Abstract
Chitosan nanoparticles (CSNPs) have attracted wide interest; however, there has been no substantial information about a direct comparison of the antimicrobial activity of CSNPs on bacteria and fungi. Thus, in this study, simple, economically feasible CSNPs were synthesized and assessed for their antimicrobial activity. This investigation indicated that the coordination inducing effect of CSNPs could dissociate the tryptophan (Trp) and tyrosine (Tyr) residue groups on the peptide chain of the bovine serum albumin (BSA) molecule, thereby increasing the absorption intensity. The growth of E. coli and S. aureus could be completely inhibited when the concentration of CSNPs in the solution was higher than 0.6 mg/mL. The CSNPs showed more potent antibacterial activity against Gram-negative bacteria (E. coli) than against Gram-positive bacteria (S. aureus). In addition, the CSNPs were effective at initiating cellular leakage of fungal mycelia and damping off fungal pathogens, and their antifungal effects were stronger on P. steckii than on A. oryzae. Furthermore, the antimicrobial activity of the CSNPs was found to be more effective against bacteria than against fungi. This study thus ascertained the antimicrobial activity of synthesized CSNPs against different microorganisms, as well as their different degrees of inhibition.
1. Introduction
Worldwide, fruits and vegetables are gaining popularity among consumers because they are healthful and rich in micronutrients. However, microbial growth and oxidative deterioration reduce the freshness, quality, and shelf-life of fresh produce [1]. Strategies are therefore needed to enhance the postharvest life of these products in order to increase their longevity and so meet rising global demand. In order to enhance the shelf-life of fresh fruits and vegetables, plastic packaging is one of the most frequently used postharvest strategies, but it could lead to severe global pollution. In light of recent food and environmental safety concerns, there is now considerable interest in antimicrobial and environmentally friendly food packaging. The employment of bio-based materials to fabricate polymeric composites, instead of petroleum-based ingredients (such as polyethylene, polyvinyl alcohol, or polystyrene), has, thus, attracted substantial attention due to their superior ecological, environmental, safety, and sustainability qualities [2,3]. In this context, chitosan films have shown great promise for their application in food preservation.
A natural biopolymer (a linear polysaccharide comprising 1–4 linked 2-amino-deoxy-β-D-glucan), chitosan is a white, hard, inelastic, and nitrogenous polysaccharide derived by partial deacetylation of chitin [4]. It has good fiber- and film-forming properties and can thus be used to form films for food and pharmaceutical applications, including edible coatings and packaging materials, or as drug-eluting carrier [4,5]. Chitosan is a biodegradable and nontoxic polysaccharose with excellent biocompatibility, making it a great antimicrobial substance approved for use in foods [6]. Furthermore, in comparison to other bio-based food packaging materials, chitosan has the added advantage of antimicrobial activity [6,7], as demonstrated in previous studies against various bacteria and fungi [8,9,10,11]. Hence, chitosan is considered a safe and efficient antimicrobial material.
Chitosan nanoparticles (CSNPs) have both the characteristics of chitosan and the advantageous properties of nanoparticles. Consequently, they can be diffused across biofilm structures and exert stronger antimicrobial effects than bulk materials because of their comparatively large surface area [12,13], while maintaining their qualities of biodegradability, biocompatibility, and non-toxicity [14,15]. According to Xing, et al. [16], CSNPs are capable of providing significant antimicrobial properties through various mechanisms. These include the interaction between positively charged CSNPs and negatively charged plasma membrane phospholipids, the metal chelating potency of CSNPs, and cell wall-penetration capability, as well as DNA inhibition and the consequent hindering of mRNA synthesis. The unique characteristics of nanoparticles, with their small size and quantum size effects, could provide chitosan nanoparticles with superior capabilities [17]. The properties of materials change at the nanoscale. This is because the properties of bulk materials remain relatively constant regardless of their size; however, as their size decreases, the percentage of surface atoms increases, thus providing nanoparticles with certain remarkable properties [18,19]. CSNPs have a broad array of applications in the fields of medicine [20,21,22] and food [23,24,25]. Although the chitosan/tripolyphosphate (TPP) system has been extensively studied, to our knowledge there have been no reports that provide a direct comparison of the antimicrobial activities of CSNPs against bacteria and those against fungi.
Thus, in this work we synthesized and characterized CSNPs and investigated their interaction with bovine serum albumin (BSA) as well as their antimicrobial activity against bacteria (Escherichia coli and Staphylococcus aureus) and fungi (Penicillium steckii and Aspergillus oryzae). The antibacterial activity of CSNPs against E. coli and S. aureus was evaluated through the calculation of minimum inhibitory concentration (MIC) and minimum bactericidal concentration (MBC), growth inhibition, and changes in bacterial membrane protein. The antifungal activity of CSNPs against P. steckii and A. oryzae was evaluated by the calculation of MIC and minimum fungicidal concentration (MFC), growth inhibition, and the conductivity of the fungal cell membrane.
2. Materials and Methods
2.1. Materials
Chitosan (CS, 85.61% deacetylated) was purchased from Jinan Haidebei Marine Biological Engineering Co. Ltd. (Jinan, China); sodium tripolyphosphate and glacial acetic acid were purchased from Chengdu Kelong Chemical Reagent Factory (Chengdu, China); and sodium hydroxide (NaOH) was purchased from Tianjin Continental Chemical Reagent Factory (Tianjin, China). BSA was the BioFroxx brand from neoFroxx (Einhausen, Germany). E. coli and S. aureus were provided by the Institute of Fruit and Vegetable Preservation and Processing, Xihua University, while P. steckii (SICC 3.1177) and A. oryzae (SICC 3.1181) were provided by the Sichuan Center of Industrial Culture Collection (SICC). Bacterial and fungal suspensions with initial densities of 2.5 × 106 and 1 × 105~106 colony-forming units per milliliter (CFU·mL−1), respectively, were prepared and refrigerated at 4 °C. All chemicals were of analytical grade and applied without further purification. Distilled water was used in all experiments.
2.2. Synthesis and Characterization of CSNPs
Synthesis of the CSNPs was conducted according to the method described by Fan, et al. [26] and Fang, et al. [27], with some modifications. The chitosan/TPP nanoparticle suspension was synthesized based on the ionotropic gelation method. First, chitosan solutions (0.76 mg/mL) and TPP solutions (0.77 mg/mL) were equilibrated at room temperature and the pH levels of the chitosan solutions were adjusted to 5.0 with NaOH. Thereafter, the solutions were homogeneously mixed at a chitosan to TPP ratio of 4.11:1 (v/v) at 600 rpm for 10 min. The resulting polymers were spray dried and pre-prepared for the subsequent experiments. The spray drying conditions were: flow rate 120 mL·h−1, drying gas flow 0.35 m3·min−1, spray pressure 130 kPa, inlet temperature 121 °C, outlet temperature 65 °C.
The CSNPs were characterized by micromorphology and thermogravimetric analysis (TGA). The micromorphology was assessed using a microscope (E200, Nanjing Nikon Jiangnan Optical Instrument Co., Ltd., Nanjing, China), while the TGA to determine the thermal stability of the CSNPs was carried out in an N2 atmosphere with a flow rate of 30 mL·min−1 using a thermogravimetric analyzer (DTG-60, Shimazu, Japan). Samples were weighed at approximately 3–5 mg of dry matter. Measurements started at 25 °C and continued up to 800 °C, with a linear increase of 10 °C·min−1 [23].
2.3. Interactions between CSNPs and BSA
The methods of Li, et al. [28] were applied to this assay, with some modifications. First, BSA solution and CSNP solution were diluted with an Na2HPO4-NaH2PO4 solution (pH 6.0) to obtain a BSA solution with a concentration of 1.0 mg/mL and CSNPs solutions with concentrations of 0, 0.25, 0.5, 0.75, 1.0, 1.25, and 1.5 mg/mL. The solutions were shaken at 30 °C and 150 rpm for 30 min in a thermostatic shaker. Thereafter, the ultraviolet-visible (UV-vis) adsorption spectra of the mixed BSA (fixed concentration 1.0 mg/mL) with increasing amounts of CSNPs were recorded in the range of 220–500 nm on an UV-vis spectrophotometer (UV2800, Shanghai Sunny Hengping Scientific Instrument Co., Ltd., Shanghai, China) at room temperature.
2.4. Evaluation of the Effects of CSNPs on Bacterial Membrane Protein
One milliliter of bacterial suspension was added to 3.0 mL of the CSNPs solutions of different concentrations, and these were then shaken at 25 °C and 120 rpm for 1 h, after which the fluorescence intensity was determined. The emission spectra of these samples were scanned from 290 to 500 nm when both of the slit widths of excitation and emission raster were 3 nm, and the excitation wavelength was fixed at 280 nm on a fluorescence spectrometer (FluoroMax-4, HORIBA Scientific, Shanghai, China) [29].
2.5. Minimal Inhibitory Concentrations (MIC) and Minimum Bactericidal Concentration (MBC)
To evaluate the antibacterial potential, the MIC of CSNPs were determined by a 2-fold serial dilution method [30]. The lowest concentration of CSNPs to inhibit the growth of bacteria was considered to be the minimum inhibitory concentration or MIC. In this method, a number of test tubes, each containing 5.0 mL of nutrient broth (NB, Oberstar, Beijing, China), were autoclaved for 15 min at 121 °C. CSNPs were dissolved in an acetic acid (HAc) solution (1.0% v/v) to prepare a CSNP solution with a concentration of 10 mg/mL. While chitosan is only soluble in acetic media, CSNPs are easily dispersed in a distilled water medium for its nice dispersity. Five milliliters of CSNP solution (10 mg/mL) was added and mixed in the first tube, after which 5.0 mL of this mixture was transferred to the next tube, and similar transfers were repeated until each tube contained a test sample solution with half the concentration of the previous one. The tubes were inoculated under aseptic conditions with 100 μL of the freshly prepared bacteria suspension (2.5 × 106 CFU·mL−1). In blank control tubes, deionized water was used instead of the CSNP solution. After mixing, the tubes were incubated in a shaker at 37 °C and 120 rpm for 24 h. The OD600 nm values were measured before and after cultivation in each tube by means of a microplate reader.
Determination of the MBC of CSNP to kill 99.9% of bacteria was performed by assaying the live organisms in those tubes from the MIC test that showed no growth. A loopful (100 μL) from the sample tubes with no obvious change in OD600 nm value was inoculated into plate count agar (PCA), and the tubes were incubated at 37 °C for 24 h and then examined for signs of growth. Growth of bacteria would demonstrate the presence of these bacteria in the original tube. On the contrary, if no growth was observed, then the original tube contained no live bacteria and, therefore, the CSNPs could be deemed bactericidal at that particular concentration [17].
2.6. Evaluation of the Effects of CSNPs on Growth of E. coli and S. aureus
CSNPs solutions were incorporated into NB to obtain various final concentrations: 0.1, 0.2, 0.4, 0.6, and 0.8 mg/mL. The solutions were then sterilized at 121 °C for 20 min. After cooling, 1.0 mL bacterial suspensions were transferred to each group. The groups were shaken at 37 °C and 120 rpm for 24 h and the OD610 nm values measured at intervals. An equal volume of sterile deionized water was used instead of the CSNPs as a control.
2.7. Evaluation of the Effects of CSNPs on the Electrical Conductivity of Mold Cell Membrane
The cellular leakage of P. steckii and A. oryzae fungi following CSNP treatment was measured by determining the electrical conductivity of potato dextrose broth (PDB) containing fungal cultures [31]. Fungal mycelial disks (4 mm) were obtained from the margin of each culture grown on potato dextrose agar (PDA) at 25 ± 1 °C for 5 days and transferred into PDB (20 mL), respectively, followed by incubation at 28 °C for 7 days. At the end of the incubation period, all of the mycelia from the PDB were collected and treated with solutions of different CSNP concentrations. The solutions with CSNPs and fungi were subjected to analysis at different times (1, 2, 3, 4, 5, 6, and 7 h) after incubation. The supernatant was examined using a DDS-11A conductivity meter (Shanghai Lei Chuang Chuangyi Instrument Co., Ltd., Shanghai, China) to analyze cellular leakage. The same volume of deionized water was used as the control [32].
2.8. MIC and Minimum Fungicidal Concentration (MFC)
Both MIC and MFC were investigated by the previously described 2-fold serial dilution method. A CSNP solution with a concentration of 10 mg/mL was prepared and incorporated into PDB resulting in the gradient CSNP solution. Each group was inoculated under aseptic conditions with 100 μL of freshly prepared fungus suspension (1 × 105~106 CFU·mL−1). The blank control tubes contained only deionized water. After mixing, the tubes were incubated at 28 °C for 3 d, after which MIC values were assessed. MFC values were determined by the flat coating method. A loopful (100 μL) from the sample tubes with no obvious change in OD600 nm value was inoculated on PCA (pH 5.6). The tubes were then incubated at 28 °C for 24 h and examined for signs of growth.
2.9. Evaluation of the Effects of CSNPs on the Mycelial Growth of P. steckii and A. oryzae
Two milliliters each of CSNP solutions of various concentrations (0.1, 0.5, 1.0, 3.0, 5.0, and 10 mg/mL) were incorporated into 18 mL PDA, and the mixtures was poured into Petri dishes. After solidification, the respective fungal mycelial disks (6 mm) were obtained from the margin of the cultures grown on PDA, transferred to the center of a solidified PDA Petri dish with the CSNPs, then incubated at 28 °C. The colony diameters of the fungi were measured after culturing for 72, 96, and 120 h. PDA without CSNPs was tested as the negative control [33].
2.10. Statistical Analysis
The tests in this investigation were carried out in triplicate. The test results were analyzed using SPSS 20.0 software (SPSS Inc., Beijing, China). The one-way analysis of variance (ANOVA), followed by the Student-Newman-Keuls test, was used to determine significant differences (p < 0.05) between treatment means.
3. Results and Discussion
3.1. Microscopic Morphology and Thermal Stability of CSNPs
A biological microscope was used to observe the microscopic morphology of CSNP powder prepared by the ion crosslinking method. As shown in Figure 1, the CSNPs were pure white and in the shape of uniform spheres with few impurities observed under the microscope. The thermal stability of the CSNPs was analyzed by TGA. Herein, the TGA curves of the CSNPs displayed two distinct stages of weight loss (Figure 2). The first stage occurred at between 30 and 120 °C and showed about 9.1% weight loss, due to the loss of free and bound water on the materials. The second stage started at 354.8 °C and continued up to 523.5 °C, during which there was a 29.7% weight loss due to polysaccharide thermal decomposition. Similarly, differential thermal analysis (DTA) of the CSNPs was performed to understand their behavior in the application of thermal energy. These first and second stages of degradation appeared as endothermic peaks at 66.5 and 332.6 °C in the DTA curve, respectively. These results confirmed that the CSNP powder had been successfully prepared by the ion crosslinking method, and that it showed good thermal stability and did not thermally decompose at a temperature lower than 354.8 °C. It was thus deemed suitable for further experimental investigation.
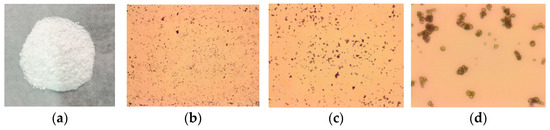
Figure 1.
Microscopic morphology of CSNPs. The first from the left are CSNPs (a), followed by 40× (b), 100× (c), 400× (d).
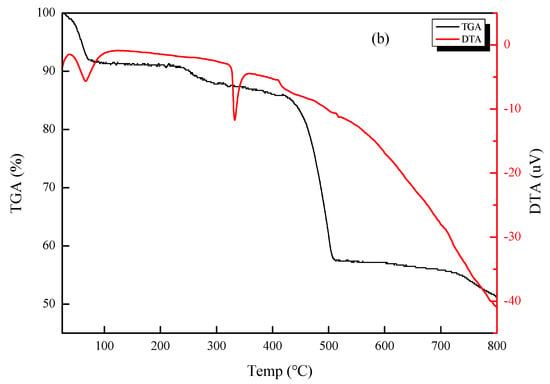
Figure 2.
TGA and DTA curves of CSNPs.
The TGA results of this study concur with those of Antoniou, et al. [23], in which values were presented for two mass loss events, with the first occurring near 100 °C and resulting in a weight loss of approximately 10%; however, the second mass loss event resulted in a weight loss of approximately 60%, a value much higher than ours. Furthermore, the DTA results in this study showed that the two endothermic peaks were compatible with the two mass loss events of the TGA. According to Rejinold et al. [34], polysaccharides usually have a strong affinity for water, and in solid state these macromolecules may have disordered structures that can be easily hydrated. The hydration properties of polysaccharides depend on primary and supramolecular structures. The endothermic peak related to the evaporation of water is expected to reflect the molecular changes brought about after cross-linking. In addition, Rejinold, et al. [34] also mentioned that the cross-linking reaction via TPP modifies the crystalline nature of chitosan, so the degradation profile of the CSNPs seems to be different to that of chitosan and more stable.
3.2. Effect of CSNPs on BSA
Bugnicourt and Ladavière [35] reported that when CSNP-loading proteins (including BSA, ovalbumin (OVA), insulin, antigens, and lysozyme) were prepared via the ionic gelation process to obtain a high encapsulation efficiency (EE), no change was observed in their biological activity. This makes them promising candidates for drug delivery applications [36]. As shown in Figure 3, the UV-vis adsorption spectra of the BSA solutions were affected by the addition of increasing quantities of CSNPs. It was clear that a large adsorption peak appeared around 278 nm, and the adsorption peak position appeared to have a weak blue shift (277.5 nm) when the concentration of the CSNP solution was higher than 0.5 mg/mL. This phenomenon was attributed to the exposure of the phenyl group of Trp and Tyr residues [37], and the shift may suggest that the hydrophobicity around Trp and Tyr residues was weakened, and the energy of π→π* transition increased. Meanwhile, it suggested the formation of the complex with BSA and CSNPs [28,38]. There was a small fluctuation around 325 nm. The control group increased from 0.0215 (324.5 nm) to 0.0237 (325 nm) and then slowly decreased to 0.0234 (325.5 nm), and the 1.5 mg/mL group increased from 0.1344 (324.5 nm) to 0.1537 (325 nm) and then slowly decreased to 0.1539 (325.5 nm). With the increases in CSNP concentrations, increases in adsorption values became more obvious, and the range was between 22–193, while the drop range was between 1–4. In addition, there was a surge between 0 and 0.25 mg/mL due to the addition of CSNPs, after which there were two substantial increases between 0.75 and 1.0 mg/mL and between 1.25 and 1.5 mg/mL. The addition of a certain amount of CSNPs thus triggered a noticeable increase in the adsorption values, suggesting a direct interaction between the CSNPs and BSA.
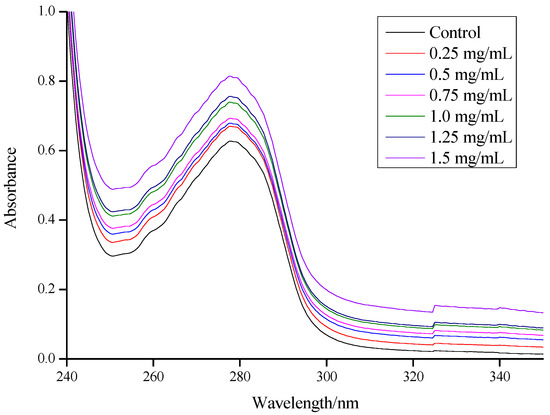
Figure 3.
UV adsorption spectroscopy of BSA and gradient concentrations of CSNP solution mixtures.
UV-vis adsorption measurement is a very simple and applicable method for exploring structural changes and understanding complex formations in ligand-protein investigations. BSA is a model globular protein with a molecular weight of about 66 kDa and one single polypeptide chain contains 583 amino acids [39]. Ascribed to Trp and Tyr residues, BSA has a strong UV adsorption peak in the range of 260–300 nm [40]. In the present study, BSA was used as a protein model and loaded onto the CSNPs via interaction. It was found that the hydrophobic interaction effect could dissociate the Trp and Tyr residue groups on the peptide chain of the BSA molecule and increase the absorption intensity. The formation of a complex between BSA and CSNPs was due to the hydrophobic interaction [28]. The complex formation induced the stretch of the peptide chain of the BSA molecule and further caused exposure of Trp and Tyr residues, so the absorption values increased [41]. As is commonly known, the carrying capacity is displayed as the highest percentage of non-compressible drug in the acceptable drug [42]. The EE is defined by the percentage of the encapsulated material (such as proteins, drug, active ingredients, etc.) detected in the formulation over the total formulation [43]. Zhang, et al. [44] mentioned that in most nanoparticle delivery systems, the drug carrying capacity is defined as an encapsulation efficiency. In our study, BSA was used as a model and carried onto the CSNPs via interaction. In water, the long hydrophilic chains of the complex could extend to the water, while some BSA might have been encapsulated among the positive hydrophilic chains, suggesting that the BSA was not only on the surface of the nanoparticles but also distributed in the outer hydrophilic layer. Therefore, the BSA carrying capacity of the nanoparticles could be termed as EE [44]. According to Yan, et al. [45], the reduced usage of TPP leads to a retarded interaction between the BSA and TPP and a reduced encapsulation of BSA into nanoparticles. Meanwhile, Abdelgawad and Hudson [46] reported that the morphology of the nanoparticles other than size would not be altered for encapsulation of BSA into nanoparticles.
3.3. Effect of CSNPs on Bacterial Membrane Protein
There are high contents of proteins in the membranes of bacteria, the residues of which could emit fluorescence. The more protein residues that are exposed in the environment, the stronger the fluorescence intensity. Changes in these fluorescence spectra could reflect changes in the position of membrane proteins and, indirectly, the conformation change of cell membranes [16]. Results in this study showed that the fluorescence intensity was positively correlated with the concentration of CSNPs, with no significant changes in wavelength of peak positions (Figure 4). The excitation peak of the Trp residues was 340 nm for E. coli, whereas the excitation peak of the Tyr residues was 305 nm and that of the Trp residues was 340 nm for S. aureus. Trp residues play a major role in fluorescence intensity. The endogenous fluorescence intensity of E. coli increased noticeably after CSNP treatment. When the concentration of CSNPs was 0.4 mg/mL, the fluorescence intensity of the E. coli increased sharply to more than 500,000; the increment was more than 80,000. However, after that, as the concentration of CSNPs increased, the fluorescence intensity increased slowly (Figure 4a). As shown in Figure 4b, the fluorescence intensity of S. aureus increased noticeably between 0.6 and 1.5 mg/mL. When the concentration of CSNPs increased from 0.6 to 0.8 mg/mL, the fluorescence intensity of S. aureus increased sharply only about 20,000; however, the rate of increase slowed after 1.5 mg/mL. After CSNPs treatment, the fluorescence intensity of the bacterial cell membrane protein increased, indicating that the CSNPs had changed the structure of the cell membrane protein, exposing Tyr and Trp residues inside the membrane, which resulted in increased fluorescence.
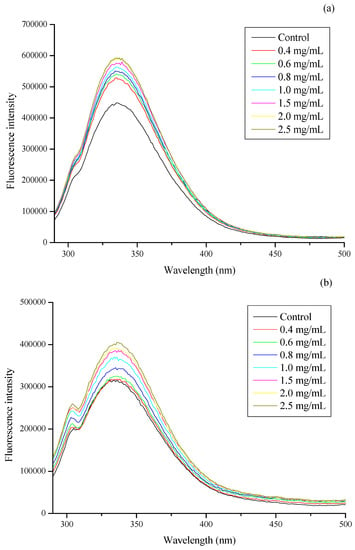
Figure 4.
Effect of gradient concentration of CSNP solutions (mg/mL) and control on the fluorescence intensity of (a) E. coli and (b) S. aureus membrane proteins. The emission spectra of these samples were scanned from 290 to 500 nm when the excitation wavelength was fixed at 280 nm.
The Trp, Tyr, and Phe residue emission peaks were 348, 303, and 282 nm, respectively. Of these, Trp had the highest fluorescence intensity, followed by Tyr, with that of Phe the weakest [47]. The excitation peaks of the Trp and Tyr residues in the membrane protein were determined by scanning the emission wavelength at 300–500 nm. In comparative analysis of the fluorescence intensities of the excitation peaks, it was found that the endogenous fluorescence of E. coli and S. aureus was mainly that of Trp, while the Tyr fluorescence was only a small part of the endogenous fluorescence of S. aureus, thus indicating that the endogenous fluorescence intensity of E. coli was higher than that of S. aureus from the beginning. The effects of CSNPs on the endogenous fluorescence intensity of the E. coli and S. aureus cell membrane proteins were, therefore, different, which may be due to CSNPs having different mechanisms against different microorganisms. In addition, the composition of the cell wall structures of Gram-positive and Gram-negative bacteria is inherently different. The cell wall of Gram-negative bacteria is thinner, but the complexity is higher, and the composition consists of peptidoglycan with various proteins, polysaccharides, and lipids, while in Gram-positive bacteria the composition of the cell wall is mainly peptidoglycan with a small number of proteins [48,49]. According to Antoniou, et al. [23], CSNPs (NH3+) might form a polymer membrane outside the cell wall of Gram-positive bacteria, thereby restraining nutrient and oxygen supplies for the metabolic activity of the bacteria. In Gram-negative bacteria, the CSNPs will diffuse through the cell wall, the disruption of which leads to leakage [50]. Therefore, while there was a sudden increase in the fluorescence intensity of E. coli with the addition of CSNPs, this phenomenon did not occur with S. aureus.
3.4. Effect of CSNPs on E. coli and S. aureus
As previously defined, the MIC is the lowest concentration at which bacteria no longer show visible growth [51]. MBC, which is complementary to MIC, is the lowest concentration at which an antibacterial agent is able to kill 99% of bacteria [52]. According to the data presented in Table 1, CSNPs showed the same antibacterial activity against E. coli and S. aureus, as reflected in the MIC and MBC values of 0.625 and 1.25 mg/mL, respectively. As shown in Figure 5a, the optical density of each experimental group was found to be closely related to the concentration of CSNPs. In the first 3 h, the OD610 nm values of each experimental group were lower than that of the control group, indicating that the CSNPs had a certain inhibitory action on the growth of E. coli at the beginning. After 6 h, the growth curve of the 0.1 mg/mL group was close to that of the control group, which indicated that the inhibitory effect of the CSNPs at this concentration on E. coli was weak. When the CSNP solution concentration reached 0.4 mg/mL, CSNPs showed an obvious antibacterial effect, and the OD610 nm value of the 0.6 mg/mL group tended to be parallel to the horizontal axis. When the concentration reached 0.8 mg/mL, E. coli had been inhibited during the whole experiment. In Figure 5b, it can be seen that, as the concentration of CSNPs increased, the inhibitory effect on S. aureus gradually increased. In the first 3 h, compared with the control group, the low-concentration groups (0.1 mg/mL group and 0.2 mg/mL group) did not inhibit S. aureus well. After 6 h, the 0.2 mg/mL group showed an obvious antibacterial effect; however, the OD610 nm value of the 0.2 mg/mL group dropped at 12 h, and then suddenly rose at 18 h It was not a stable antibacterial trend. The 0.4 mg/mL group showed a downward convex curve, indicating that the antibacterial effect of the CSNP became weaker with the increase of treatment time and the time would be much shorter than other experimental groups. When the concentration of CSNPs was higher than 0.6 mg/mL, the growth of S. aureus was completely inhibited. In addition, after 6 h, both E. coli and S. aureus were completely inhibited when the concentration was higher than 0.6 mg/mL and this result was consistent with the results for the MIC of the CSNPs against E. coli and S. aureus.

Table 1.
The MIC and MBC of CSNPs against E. coli and S. aureus.
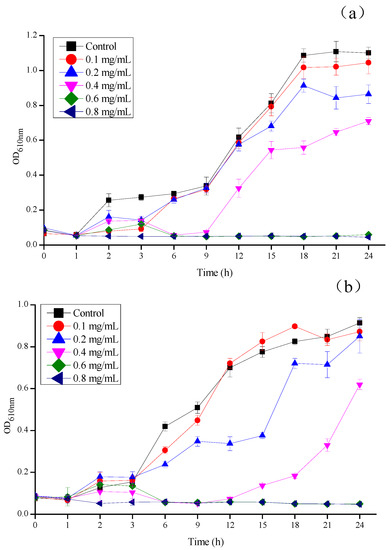
Figure 5.
Growth inhibition of (a) E. coli and (b) S. aureus under different concentrations of CSNP solutions (mg/mL). The growth inhibition was measured as turbidity (OD610 nm) for 24 h at 1 h intervals.
The CSNPs showed strong and stable bacteriostasis during the entire experiment against E. coli. However, against S. aureus, the low-concentration CSNP (0.1 mg/mL group and 0.2 mg/mL group) solutions did not work in a short time and the concentration CSNP solution of 0.4 mg/mL did not have a long antibacterial time. This indicated that the CSNPs showed more potent antibacterial activity against Gram-negative bacteria (E. coli) than against Gram-positive bacteria (S. aureus), whereas the different mechanism led to the different effects. Although there have been numerous attempts to explain the microbial killing mechanism of CSNPs, it remains unclear. Some studies have reported that the interaction of the negatively charged microbial cell wall with chitosan cationic charges leads to the leakage of the intracellular components [17,53,54]. Qi, et al. [17] reported that chitosan has a chelating effect in microorganisms containing media, thus restricting the growth of microorganisms, and also that they can form bonds on the surface of bacterial cells via ionic groups, resulting in the interruption of micronutrient transportation into bacterial cells. According to Hussein, et al. [55], the thickness of the bacterial peptidoglycan layer plays a crucial role in the susceptibility of bacterial cell walls to CSNPs. The peptidoglycan layer of Gram-positive bacteria (7–8 nm) is thinner in comparison to that of Gram-negative bacteria (20–80 nm) [56]. Moreover, Gram-positive bacteria have more anionic binding sites on the outer surface of their cells walls (e.g., teichoic acid, phospholipids, and cardiolipin) [57]. Based on the above statements and results, we suggest that CSNPs might form a polymer membrane outside Gram-positive bacterial cell walls. The polymer membrane could function as a barrier that results in the interruption of essential nutrients and oxygen transport into bacterial cells, and the lack of nutrients and oxygen in the bacterial cells slows down the activities of the microorganisms, thereby inhibiting their growth. By contrast, CSNPs could penetrate the cell walls of Gram-negative bacteria, leading to the leakage of intracellular components.
3.5. Effect on the Conductivity of Mold Cell Membrane
An extracellular conductivity study was carried out to analyze the effect of CSNPs on mold cell membrane. It was observed that cellular electrolyte leakage in the fungal mycelia increased significantly after all CSNP treatments, compared to the control. Moreover, the extracellular conductivity continued to increase as the CSNP concentration in the solutions increased. At the end of the 2.5 mS·cm−1 treatment (7 h), P. steckii and A. oryzae were found to be at 10.50 and 10.84 mS·cm−1, respectively, while the control groups of P. steckii and A. oryzae were measured at 9.77 and 10.27 mS·cm−1, respectively (Figure 6a,b), due to the loss of cell wall/membrane integrity. Furthermore, for P. steckii (Figure 6a), 3 h was found to be the optimal treatment time, whereas 4 h was best for A. oryzae (Figure 6b). After that, the extracellular conductivity curve changed little with the increase of time.

Figure 6.
Effect of different concentration of CSNP solutions (mg/mL) on the extracellular conductivity of (a) P. steckii and (b) A. oryzae after 1, 2, 3, 4, 5, 6, and 7 h of incubation.
The present results indicate that the CSNPs were effective in breaking down the fungal cell membranes. Qi, et al. [17] and Orellano, et al. [53] reported that CSNPs can disrupt microbial cell membranes, leading to the leakage of cytoplasm. Abdallah, et al. [54] reported that the antibacterial activity of CSNPs can, at least partially, be linked to apoptosis, the generation of reactive oxygen species, the destruction or disintegration of cell walls, reductions in biofilm formation, and swimming and leakage of the intracellular contents, and eventually cause cell death. In this study, as shown in Figure 4, CSNPs were found to have an impact on the cell walls and cytoplasmic membranes of bacteria. According to Kritchenkov, et al. [58], the antibacterial activity of all studied chitosan derivatives correlates with that of their antifungal activity, which could be explained by their mechanisms being the same. Moreover, as shown in Figure 6, it is evident that the effect of CSNPs on fungal cell membranes is that of rapid destruction, rather than a gradual process. The leakage of the fungal intracellular contents could thus be attributed to the impact of CSNPs on the fungal cell wall, and may be similar to the mechanism of action of CSNPs on Gram-negative bacteria.
3.6. Effect of CSNPs on P. steckii and A. oryzae
This study analyzed not only the antibacterial properties of CSNPs, but also their antifungal properties. According to the data presented in Table 2, in the concentration range investigated, the MIC value of CSNPs against P. steckii was 5 mg/mL; however, the CSNPs had little antifungal effect on A. oryzae. There was an obvious correspondence between the colony diameters of the fungi and the concentration of the CSNPs (Figure 7). In Figure 7a, it can be seen that all concentrations of CSNPs reduced the fungal mycelial growth in comparison to the control. As the concentration of CSNPs in the solutions increased, so the inhibitory effect of the CSNPs on mycelial growth became more severe. After culturing for 120 h, the CSNPs still had an inhibitory effect on the growth of P. steckii, which changed significantly under different concentrations of treatment. Figure 7b shows that the CSNPs had a slight inhibitory effect on the mycelial growth of A. oryzae. These results are consistent with those of the MIC and MFC tests. In the investigated concentration range, the CSNPs showed more potent antifungal activity against P. steckii than A. oryzae. Furthermore, the results of the MIC and MFC/MBC of the CSNPs showed clearly that the antimicrobial activity of CSNPs is stronger against bacteria than it is against fungi. Kim, et al. [59] also reported that bacteria were more susceptible to CSNPs than fungi. According to Verlee, et al. [60], in contrast to Gram-positive and Gram-negative bacteria, the difference between chitosan-sensitive and chitosan-resistant fungi is less pronounced.

Table 2.
The MIC and MFC of CSNPs against P. steckii and A. oryzae.
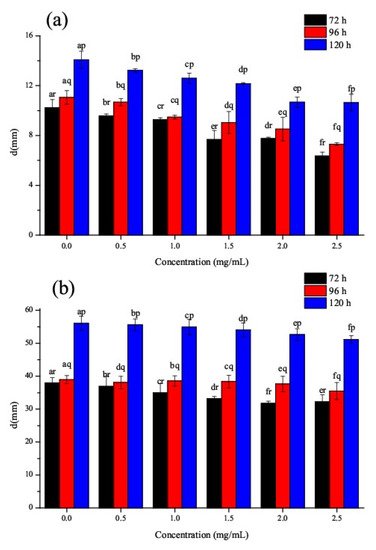
Figure 7.
Effect of gradient concentrations of CSNP solutions (mg/mL) on mycelial growth of (a) P. steckii and (b) A. oryzae after 72, 96, and 120 h of incubation.
According to MubarakAli, et al. [61], the negative charge on the surface of fungal membranes might decrease due to the strong binding of chitosan to fungal cells. This change mainly decreases the actual concentration of K+, thus altering the state of balance, resulting in a K+ efflux. Firstly, chitosan interacts electrostatically with negatively charged phospholipids to affect cell membranes [62]. Then, the cell membrane is disrupted by chitosan, causing it to enter the cell [62,63,64], which could lead to the inhibition of DNA/RNA synthesis [65] and the disruption of protein synthesis [66]. This explains why a higher concentration of CSNPs is required for the fungicidal activity against P. steckii and A. oryzae. Furthermore, chitosan or chitosan derivatives have also shown antifungal activity against other fungi, such as C. albicans [13,53], A. fumigatus [58], and Xanthomonas oryzae pv. oryzae [54].
4. Conclusions
In this study, CSNPs were synthesized and characterized in order to investigate their interaction with BSA cell membrane protein and their in vitro antimicrobial activity against bacteria (E. coli and S. aureus) and fungi (P. steckii and A. oryzae). The results suggested a direct interaction between CSNPs and BSA. In bacterial tests, the CSNPs showed more potent antibacterial activity against Gram-negative bacteria (E. coli) than against Gram-positive bacteria (S. aureus). In fungal tests, the CSNPs showed a stronger antifungal effect on P. steckii than on A. oryzae and could disrupt the fungal cell membranes, causing the leakage of cytoplasm and, eventually, cell death. In addition, a comparative analysis of the MIC and MFC/MBC of the CSNPs showed clearly that their antimicrobial activity was stronger against bacteria than it was against fungi. It remains unclear, however, why bacteria are more susceptible to CSNPs than fungi; a more detailed investigation into the mechanism involved in the antimicrobial action of CSNPs is required. Goy, et al. [65] mentioned that chitosan might lead to the inhibition of DNA/RNA synthesis once chitosan enters the cell. Therefore, we have considered studying the antimicrobial mechanism of CSNPs at the genetic level. For example, could CSNPs lead to the inhibition of DNA/RNA synthesis or the expression of genes involved in essential nutrient transport and metabolism of microorganisms, and what pathways are involved?
Author Contributions
Administration: J.Y. and Y.X.; funding acquisition: J.Y. and Y.X.; conceptualization: C.C. and X.B.; investigation: Y.S., Q.X., and X.L. (Xiaocui Liu); methodology: X.W., X.G., and X.L. (Xuanlin Li); data curation: X.G., P.Y., and L.X.; writing: X.W. and Y.X. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by Key Laboratory of Storage of Agricultural Products, Ministry of Agriculture and Rural Affairs (kf202006), the Science and technology support program of Sichuan (2019YFN0174, 2018NZ0090, 2019NZZJ0028, and 2017NFP0030), Science and technology support program of Yibin (2018ZSF002), Chengdu Science and Technology Project- key research and development program (2019-YF05-00628-SN and 2019-YF05-00190-SN), Xihua University Graduate Innovation Fund Project (ycjj2017153 and ycjj2019082), College Students innovation and entrepreneurship training program of Xihua University (201710623081), and Innovation Team Construction Program of Sichuan Education Department (15TD0017).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
The data presented in this study are available in the article.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Kumar, S.; Mukherjee, A.; Dutta, J. Chitosan based nanocomposite films and coatings: Emerging antimicrobial food packaging alternatives. Trends Food Sci. Technol. 2020, 97, 196–209. [Google Scholar] [CrossRef]
- Garavand, F.; Rouhi, M.; Razavi, S.H.; Cacciotti, I.; Mohammadi, R. Improving the integrity of natural biopolymer films used in food packaging by crosslinking approach: A review. Int. J. Biol. Macromol. 2017, 104, 687–707. [Google Scholar] [CrossRef] [PubMed]
- Mirzaei-Mohkam, A.; Garavand, F.; Dehnad, D.; Keramat, J.; Nasirpour, A. Optimisation, antioxidant attributes, stability and release behaviour of carboxymethyl cellulose films incorporated with nanoencapsulated vitamin E. Prog. Org. Coat. 2019, 134, 333–341. [Google Scholar] [CrossRef]
- Dedloff, M.R.; Effler, C.S.; Holban, A.M.; Gestal, M.C. Use of biopolymers in mucosally-administered vaccinations for respiratory disease. Materials 2019, 12, 2445. [Google Scholar] [CrossRef] [Green Version]
- Kaewklin, P.; Siripatrawan, U.; Suwanagul, A.; Lee, Y.S. Active packaging from chitosan-titanium dioxide nanocomposite film for prolonging storage life of tomato fruit. Int. J. Biol. Macromol. 2018, 112, 523–529. [Google Scholar] [CrossRef] [PubMed]
- Lei, J.; Yang, L.; Zhan, Y.; Wang, Y.; Ye, T.; Li, Y.; Deng, H.; Li, B. Plasma treated polyethylene terephthalate/polypropylene films assembled with chitosan and various preservatives for antimicrobial food packaging. Colloids Surf. B Biointerfaces 2014, 114, 60–66. [Google Scholar] [CrossRef] [PubMed]
- Dutta, P.K.; Tripathi, S.; Mehrotra, G.K.; Dutta, J. Perspectives for chitosan based antimicrobial films in food applications. Food Chem. 2009, 114, 1173–1182. [Google Scholar] [CrossRef]
- Nguyen, T.V.; Nguyen, T.T.H.; Wang, S.-L.; Vo, T.P.K.; Nguyen, A.D. Preparation of chitosan nanoparticles by TPP ionic gelation combined with spray drying, and the antibacterial activity of chitosan nanoparticles and a chitosan nanoparticle–amoxicillin complex. Res. Chem. Intermed. 2016, 43, 3527–3537. [Google Scholar] [CrossRef]
- Li, B.; Zhang, Y.; Yang, Y.; Qiu, W.; Wang, X.; Liu, B.; Wang, Y.; Sun, G. Synthesis, characterization, and antibacterial activity of chitosan/TiO2 nanocomposite against Xanthomonas oryzae pv. oryzae. Carbohydr. Polym. 2016, 152, 825–831. [Google Scholar] [CrossRef] [PubMed]
- Huang, W.; Xu, H.; Xue, Y.; Huang, R.; Deng, H.; Pan, S. Layer-by-layer immobilization of lysozyme–chitosan–organic rectorite composites on electrospun nanofibrous mats for pork preservation. Food Res. Int. 2012, 48, 784–791. [Google Scholar] [CrossRef]
- Rabea, E.I.; Badawy, M.E.-T.; Stevens, C.V.; Smagghe, G.; Steurbaut, W. Chitosan as antimicrobial agent: Applications and mode of action. Biomacromolecules 2003, 4, 1457–1465. [Google Scholar] [CrossRef]
- Shukla, S.K.; Mishra, A.K.; Arotiba, O.A.; Mamba, B.B. Chitosan-based nanomaterials: A state-of-the-art review. Int. J. Biol. Macromol. 2013, 59, 46–58. [Google Scholar] [CrossRef] [PubMed]
- Ing, L.Y.; Zin, N.M.; Sarwar, A.; Katas, H. Antifungal activity of chitosan nanoparticles and correlation with their physical properties. Int. J. Biomater. 2012, 2012, 632698. [Google Scholar] [CrossRef] [PubMed]
- Das, R.K.; Kasoju, N.; Bora, U. Encapsulation of curcumin in alginate-chitosan-pluronic composite nanoparticles for delivery to cancer cells. Nanomedicine 2010, 6, 153–160. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Li, X.; Li, K.; Tao, T. Plasmas ozone inactivation of Legionella in deionized water and wastewater. Environ. Sci. Pollut. Res. Int. 2018, 25, 9697–9707. [Google Scholar] [CrossRef]
- Xing, K.; Chen, X.G.; Liu, C.S.; Cha, D.S.; Park, H.J. Oleoyl-chitosan nanoparticles inhibits Escherichia coli and Staphylococcus aureus by damaging the cell membrane and putative binding to extracellular or intracellular targets. Int. J. Food Microbiol. 2009, 132, 127–133. [Google Scholar] [CrossRef] [PubMed]
- Qi, L.; Xu, Z.; Jiang, X.; Hu, C.; Zou, X. Preparation and antibacterial activity of chitosan nanoparticles. Carbohydr. Res. 2004, 339, 2693–2700. [Google Scholar] [CrossRef] [PubMed]
- Divya, K.; Jisha, M.S. Chitosan nanoparticles preparation and applications. Environ. Chem. Lett. 2017, 16, 101–112. [Google Scholar] [CrossRef]
- Hosseinnejad, M.; Jafari, S.M. Evaluation of different factors affecting antimicrobial properties of chitosan. Int. J. Biol. Macromol. 2016, 85, 467–475. [Google Scholar] [CrossRef] [PubMed]
- Jarmila, V.; Vavrikova, E. Chitosan derivatives with antimicrobial, antitumour and antioxidant activities—A review. Curr. Pharm. Des. 2011, 17, 3596–3607. [Google Scholar] [CrossRef]
- Ghadi, A.; Mahjoub, S.; Tabandeh, F.; Talebnia, F. Synthesis and optimization of chitosan nanoparticles: Potential applications in nanomedicine and biomedical engineering. Caspian J. Intern. Med. 2014, 5, 156–161. [Google Scholar]
- Janes, K.A.; Fresneau, M.P.; Marazuela, A.; Fabra, A.; Alonso, M.J. Chitosan nanoparticles as delivery systems for doxorubicin. J. Control. Release 2001, 73, 2–3. [Google Scholar] [CrossRef]
- Antoniou, J.; Liu, F.; Majeed, H.; Zhong, F. Characterization of tara gum edible films incorporated with bulk chitosan and chitosan nanoparticles: A comparative study. Food Hydrocoll. 2015, 44, 309–319. [Google Scholar] [CrossRef]
- Ramezani, Z.; Zarei, M.; Raminnejad, N. Comparing the effectiveness of chitosan and nanochitosan coatings on the quality of refrigerated silver carp fillets. Food Control. 2015, 51, 43–48. [Google Scholar] [CrossRef]
- Amjadi, S.; Nazari, M.; Alizadeh, S.A.; Hamishehkar, H. Multifunctional betanin nanoliposomes-incorporated gelatin/chitosan nanofiber/ZnO nanoparticles nanocomposite film for fresh beef preservation. Meat Sci. 2020, 167, 108161. [Google Scholar] [CrossRef] [PubMed]
- Fan, W.; Yan, W.; Xu, Z.; Ni, H. Formation mechanism of monodisperse, low molecular weight chitosan nanoparticles by ionic gelation technique. Colloids Surf. B Biointerfaces 2012, 90, 21–27. [Google Scholar] [CrossRef]
- Fang, Y.; Xing, C.; Liu, J.; Zhang, Y.; Li, M.; Han, Q. Supermolecular film crosslinked by polyoxometalate and chitosan with superior antimicrobial effect. Int. J. Biol. Macromol. 2020, 154, 732–738. [Google Scholar] [CrossRef]
- Li, G.; Huang, J.; Chen, T.; Wang, X.; Zhang, H.; Chen, Q. Insight into the interaction between chitosan and bovine serum albumin. Carbohydr. Polym. 2017, 176, 75–82. [Google Scholar] [CrossRef]
- Mei, L.; Xu, Z.; Shi, Y.; Lin, C.; Jiao, S.; Zhang, L.; Li, P. Multivalent and synergistic chitosan oligosaccharide-Ag nanocomposites for therapy of bacterial infection. Sci. Rep. 2020, 10, 10011. [Google Scholar] [CrossRef] [PubMed]
- Lee, D.S.; Woo, J.Y.; Ahn, C.B.; Je, J.Y. Chitosan-hydroxycinnamic acid conjugates: Preparation, antioxidant and antimicrobial activity. Food Chem. 2014, 148, 97–104. [Google Scholar] [CrossRef] [PubMed]
- Manhas, R.K.; Kaur, T. Biocontrol potential of Streptomyces hydrogenans strain DH16 toward Alternaria brassicicola to control damping off and black leaf spot of Raphanus sativus. Front. Plant Sci. 2016, 7, 1869. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vanti, G.L.; Masaphy, S.; Kurjogi, M.; Chakrasali, S.; Nargund, V.B. Synthesis and application of chitosan-copper nanoparticles on damping off causing plant pathogenic fungi. Int. J. Biol. Macromol. 2020, 156, 1387–1395. [Google Scholar] [CrossRef] [PubMed]
- Gomes, A.C.A.; da Costa Lima, M.; de Oliveira, K.A.R.; Dos Santos Lima, M.; Magnani, M.; Camara, M.P.S.; de Souza, E.L. Coatings with chitosan and phenolic-rich extract from acerola (Malpighia emarginata D.C.) or jabuticaba (Plinia jaboticaba (Vell.) Berg) processing by-product to control rot caused by Lasiodiplodia spp. in papaya (Carica papaya L.) fruit. Int. J. Food Microbiol. 2020, 331, 108694. [Google Scholar] [CrossRef] [PubMed]
- Rejinold, N.S.; Muthunarayanan, M.; Muthuchelian, K.; Chennazhi, K.P.; Nair, S.V.; Jayakumar, R. Saponin-loaded chitosan nanoparticles and their cytotoxicity to cancer cell lines in vitro. Carbohydr. Polym. 2011, 84, 407–416. [Google Scholar] [CrossRef]
- Bugnicourt, L.; Ladavière, C. Interests of chitosan nanoparticles ionically cross-linked with tripolyphosphate for biomedical applications. Prog. Polym. Sci. 2016, 60, 1–17. [Google Scholar] [CrossRef]
- Almalik, A.; Alradwan, I.; Kalam, M.A.; Alshamsan, A. Effect of cryoprotection on particle size stability and preservation of chitosan nanoparticles with and without hyaluronate or alginate coating. Saudi Pharm. J. 2017, 25, 861–867. [Google Scholar] [CrossRef]
- Kandori, K.; Uoya, Y.; Ishikawa, T. Effects of acetonitrile on adsorption behavior of bovine serum albumin onto synthetic calcium hydroxyapatite particles. J. Colloid Interface Sci. 2002, 252, 269–275. [Google Scholar] [CrossRef]
- Li, X.F.; Feng, X.Q.; Yang, S. Synthesis, antibacterial activity and effect of O-carboxymethyl chitosan on cell membrane protein. Food Ind. Tech. 2010, 31, 143–145. [Google Scholar] [CrossRef]
- Ru, Q.; Wang, Y.; Lee, J.; Ding, Y.; Huang, Q. Turbidity and rheological properties of bovine serum albumin/pectin coacervates: Effect of salt concentration and initial protein/polysaccharide ratio. Carbohyd. Polym. 2012, 88, 838–846. [Google Scholar] [CrossRef]
- Wang, Z.H.; Zeng, R.; Tu, M.; Zhao, J.H. A novel biomimetic chitosan-based nanocarrier with suppression of the protein-nanocarrier interactions. Mater. Lett. 2012, 77, 38–40. [Google Scholar] [CrossRef]
- Huang, Y.; Liu, Y.; Zhang, Q.; Jia, C.M.; Hua, M.Q.; Cao, J.; Zhu, W.H. Interaction between low molecular weight chitosan derivatives and bovine serum album. Spectrosc. Spect. Anal. 2017, 37, 1814–1818. [Google Scholar] [CrossRef]
- Mitrevej, A.; Sinchaipanid, N.; Faroongsarng, D. Spray-dried rice starch: Comparative evaluation of direct compression fillers. Drug Dev. Ind. Pharm. 1996, 22, 587–594. [Google Scholar] [CrossRef]
- Piacentini, E. Encapsulation Efficiency. In Encyclopedia of Membranes; Drioli, E., Giorno, L., Eds.; Springer: Berlin/Heidelberg, Germany, 2016; pp. 706–707. [Google Scholar]
- Zhang, H.-L.; Wu, S.-H.; Tao, Y.; Zang, L.-Q.; Su, Z.-Q. Preparation and characterization of water-soluble chitosan nanoparticles as protein delivery system. J. Nanomater. 2010, 2010, 898910. [Google Scholar] [CrossRef]
- Yan, Q.; Weng, J.; Wu, X.; Wang, W.; Yang, Q.; Guo, F.; Wu, D.; Song, Y.; Chen, F.; Yang, G. Characteristics, cryoprotection evaluation and in vitro release of BSA-loaded chitosan nanoparticles. Mar. Drugs 2020, 18, 315. [Google Scholar] [CrossRef] [PubMed]
- Abdelgawad, A.M.; Hudson, S.M. Chitosan nanoparticles: Polyphosphates cross-linking and protein delivery properties. Int. J. Biol. Macromol. 2019, 136, 133–142. [Google Scholar] [CrossRef]
- Tao, W.S.; Li, W.; Jiang, Y.M. Protein Molecular Foundation; Higher Education Press: Beijing, China, 1995; pp. 260–262. [Google Scholar]
- Burt, S. Essential oils: Their antibacterial properties and potential applications in foods—A review. Int. J. Food Microbiol. 2004, 94, 223–253. [Google Scholar] [CrossRef]
- Vahedikia, N.; Garavand, F.; Tajeddin, B.; Cacciotti, I.; Jafari, S.M.; Omidi, T.; Zahedi, Z. Biodegradable zein film composites reinforced with chitosan nanoparticles and cinnamon essential oil: Physical, mechanical, structural and antimicrobial attributes. Colloids Surf. B Biointerfaces 2019, 177, 25–32. [Google Scholar] [CrossRef]
- Shapi’i, R.A.; Othman, S.H.; Nordin, N.; Kadir Basha, R.; Nazli Naim, M. Antimicrobial properties of starch films incorporated with chitosan nanoparticles: In vitro and in vivo evaluation. Carbohydr. Polym. 2020, 230, 115602. [Google Scholar] [CrossRef]
- Diao, Y.; Yu, X.; Zhang, C.; Jing, Y. Quercetin-grafted chitosan prepared by free radical grafting: Characterization and evaluation of antioxidant and antibacterial properties. J. Food Sci. Technol. 2020, 57, 2259–2268. [Google Scholar] [CrossRef]
- Abedian, Z.; Jenabian, N.; Moghadamnia, A.A.; Zabihi, E.; Tashakorian, H.; Rajabnia, M.; Sadighian, F.; Bijani, A. Antibacterial activity of high-molecular-weight and low-molecular-weight chitosan upon oral pathogens. J. Conserv. Dent. 2019, 22, 169–174. [Google Scholar] [CrossRef] [PubMed]
- Orellano, M.S.; Isaac, P.; Breser, M.L.; Bohl, L.P.; Conesa, A.; Falcone, R.D.; Porporatto, C. Chitosan nanoparticles enhance the antibacterial activity of the native polymer against bovine mastitis pathogens. Carbohydr. Polym. 2019, 213, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Abdallah, Y.; Liu, M.; Ogunyemi, S.O.; Ahmed, T.; Fouad, H.; Abdelazez, A.; Yan, C.; Yang, Y.; Chen, J.; Li, B. Bioinspired green synthesis of chitosan and zinc oxide nanoparticles with strong antibacterial activity against rice pathogen Xanthomonas oryzae pv. oryzae. Molecules 2020, 25, 4795. [Google Scholar] [CrossRef]
- Mohamady Hussein, M.A.; Banos, F.G.D.; Grinholc, M.; Abo Dena, A.S.; El-Sherbiny, I.M.; Megahed, M. Exploring the physicochemical and antimicrobial properties of gold-chitosan hybrid nanoparticles composed of varying chitosan amounts. Int. J. Biol. Macromol. 2020, 162, 1760–1769. [Google Scholar] [CrossRef] [PubMed]
- Eaton, P.; Fernandes, J.C.; Pereira, E.; Pintado, M.E.; Xavier Malcata, F. Atomic force microscopy study of the antibacterial effects of chitosans on Escherichia coli and Staphylococcus aureus. Ultramicroscopy 2008, 108, 1128–1134. [Google Scholar] [CrossRef]
- Matica, M.A.; Aachmann, F.L.; Tondervik, A.; Sletta, H.; Ostafe, V. Chitosan as a wound dressing starting material: Antimicrobial properties and mode of action. Int. J. Mol. Sci. 2019, 20, 5889. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kritchenkov, A.S.; Zhaliazniak, N.V.; Egorov, A.R.; Lobanov, N.N.; Volkova, O.V.; Zabodalova, L.A.; Suchkova, E.P.; Kurliuk, A.V.; Shakola, T.V.; Rubanik, V.V., Jr.; et al. Chitosan derivatives and their based nanoparticles: Ultrasonic approach to the synthesis, antimicrobial and transfection properties. Carbohydr. Polym. 2020, 242, 116478. [Google Scholar] [CrossRef]
- Kim, Y.H.; Kim, G.H.; Yoon, K.S.; Shankar, S.; Rhim, J.W. Comparative antibacterial and antifungal activities of sulfur nanoparticles capped with chitosan. Microb. Pathog. 2020, 144, 104178. [Google Scholar] [CrossRef] [PubMed]
- Verlee, A.; Mincke, S.; Stevens, C.V. Recent developments in antibacterial and antifungal chitosan and its derivatives. Carbohydr. Polym. 2017, 164, 268–283. [Google Scholar] [CrossRef]
- MubarakAli, D.; LewisOscar, F.; Gopinath, V.; Alharbi, N.S.; Alharbi, S.A.; Thajuddin, N. An inhibitory action of chitosan nanoparticles against pathogenic bacteria and fungi and their potential applications as biocompatible antioxidants. Microb. Pathog. 2018, 114, 323–327. [Google Scholar] [CrossRef]
- Palma-Guerrero, J.; Lopez-Jimenez, J.A.; Perez-Berna, A.J.; Huang, I.C.; Jansson, H.B.; Salinas, J.; Villalain, J.; Read, N.D.; Lopez-Llorca, L.V. Membrane fluidity determines sensitivity of filamentous fungi to chitosan. Mol. Microbiol. 2010, 75, 1021–1032. [Google Scholar] [CrossRef]
- Palma-Guerrero, J.; Jansson, H.B.; Salinas, J.; Lopez-Llorca, L.V. Effect of chitosan on hyphal growth and spore germination of plant pathogenic and biocontrol fungi. J. Appl. Microbiol. 2008, 104, 541–553. [Google Scholar] [CrossRef] [PubMed]
- Palma-Guerrero, J.; Huang, I.C.; Jansson, H.B.; Salinas, J.; Lopez-Llorca, L.V.; Read, N.D. Chitosan permeabilizes the plasma membrane and kills cells of Neurospora crassa in an energy dependent manner. Fungal Genet. Biol. 2009, 46, 585–594. [Google Scholar] [CrossRef] [PubMed]
- Goy, R.C.; de Britto, D.; Assis, O.B.G. A Review of the antimicrobial activity of chitosan. Polimeros 2009, 19, 241–247. [Google Scholar] [CrossRef]
- Galvan Marquez, I.; Akuaku, J.; Cruz, I.; Cheetham, J.; Golshani, A.; Smith, M.L. Disruption of protein synthesis as antifungal mode of action by chitosan. Int. J. Food Microbiol. 2013, 164, 108–112. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).