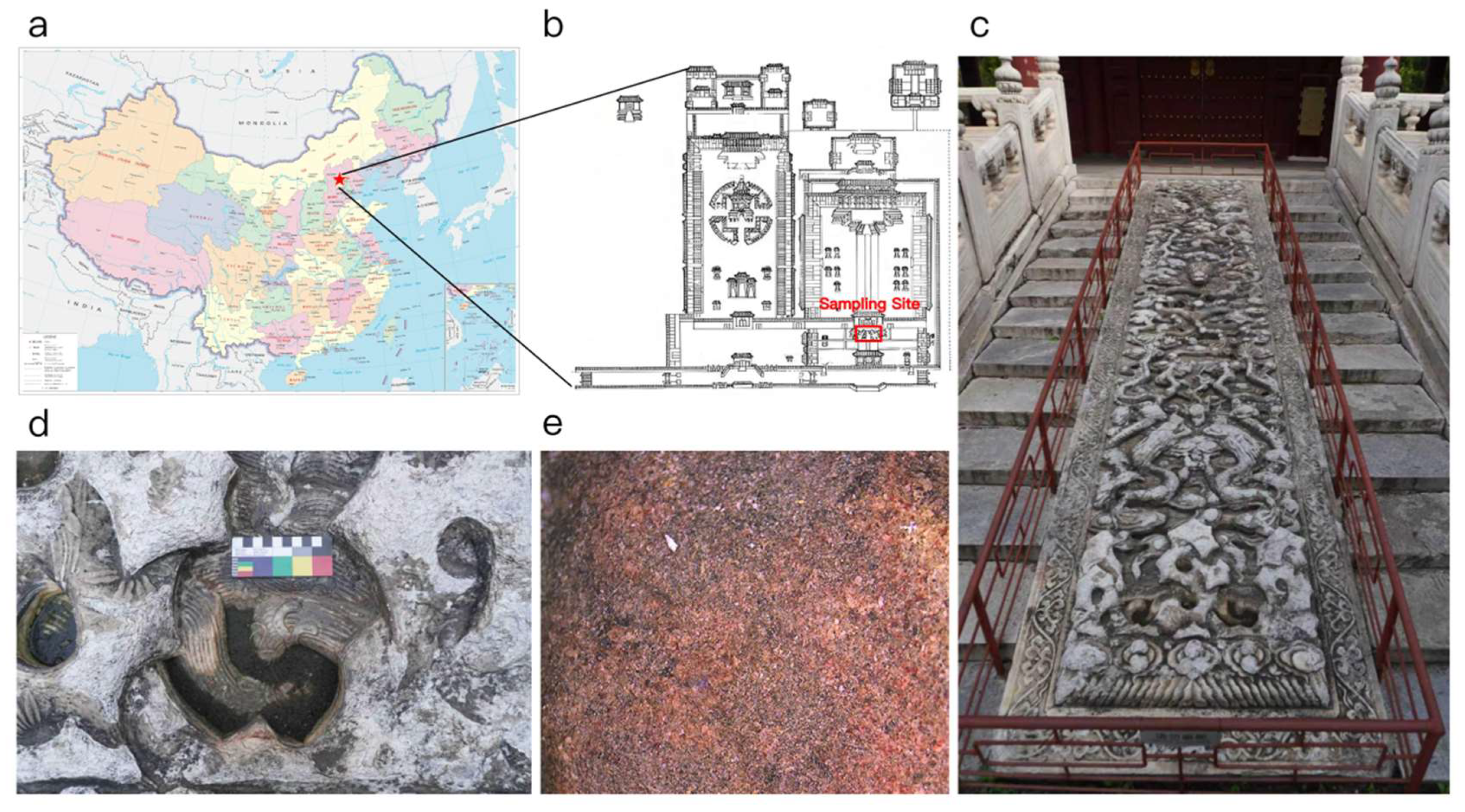
2.1. Sampling Method and Sampling Site Conditions
2.2. Extended Depth-of-Field Microscopy
2.3. Micro-Raman Spectroscopy Analysis
2.4. Scanning Electron Microscopy with Energy-Dispersive X-Ray Spectroscopy
2.5. Microbial Cultivation and Molecular Identification
2.6. Extraction of Fungal Red Pigments
2.7. UHPLC-HRMS Analysis of Pigment Components
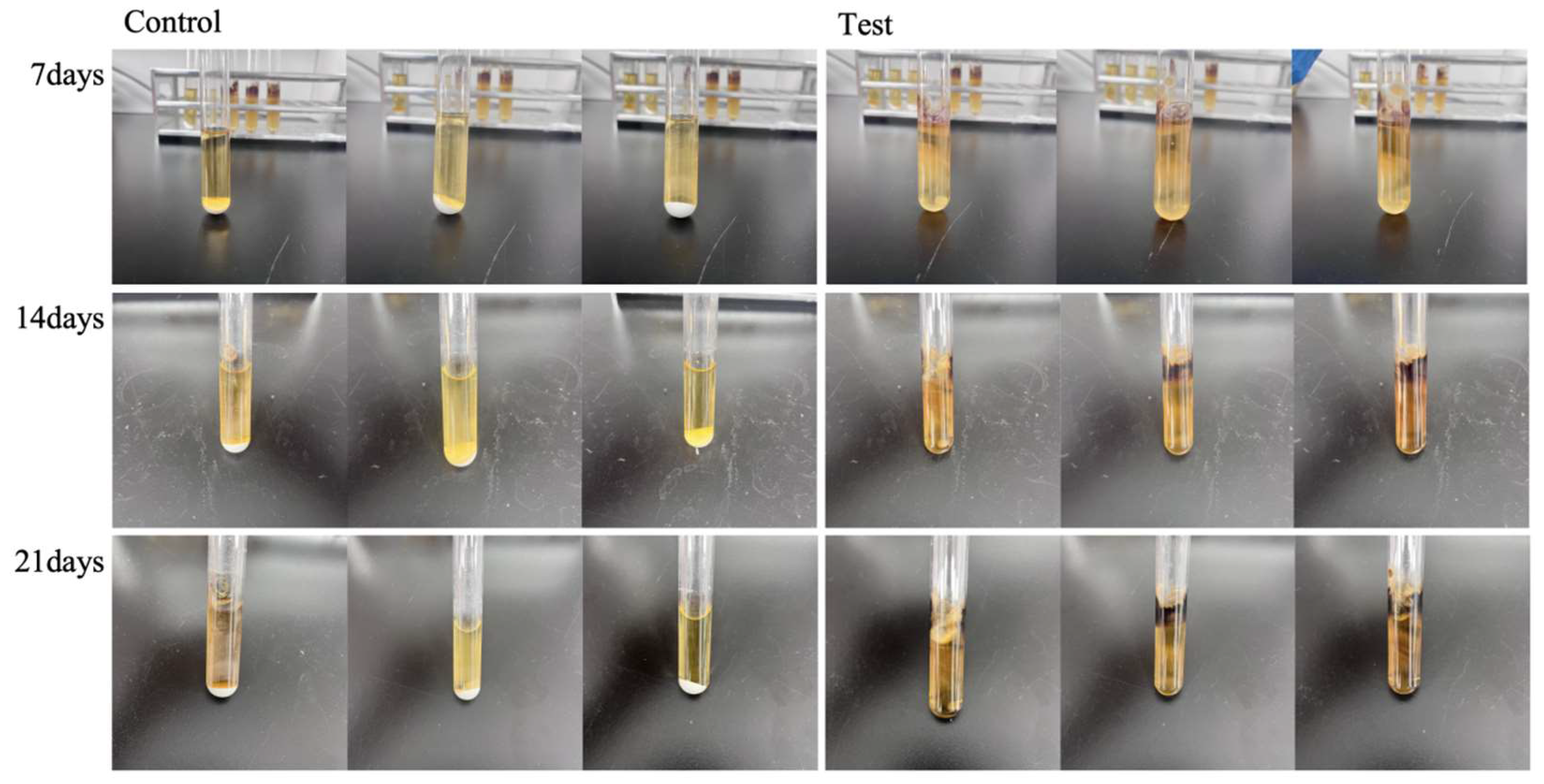
2.8. Infection Experiment
2.9. Nucleotide Sequence Deposition
3. Results
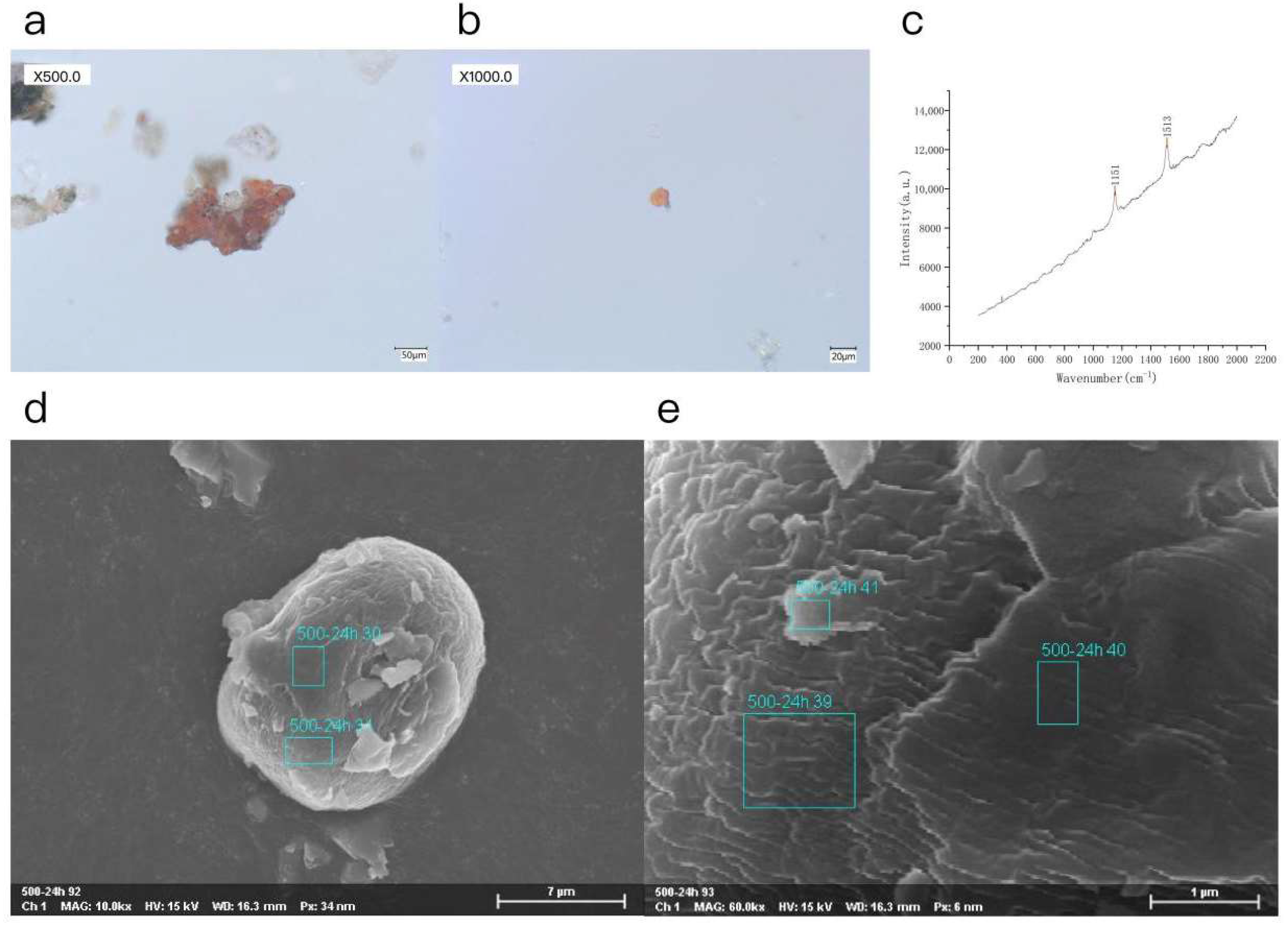
3.1. Compositional Analysis of Red Contaminants on Stone Carvings
| Analyzed Point | C | N | O | Na | P | K | Mg |
|---|---|---|---|---|---|---|---|
| 50,024h 30 | 77.11 | 4.74 | 17.23 | 0.33 | 0.42 | 0.15 | |
| 50,024h 31 | 88.22 | 10.94 | 0.55 | 0.29 | |||
| 50,024h 39 | 87.96 | 11.80 | 0.60 | 0.32 | |||
| 50,024h 40 | 79.00 | 3.58 | 16.29 | 0.39 | 0.49 | 0.26 | |
| 50,024h 41 | 80.14 | 17.28 | 0.10 | 0.09 | 0.46 |
3.2. Microbial Community Composition Analysis of the Danbi Stone Carvings at Beijing Confucian Temple and the Imperial College
| Strain Number | Closet Relative Strain | Similarity | Accession Number | |
|---|---|---|---|---|
| Bacteria | KG-B1 | Lysinibacillus fusiformis | 99.63% | KY569483.1 |
| KG-B2 | Bacillus cereus | 100.00% | MH458752.1 | |
| Fungi | KG-F1 | Didymella exigua | 97.96% | KAF1928462.1 |
| KG-F2 | Trichoderma reesei | 97.62% | EGR48663.1 | |
| KG-F3 | Lizonia empirigonia | 100.00% | KAF1343396.1 |
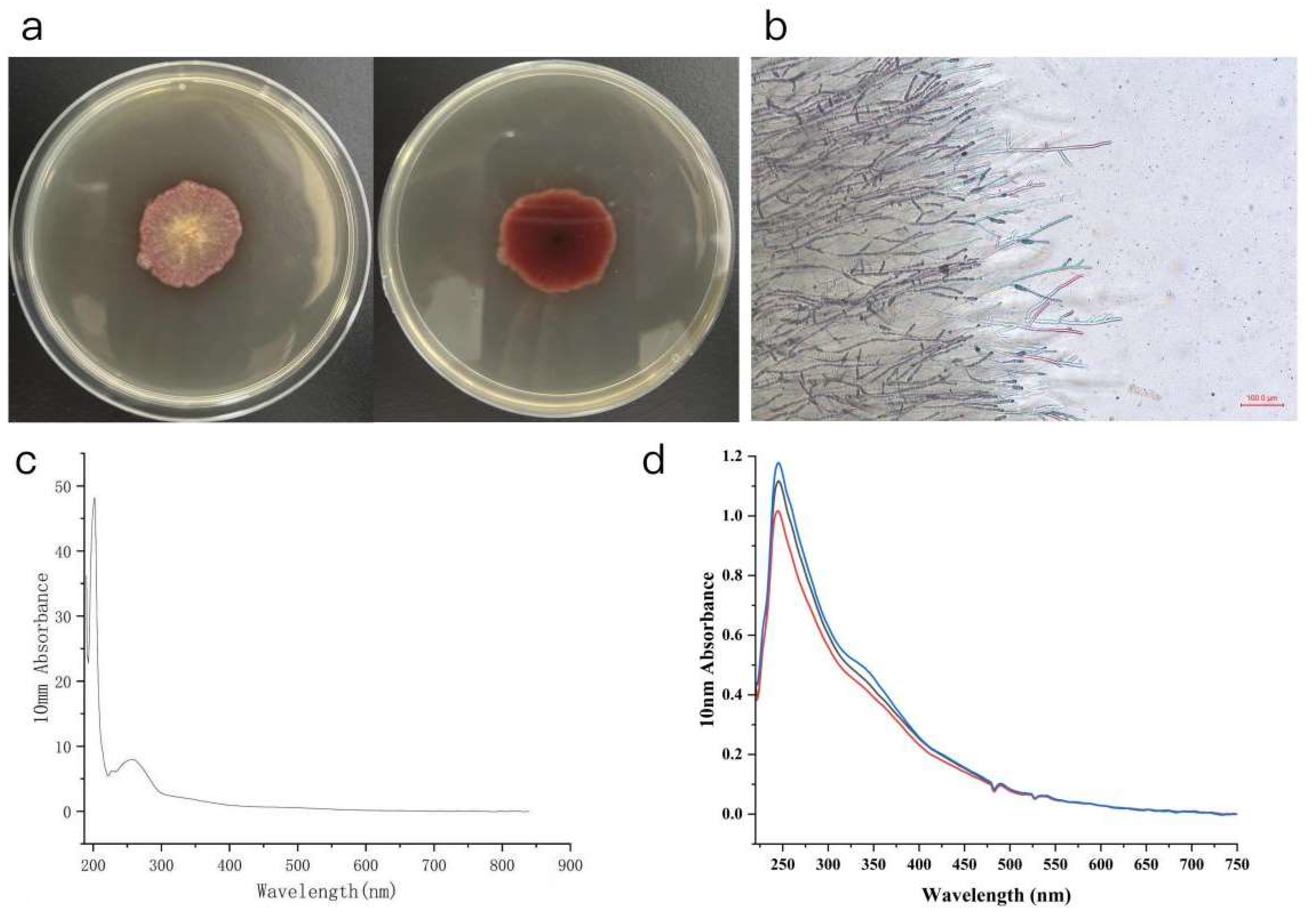
3.3. Extraction and Analysis of Red Pigments from the Pigment-Producing Fungus Lizonia empirigonia
3.4. Infection Experiment of Pigment-Producing Fungus L. empirigonia on Stone Materials
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Pu, H.; Wang, X. The Impact of Environment on Cultural Relics. Sci. Cult. 2023, 9, 49–62. [Google Scholar] [CrossRef]
- Cantisani, E.; Cuzman, O.A.; Vettori, S.; Chelazzi, L.; Ciattini, S.; Ricci, M.; Manganelli Del Fá, R.; Chiarantini, L.; Garzonio, C.A. A Multi-Analytical Approach for the Study of Red Stains on Heritage Marble. Analyst 2019, 144, 2375–2386. [Google Scholar] [CrossRef]
- Abdel-Maksoud, G.; Awad, H.; Rashed, U. Different Cleaning Techniques for Removal of Iron Stain from Archaeological Bone Artifacts: A Review. Egypt. J. Chem. 2021, 65, 73–87. [Google Scholar] [CrossRef]
- Zanardini, E.; Andreoni, V.; Borin, S.; Cappitelli, F.; Daffonchio, D.; Talotta, P.; Sorlini, C.; Ranalli, G.; Bruni, S.; Cariati, F. Lead-Resistant Microorganisms from Red Stains of Marble of the Certosa of Pavia, Italy and Use of Nucleic Acid-Based Techniques for Their Detection. Int. Biodeterior. Biodegrad. 1997, 40, 171–182. [Google Scholar] [CrossRef]
- Villani, E.; Suzuki, A.; Ricci, M.; Salvadori, B.; Vettori, S.; Cantisani, E. Red Stains on Heritage Marbles: Application of Micro-Scale Analyses to Assess the Presence and Distribution of Lead Compounds. Analyst 2024, 149, 4872–4880. [Google Scholar] [CrossRef]
- Gaylarde, C.; Little, B. Biodeterioration of Stone and Metal—Fundamental Microbial Cycling Processes with Spatial and Temporal Scale Differences. Sci. Total Environ. 2022, 823, 153193. [Google Scholar] [CrossRef]
- Konkol, N.; McNamara, C.; Sembrat, J.; Rabinowitz, M.; Mitchell, R. Enzymatic Decolorization of Bacterial Pigments from Culturally Significant Marble. J. Cult. Herit. 2009, 10, 362–366. [Google Scholar] [CrossRef]
- Hallmann, C.; Rüdrich, J.; Enseleit, M.; Friedl, T.; Hoppert, M. Microbial Diversity on a Marble Monument: A Case Study. Environ. Earth Sci. 2011, 63, 1701–1711. [Google Scholar] [CrossRef]
- Bruni, S.; Cariati, F.; Bianchi, C.L.; Zanardini, E.; Sorlini, C. Spectroscopic Investigation of Red Stains Affecting the Carrara Marble Façade of the Certosa of Pavia. Archaeometry 1995, 37, 249–255. [Google Scholar] [CrossRef]
- Palla, F.; Tartamella, E. Chromatic Alteration on Marble Surfaces Analysed by Molecular Biology Tools. Conserv. Sci. Cult. Herit. 2007, 7, 111–121. [Google Scholar] [CrossRef]
- Tang, L.K. Reinventing Confucius and His Shrine: The Temple of Culture (Wenmiao) in Late Imperial China. Ph.D. Thesis, University of Pennsylvania, Philadelphia, PA, USA, 2018. [Google Scholar]
- Barbosa, S.; Moura, P.C.; Dias, A.; Haneklaus, N.; Bellefqih, H.; Kiegiel, K.; Canovas, C.R.; Nieto, J.M.; Bilal, E.; Pessanha, S. An Advanced Image Processing and Multivariate Statistical Methodology to Interpret Micro-EDXRF 2D Maps: Uncovering Heterogeneity and Spatial Distribution Patterns of Rare Earth Elements in Phosphogypsum. Chemosphere 2025, 381, 144478. [Google Scholar] [CrossRef]
- Amadi, O.C. Use of Starch Containing Tubers for the Formulation of Culture Media for Fungal Cultivation. Afr. J. Microbiol. Res. 2012, 6, 4527–4532. [Google Scholar] [CrossRef]
- Li, P.; Han, S.; Wang, M.; Zhang, X.; Zhi, S.; Jin, M.; Jokela, J.; He, S.; Liu, L. Elucidation of Novel Turnagainolides and Their Biosynthetic Gene Cluster in Bacillus subtilis. Appl. Env. Microbiol. 2025, 91, e02574-24. [Google Scholar] [CrossRef]
- Li, Y.; Ma, J.; Zhang, J.; Liang, C.; Yang, B.; Zhao, N. Optimization of Extraction Process of Chaetomium Aureum Pigment by Response Surface Methodology. Food Chem. 2021, 42, 295–299. [Google Scholar]
- Li, C.; Zhang, X.; Liu, H.; Qu, L.; Zhu, M.; Duan, H. Application of Nanosecond Laser Cleaning Technology on Marble in the Forbidden City. Opt. Tech. 2024, 50, 500–507. [Google Scholar]
- Chen, C.-Y.; Huang, W.-Z.; Gao, Q.; Fan, L.-W.; Andy, H.S. Assignments on Raman Peaks of Red Coral Based on Experimental RamanSpectroscopy and Density Functional Theory Calculation. Spectrosc. Spectr. Anal. 2021, 41, 127–130. [Google Scholar]
- Liu, Y.; He, Q.; Wang, Z.; Jiao, X.; Zhang, Y. The Optical Properties of Nano-structural α-Fe2O3 Dependence on the Shape. Microsc. Res. Tech. 2025, 88, 154–162. [Google Scholar] [CrossRef] [PubMed]
- Salagram, M.; Prasad, V.K.; Subrahmanyam, K. IR and Optical Study of Pb3O4 (2PbO.PbO2) Glass Containing a Small Amount of Silica. J. Alloys Compd. 2002, 335, 228–232. [Google Scholar] [CrossRef]
- Grasso, D.; Strevett, K.; Pesari, H. Impact of Sodium and Potassium on Environmental Systems. J. Environ. Syst. 1992, 22, 297–323. [Google Scholar] [CrossRef]
- Liu, L.; Yang, C.; Liang, F.; Li, C.; Zeng, Q.; Han, S.; Li, S.; Liu, Y. Genome-Wide Survey of the Bipartite Structure and Pathogenesis-Related Genes of Neostagonosporella Sichuanensis, a Causal Agent of Fishscale Bamboo Rhombic-Spot Disease. Front. Microbiol. 2024, 15, 1456993. [Google Scholar] [CrossRef]
- Giusti, M.M.; Miyagusuku-Cruzado, G.; Wallace, T.C. Flavonoids as Natural Pigments. In Handbook of Natural Colorants; Stevens, C., Bechtold, T., Manian, A., Pham, T., Eds.; Wiley: Hoboken, NJ, USA, 2023; pp. 371–390. ISBN 978-1-119-81171-8. [Google Scholar]
- Iwashina, T. Contribution to Flower Colors of Flavonoids Including Anthocyanins: A Review. Nat. Prod. Commun. 2015, 10, 529–544. [Google Scholar] [CrossRef]
- Rani, A.; Saini, K.; Bast, F.; Mehariya, S.; Bhatia, S.; Lavecchia, R.; Zuorro, A. Microorganisms: A Potential Source of Bioactive Molecules for Antioxidant Applications. Molecules 2021, 26, 1142. [Google Scholar] [CrossRef] [PubMed]
- Rukmana, R.M.; Silfarohana, R.; Putra, A.D.P.; Safrina, D.; Susanti, D.; Wijaya, N.R.; Adi, M.B.S.; Prastiyanto, M.E.; Yanuar Cahyaningrum, A.; Nurmaulawati, R.; et al. Phytochemical Profile, Antioxidant Activity and Anticancer Activity of Gamma-Irradiated Black Rice Bran (Oryza sativa L.) Ethanolic Extract: In-Vitro and In-Silico Study. Sci. Technol. Indones. 2025, 10, 628–643. [Google Scholar] [CrossRef]
- Jahangiri, A.; Møller, A.H.; Madsen, B.; Joernsgaard, B.; Vaerbak, S.; Danielsen, M.; Dalsgaard, T.K. Incorporation of Bixin in Aqueous Media: Self-Formulation with Sorbitol Ester of Norbixin. Food Chem. 2019, 294, 433–439. [Google Scholar] [CrossRef] [PubMed]
- Cui, H.; Li, X.; Ji, Y.; Zhao, S.; Yang, J. Effects of L-Proline on the Stability of Mulberry Anthocyanins and the Mechanism of Interaction between L-Proline and Cyanidin-3-O-Glycoside. Molecules 2024, 29, 4544. [Google Scholar] [CrossRef]
- El-Metwally, I.M.; Sadak, M.S.; Saudy, H.S. Stimulation Effects of Glutamic and 5-Aminolevulinic Acids On Photosynthetic Pigments, Physio-Biochemical Constituents, Antioxidant Activity, and Yield of Peanut. Gesunde Pflanz. 2022, 74, 915–924. [Google Scholar] [CrossRef]
- Hari, R.K.; Patel, T.R.; Martin, A.M. An Overview of Pigment Production in Biological Systems: Functions, Biosynthesis, and Applications in Food Industry. Food Rev. Int. 1994, 10, 49–70. [Google Scholar] [CrossRef]
- Gadd, G.M.; Fomina, M.; Pinzari, F. Fungal Biodeterioration and Preservation of Cultural Heritage, Artwork, and Historical Artifacts: Extremophily and Adaptation. Microbiol. Mol. Biol. Rev. 2024, 88, e00200-22. [Google Scholar] [CrossRef]
- Graziani, G.; Sassoni, E.; Franzoni, E.; Scherer, G.W. Hydroxyapatite Coatings for Marble Protection: Optimization of Calcite Covering and Acid Resistance. Appl. Surf. Sci. 2016, 368, 241–257. [Google Scholar] [CrossRef]
- Bhattacharjee, A.; Thompson, A.M.; Schwarz, K.C.; Burnet, M.C.; Kim, Y.-M.; Nunez, J.R.; Fansler, S.J.; Farris, Y.; Brislawn, C.J.; Metz, T.O.; et al. Soil Microbial EPS Resiliency Is Influenced by Carbon Source Accessibility. Soil Biol. Biochem. 2020, 151, 108037. [Google Scholar] [CrossRef]
- Maravelaki, P.N. Surface Cleaning: Implications from Choices & Future Perspectives. In Conserving Stone Heritage;Cultural Heritage Science; Gherardi, F., Maravelaki, P.N., Eds.; Springer International Publishing: Cham, Switzerland, 2022; pp. 37–74. ISBN 978-3-030-82941-4. [Google Scholar]
- Sierra-Fernandez, A.; Gomez-Villalba, L.S.; De La Rosa-García, S.C.; Gomez-Cornelio, S.; Quintana, P.; Rabanal, M.E.; Fort, R. Inorganic Nanomaterials for the Consolidation and Antifungal Protection of Stone Heritage. In Advanced Materials for the Conservation of Stone; Hosseini, M., Karapanagiotis, I., Eds.; Springer International Publishing: Cham, Switzerland, 2018; pp. 125–149. ISBN 978-3-319-72259-7. [Google Scholar]
| No. | RT [min] | Formula | Name | Calc. MW | mzVault Best Match | Group Area |
|---|---|---|---|---|---|---|
| 1 | 52.475 | C22H43NO | Erucamide | 337.33428 | 89.8 | 820,227,807 |
| 2 | 47.428 | C16H33NO | Hexadecanamide | 255.25616 | 89.9 | 68,811,881.67 |
| 3 | 1.686 | C12H22O11 | α,αTrehalose | 342.11585 | 97.6 | 159,702,808,1 |
| 4 | 1.626 | C5H11NO2 | Betaine | 117.07903 | 83.4 | 723,338,274.4 |
| 5 | 1.624 | C6H14O6 | D-(-)-Mannitol | 182.07885 | 96.3 | 325,425,455.2 |
| 6 | 56.348 | C22H45NO | Docosanamide | 339.35012 | 88.3 | 27,854,718.91 |
| 7 | 1.662 | C6H12O6 | D-(-)-Fructose | 180.06314 | 87 | 256,867,999 |
| 8 | 1.634 | C7H15NO3 | DLCarnitine | 161.10526 | 95.1 | 204,158,667.1 |
| 9 | 1.615 | C5H9NO4 | LGlutamic acid | 147.05305 | 96.7 | 199,373,258.1 |
| 10 | 1.723 | C9H17NO4 | Acetyl-L-carnitine | 203.11596 | 89.9 | 125,104,158.3 |
| 11 | 9.585 | C10H13N5O4 | Adenosine | 267.09675 | 85.3 | 191,134,604 |
| 12 | 44.672 | C18H30O3S | 4-Dodecylbenzenesulfonic acid | 326.19115 | 90.5 | 75,398,867.34 |
| 13 | 1.524 | C6H14N4O2 | DL-Arginine | 174.11177 | 96.2 | 184,175,321.4 |
| 14 | 1.57 | C5H13NO | Choline | 103.09979 | 88.8 | 169,656,092.2 |
| 15 | 1.631 | C6H12O7 | Gluconic acid | 196.05789 | 95.8 | 132,819,050.8 |
| 16 | 36.801 | C24H30O6 | Bis(4-ethylbenzylidene)sorbitol | 414.20433 | 87.2 | 86,732,885.87 |
| 17 | 1.595 | C5H9NO3 | cis-4-Hydroxy-D-proline | 131.05831 | 86.2 | 100,709,979.1 |
| 18 | 21.662 | C11H18N2O2 | Cyclo(leucylprolyl) | 210.13691 | 91.1 | 90,683,092.63 |
| 19 | 1.735 | C4H6O5 | L-(-)-Malic acid | 134.02131 | 94.5 | 76,300,324.43 |
| 20 | 1.607 | C5H7NO3 | D-(+)-Pyroglutamic Acid | 129.04271 | 72.2 | 65,357,423.24 |
| 21 | 3.545 | C6H8O7 | Citric acid | 192.02679 | 91.1 | 63,419,235.77 |
| 22 | 11.597 | C10H13N5O5 | Guanosine | 283.09151 | 96.5 | 52,569,033.3 |
| 23 | 19.064 | C7H8O3 | 2-Methoxyresorcinol | 140.04743 | 60.3 | 51,097,512 |
| 24 | 1.602 | C5H10N2O3 | L-Glutamine | 146.06928 | 88.9 | 2,705,521.505 |
| 25 | 22.517 | C14H16N2O2 | Cyclo(phenylalanyl-prolyl) | 244.12129 | 89.4 | 47,292,888.72 |
| 26 | 1.712 | C5H9NO2 | 2-Pyrrolidinecarboxylic acid | 115.06343 | 76.8 | 40,601,399.4 |
| 27 | 1.789 | C6H11N3O2 | 2-oxopiperidine-3-carbohydrazide | 157.08533 | 52.6 | 39,392,996.08 |
| 28 | 1.639 | C5H12O5 | D-(+)-Arabitol | 152.06817 | 94.7 | 38,666,127.63 |
| 29 | 1.467 | C9H20N2O2 | N6,N6,N6-Trimethyl-L-lysine | 188.1526 | 80.8 | 34,726,184.93 |
| 30 | 1.485 | C6H9N3O2 | L-Histidine | 155.06964 | 83.5 | 25,785,807.45 |
| 31 | 1.716 | C24H42O21 | Stachyose | 666.22144 | 89.2 | 23,797,225.01 |
| 32 | 25.528 | C15H10O4 | Daidzein | 254.05776 | 90.1 | 15,109,429.33 |
| 33 | 17.986 | C11H9NO2 | trans-3-Indoleacrylic acid | 187.06359 | 93.4 | 19,691,620.98 |
| 34 | 18.098 | C11H15N5O3S | 5′-Methylthioadenosine | 297.08986 | 73.9 | 7,012,109.486 |
| 35 | 18.787 | C7H12O5 | 2-Isopropylmalic acid | 176.06824 | 91.5 | 16,645,708.84 |
| 36 | 28.332 | C15H10O5 | Genistein | 270.05258 | 93.4 | 8,445,606.609 |
| 37 | 5.464 | C9H12N2O6 | Uridine | 244.06917 | 90.5 | 15,068,491.57 |
| 38 | 18.1 | C11H20N2O3 | Leucylproline | 228.14751 | 88.6 | 13,137,288.88 |
| 39 | 7.125 | C6H11NO3 | 4-Acetamidobutanoic acid | 145.07374 | 85.6 | 11,930,711.72 |
| 40 | 17.985 | C11H12N2O2 | L-Tryptophan | 204.09014 | 79.6 | 11,520,779.13 |
| 41 | 16.265 | C7H13NO3 | N-Acetyl-DL-norvaline | 159.08952 | 86.5 | 7,278,669.25 |
| 42 | 1.639 | C10H18N4O6 | Argininosuccinic acid | 290.12375 | 69.6 | 9,767,673.54 |
| 43 | 1.739 | C4H4O4 | Fumaric acid | 116.01068 | 61.6 | 9,065,606.105 |
| 44 | 19.573 | C10H13NO2 | N-Acetyltyramine | 179.09476 | 83.3 | 8,816,265.808 |
| 45 | 1.711 | C12H24O11 | Lactitol | 344.13222 | 83.3 | 8,137,293.08 |
| 46 | 24.416 | C10H9N | 2-Naphthylamine | 143.07359 | 57.6 | 6,885,824.755 |
| 47 | 26.445 | C11H12O5 | Trinexapac | 224.06813 | 88.2 | 6,702,966.477 |
| 48 | 17.986 | C8H7N | Indole | 117.058 | 82.2 | 6,480,463.816 |
| 49 | 32.415 | C10H15NO2S | N-Butylbenzenesulfonamide | 213.08235 | 81.7 | 5,415,251.221 |
| 50 | 1.615 | C6H12O5 | D-(+)-Fucose | 164.06852 | 82.7 | 5,344,430.341 |
| 51 | 25.954 | C16H12O5 | Glycitein | 284.0685 | 92.3 | 5,188,871.246 |
| 52 | 46.771 | C16H32O3 | 16-Hydroxyhexadecanoic acid | 272.23483 | 80.8 | 2,012,426.871 |
| 53 | 3.548 | C5H4O3 | 2-Furoic acid | 112.01576 | 79.4 | 3,584,579.635 |
| 54 | 23.083 | C10H10O4 | Dimethyl Phthalate | 194.05763 | 3,439,623.277 | |
| 55 | 23.469 | C25H24O12 | 4,5-Dicaffeoylquinic acid | 516.1263 | 91.3 | 2,460,159.063 |
| 56 | 20.355 | C6H12O3 | 2-Hydroxycaproic acid | 132.07841 | 93.9 | 2,990,587.119 |
| 57 | 2.719 | C9H13N3O5 | Cytidine | 243.08531 | 64.3 | 2,907,505.221 |
| 58 | 20.207 | C14H18N2O6 | gamma-Glu-Tyr | 310.1164 | 54.3 | 2,736,273.22 |
| 59 | 16.791 | C9H17NO5 | Pantothenic acid | 219.11052 | 89.8 | 2,636,928.59 |
| 60 | 15.798 | C9H11NO2 | Phenylalanine | 165.07873 | 58.7 | 2,552,333.643 |
| 61 | 4.477 | C4H6O4 | Succinic acid | 118.02643 | 90.1 | 2,273,761.998 |
| 62 | 21.068 | C8H8O3 | 3-Methylsalicylic acid | 152.0471 | 89.3 | 1,942,393.343 |
| 63 | 21.895 | C8H14O4 | Suberic acid | 174.08894 | 84 | 1,697,284.831 |
| 64 | 6.062 | C10H15N3O5 | 3′-O-Methylcytidine | 257.10039 | 58.2 | 1,238,264.001 |
| 65 | 2.489 | C8H14O7 | Ethyl-β-Dglucuronide | 222.07362 | 52.4 | 568,154.5422 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, Y.; Liu, J.; Zhou, Y.; Hu, W.; Pan, J.; Zha, J. Research on the Formation Mechanisms of Red Stains on Outdoor Marble Cultural Relics at Beijing Confucian Temple and the Imperial College. Coatings 2025, 15, 1488. https://doi.org/10.3390/coatings15121488
Wang Y, Liu J, Zhou Y, Hu W, Pan J, Zha J. Research on the Formation Mechanisms of Red Stains on Outdoor Marble Cultural Relics at Beijing Confucian Temple and the Imperial College. Coatings. 2025; 15(12):1488. https://doi.org/10.3390/coatings15121488
Chicago/Turabian StyleWang, Yuanyuan, Jiaru Liu, Yi Zhou, Wenjia Hu, Jiao Pan, and Jianrui Zha. 2025. "Research on the Formation Mechanisms of Red Stains on Outdoor Marble Cultural Relics at Beijing Confucian Temple and the Imperial College" Coatings 15, no. 12: 1488. https://doi.org/10.3390/coatings15121488
APA StyleWang, Y., Liu, J., Zhou, Y., Hu, W., Pan, J., & Zha, J. (2025). Research on the Formation Mechanisms of Red Stains on Outdoor Marble Cultural Relics at Beijing Confucian Temple and the Imperial College. Coatings, 15(12), 1488. https://doi.org/10.3390/coatings15121488







