1. Introduction
Extensive laboratory evidence has demonstrated the antibacterial properties of copper alloys, which has led to several field trials aimed at providing a proof of concept, particularly in clinical settings [
1,
2,
3]. A multihospital clinical trial of six US Environmental Protection Agency (EPA)-registered antimicrobial copper alloys found that the microbial burden of copper alloy surfaces was six times lower (465 CFU/100 cm
2) than that of conventional surfaces such as plastics, coated carbon steel, aluminum and stainless steel (2674 CFU/100 cm
2) [
4]. The microbial burdens on both the copper alloy and conventional surfaces were above those proposed as harmless on a surface immediately after cleaning (250 CFU/100 cm
2) [
5]. However, the results of the trial showed a reduction in the rate of infections of 58% in “copper” rooms, compared to the “non-copper” rooms.
In a Finnish study, door handles made of copper alloys (99.8 wt.% Cu) and brass (60.5 wt.% Cu, 36.5 wt.% Zn) were installed in a hospital, a kindergarten, a retirement home and an office building, and microbial levels were compared to reference chromed door handles [
3]. In terms of total aerobic plate count, door handles made of copper alloys outperformed those made of brass, which, in turn, did not (on average) show significant differences with the chromed material [
3]. Lower levels of both Gram-negative bacteria and
Staphylococcus aureus were found on copper alloy surfaces than on brass and reference surfaces.
Door handle surfaces have the highest levels of bacterial contamination in clinical environments [
6]. Recently published studies have tested a newly developed copper-silver alloy coating and found high antibacterial efficacy against
S. aureus MSSA and MRSA,
Pseudomonas aeruginosa,
Escherichia coli and
Enterobacter aerogenes [
7,
8]. Based on these findings, the present study aims to investigate the antibacterial efficacy of this copper-silver alloy coating on door handles in a private clinic, FamilieLægerne Espergærde, and a wound care center, Southwest Regional (SWR) Wound Care Center. In addition, it isolates and identifies microorganisms from the surfaces of interest to determine any species-specific effects. Finally, it evaluates the durability of the copper-silver alloy coating under the conditions tested.
2. Materials and Methods
2.1. Manufacturing and Installation of Door Handles
The stainless-steel door handles (Ruko Assa Abloy, Stockholm, Sweden) were electroplated with a copper-silver alloy coating at Elplatek A/S [
7]. In the private clinic FamilieLægerne Espergærde (Egeskovvej 20, 3490 Kvistgård, DK), copper-silver alloy coated door handles (hereafter referred to as “test door handles”) were installed on the doors of two doctors’ and two nurses’ exam rooms. Stainless steel door handles of four other offices were used as a reference material and sampled for microorganisms. Weekly sampling was performed for six weeks. At the SWR Wound Care Center (2002 Oxford Ave, Lubbock, TX 79410, USA), test door handles were installed on the doors of seven exam rooms, one public restroom and two laboratory rooms. The original satin brass door handles of six other exam rooms, three public restrooms and one laboratory room were sampled as reference material. Weekly sampling was performed for six weeks. All reference and test door handles were disinfected with 70% ethanol when the field trials started. Samplings were performed by the same person between 7.45 am and 8.00 am on Thursdays. The routine cleaning of door handles at the private clinic and the wound care center was carried out by wiping the surfaces with a dry cloth on Mondays.
2.2. Microbiological Sampling and Matrix-Assisted Laser Desorption Ionization Time-of-Flight Mass Spectrometry (MALDI-TOF MS) Analysis
In the private clinic in Denmark, every week the door handle surfaces (100 cm2) were swabbed thoroughly (horizontal and vertical sweeps) with a flocked, sterile swab applicator (BD™ ESwab Regular Collection Kit, Franklin Lakes, NJ, USA). The swab was inserted into a sampling tube, which contained 1 mL of Liquid Amies Medium, and samples were transported to the laboratory at the Technical University of Denmark within 1 h. The sampling tubes were sonicated for 2 min at 28 kHz (Delta 220; Deltasonic, Meaux, France) and vortexed for 15 s. Four-hundred µL of the sampling suspensions were plated in duplicates on 5% blood agar (BA) plates (BD™, Franklin Lakes, NJ, USA). The plates were incubated at 37 ± 1 °C, and a total aerobic plate count was performed after 48 h. Colony-forming units (CFU) per plate corresponded to CFU per door handle surface (100 cm2). Average values of CFU/100 cm2 for test and reference door handles were log-transformed and presented as total average values ± standard deviation (SD). All isolates collected from the test door handles at the third and last sampling were re-streaked on BA plates, and single colonies were stored for later identification at −80 °C in a freezing medium (Tryptone Soy Broth 30 g/L, Glucose 5 g/L, Skim milk powder 20 g/L, Glycerol 40 g/L in distilled water). Single colonies were also randomly selected from the reference door handles and stored in the same way. Identification of microbial species was performed using MALDI-TOF MS on a Microflex LT instrument (Bruker Daltonik GmbH, Bremen, Germany). Protein profiles were acquired with the FlexControl 3.3 software (Bruker Daltonik GmbH, Bremen, Germany) and analyzed with FlexAnalysis 3.3 (Bruker Daltonik GmbH, Bremen, Germany). The database used to match spectra was Bruker Taxonomy (7311 MSPs). MALDI-TOF MS scores x > 2.0 were used to identify isolates to species level, while scores of x = 1.8–2.0 were used to identify them to genus level. Scores of x < 1.8 were not considered in this study.
2.3. Microbiological Sampling and Analyses of Bacterial Load by Direct Sequence Analysis
Microbiological sampling and analyses were performed on-site at the medical laboratory at the SWR Wound Care Center, which is accredited by the College of American Pathologists (CAP). Every week, the door handle surfaces (100 cm2) were swabbed thoroughly (horizontal and vertical sweeps) with a sterile cotton swab. Cotton swabs were inserted into 2 mL sterile screw cap microtubes, and 500 µL of phosphate-buffered saline solution (PBS; Dulbecco A; Oxoid) was added. Bacteria were detached from the cotton swabs by shaking at 20 Hz for 2 min using a Qiagen TissueLyser (Qiagen Inc., Valencia, CA, USA). In the first two weekly samplings (out of six), 500 µL was added to sterile screw cap microtubes and genomic DNA was extracted using the Roche High Pure PCR Template Preparation kit (Roche Life Sciences, Indianapolis, IN, USA) according to the manufacturer’s specifications. Sample lysates for DNA extraction were prepared using the Qiagen TissueLyser and 0.5 mm zirconium oxide beads (Next Advance, Averill Park, NY, USA). Targeting the universal 16S rRNA gene sequence, a semi-quantitative determination of bacterial load was performed using TaqMan real-time PCR Assay with the LightCycler® 480 (Roche Life Sciences). Forward (5′-CCATGAAGTCGGAATCGCTAG-3′) and reverse (5′-GCTTGACGGGCGGTGT-3′) 16S rDNA primers (20 µM each) were used with a 16S rDNA probe (5′-TACAAGGCCCGGGAACGTATTCACCG-3′) in Quanta PerfeCTa® qPCR ToughMix (Quanta Biosciences, Beverly, MA, USA). The template DNA (2.5 µL) was added to the master mix containing primers and probe (10 µL each), and the reaction was run with the following thermal cycling profile: 50 °C for 2 min, 95 °C for 10 min, 35 cycles at 95 °C for 15 s, 60 °C for 1 min, and 40 °C for 30 s. E. coli c600 (ATCC 23724, Manassas, VA, USA) genomic DNA was used as a positive 16S rDNA control, and molecular grade water (Phenix Research Products, Chandler, NC, USA) was used as a no-template control.
2.4. Microbiological Sampling and Analyses of Bacterial Load by Plating and Sequence Analysis
For the remaining four weekly samplings (out of six) at the SWR Wound Care Center, 500 µL from the swab collection tubes was plated on Tryptone Soy Agar (TSA) (Oxoid CM0131) plates. Plates were incubated at 37 ± 1 °C and counted after 48 h. For each sampling, the average values of CFU/100 cm2 for test and reference door handles were log-transformed and presented as total average values ± SD. Plates were then washed using 1 mL PBS, and bacterial material was collected into sterile Eppendorf tubes.
2.5. Microbiological Sampling, Identification of Pathogens and Determination of Resistance Genes by Sequence Analysis
The possible presence of pathogenic bacteria or resistance genes, or both, was tested using the TaqMan real-time PCR Assay for P. aeruginosa, Serratia marcescens, S. aureus, Streptococcus pyrogenes, Streptococcus agalactiae and the mecA and vanA genes. This was done using primer sequences that were the property of the CAP-accredited medical laboratory at the SWR Wound Care Center. This analysis was performed on all bacterial DNA extracted directly from the swabs and on DNA extracted from bacterial colonies on the agar plates. In the third sampling, four DNA extracts from the test door handles had Ct (cycle threshold) values for the 16S rRNA gene below 30 (the cut-off value), and were selected for further screening to detect the possible presence of pathogenic bacteria or resistance genes, or both. In the same sampling, eight DNA extracts from the reference door handles (control) had Ct-values below 30, of which six samples were randomly chosen as representatives of the control group. For the remaining samplings, the same number of DNA extracts were selected (four for the test and six for the control), and they were randomly chosen from the test and control DNA extracts with Ct-values below 30. The remaining 500 µL of the 1 mL washing suspensions from the agar plates was spread on TSA plates and incubated at 37 ± 1 °C for 24 h, and single colonies were re-streaked on Mannitol salt agar (selective for staphylococci and micrococcaceae) and Cetrimide agar plates (selective for P. aeruginosa). This was done to further verify the presence or absence of these species.
2.6. Energy Dispersive X-Ray Spectroscopy (EDS) Analysis on Copper-Silver Alloy Coated Door Handles
The chemical compositions of selected copper-silver alloy coated door handles were checked prior to and after field testing, using the Hitachi TM3030 Plus Tabletop Microscope (Hitachi, Krefeld, Germany) operated at 15 kV and equipped with the Oxford Inca software and the Bruker Quantax 70 EDS System. EDS analysis was performed on three different spots at the surface of each sample. The output values (normalized weight percentage) were averaged and re-calculated with respect to the total content of copper and silver, to evaluate the difference before and after installation of the door handles. The presence of other elements (C, O) was also reported in normalized weight percentage if greater than 5 wt.%, to evaluate possible changes in surface composition of the coating during usage.
2.7. Statistical Analysis
Average values of CFU/100 cm2 for test and reference door handles at each sampling were log-transformed. Values were tested for equal or unequal variance with the F-test, and statistical significance of the difference between test and reference door handles was verified using the t-test.
3. Results and Discussion
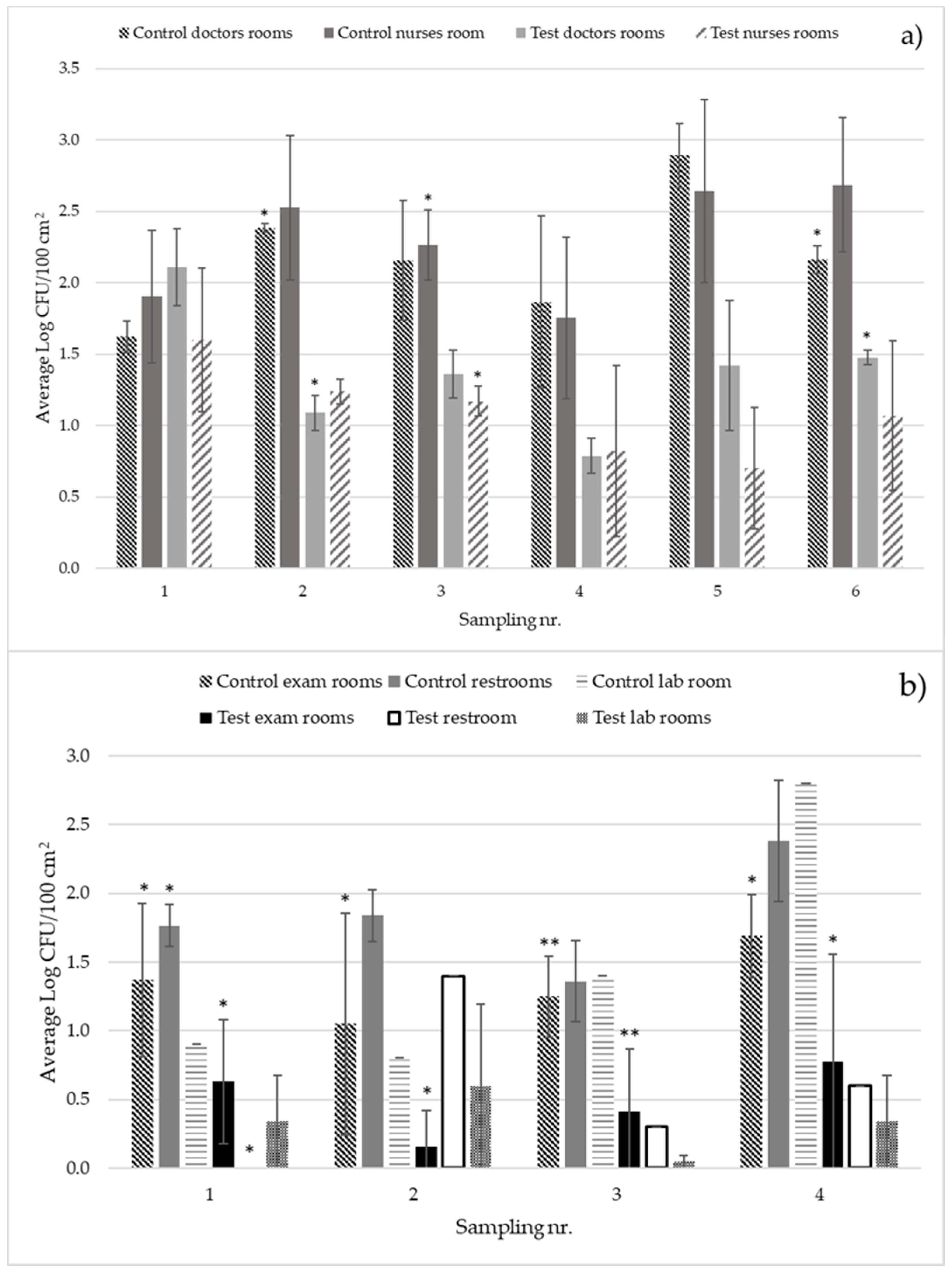
Both the reference stainless steel door handles and satin brass door handles had a microbial load that was approximately twice as large as the copper-silver alloy coated door handles. At FamilieLægerne Espergærde, the averaged total aerobic plate count from copper-silver alloy coated and uncoated reference stainless steel door handles were, respectively, 1.3 ± 0.4 and 2.4 ± 0.4 Log CFU/100 cm
2 (
p-value 0.0008). At the SWR Wound Care Center, these values were, respectively, 0.8 ± 0.3 and 1.7 ± 0.4 Log CFU/100 cm
2 (
p-value 0.0068) (
Figure 1). All surfaces in the field tests, except for stainless steel, had a microbial load below 2.4 Log CFU/100 cm
2 (which is the standard for acceptable microbial level on a surface immediately after terminal cleaning) [
5]. The microbial load on the satin brass reference door handles (1.7 ± 0.4 Log CFU/cm
2) was lower than that of the stainless-steel reference door handles (2.4 ± 0.4 Log CFU/cm
2), probably due to the antibacterial activity of brass (
Figure 1).
Interestingly, when DNA was directly extracted from the swabs, there was no difference in bacterial load between the coated and reference door handles (Ct-values of 24.5 and 24.6, respectively), as estimated by qPCR of the 16S rRNA gene (
Table 1). This might be due to the fact that DNA was recovered from both living and dead bacterial cells on the surfaces. However, bacteria with fewer alleles of the 16S rRNA gene could also have been selectively targeted.
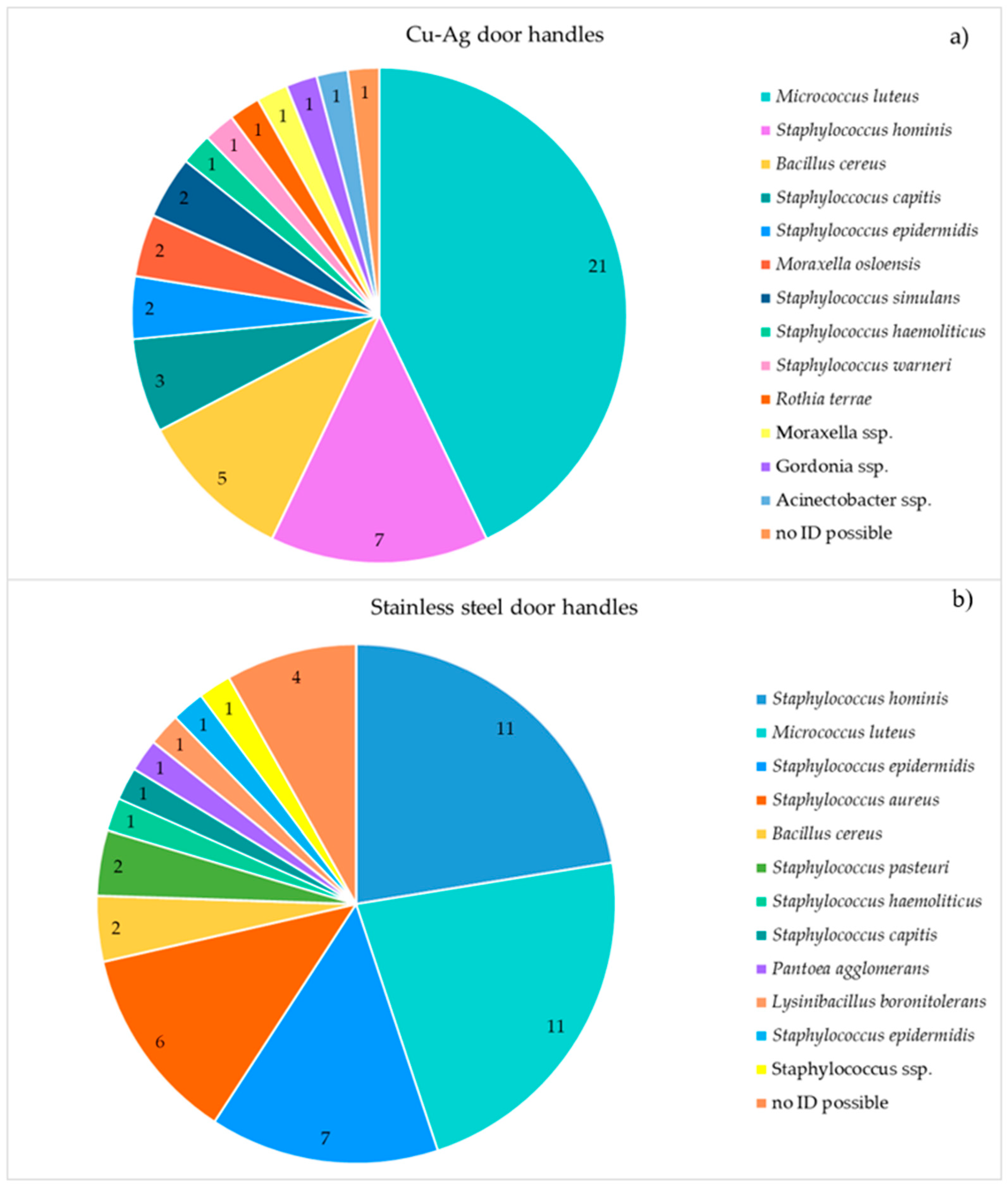
In the Danish clinic, MALDI-TOF MS analysis was performed on randomly chosen isolates from both copper-silver alloy coated and uncoated stainless steel surfaces. There was no marked difference among the surfaces in terms of surviving bacterial species. The most abundant bacterial species were
Micrococcus luteus and staphylococci (
S. hominis,
S. epidermidis and
S. capitis) on both copper-silver alloy coated and uncoated door handles.
S. aureus was found on the stainless steel but not on the copper-silver alloy coated door handles (
Figure 2).
DNA extracted directly from the swabs was tested by qPCR for the presence of six pathogenic bacteria and two resistance genes, and none of the samples were positive. DNA from colonies on agar-plates from 16 of the test samples and from 24 control samples at the SWR Wound Care Center was tested for the presence of
S. aureus by PCR. Three out of the sixteen test samples (19%) and 3 out of the 24 reference samples (13%) were positive (
Table 1).
The presence of S. aureus was also confirmed by the yellow discoloration on selective (mannitol salt) agar plates. P. aeruginosa, S. marcescens, S. pyrogenes and S. agalactiae were not detected in any of the biomass-plate samples.
Resistance genes were detected in the biomass-plate samples, possibly because of their much higher bacterial load than in the amount extracted directly from the swabs. Seven out of the sixteen test samples (44%) and 22 out of the 24 reference samples (92%) were positive for the
mecA gene (
Table 1). In the last sampling, the
vanA gene was detected in one biomass-plate sample from the control group. The greater occurrence of the
mecA gene in the control group could be due to the larger bacterial counts on the plates and hence a larger amount of biomass. Selective antibacterial efficacy of copper surfaces against Gram-negative bacteria and
S. aureus, as previously suggested, was not observed [
3].
The EDS analysis on coated door handles prior to and after field testing at FamilieLægerne Espergærde revealed a 5 ± 1 wt.% relative difference in terms of copper and silver content, whereas there was basically no change in relative composition of copper and silver prior to and after installation at the SWR Wound Care Center. Carbon and oxygen could be detected on the surfaces after the field tests, but only the amount of carbon was above 5 wt.% (11.4 ± 2.6 wt.% and 12.3 ± 1.9 wt.%). No significant surface oxidation was observed on the surface prior to and after field testing. It is likely that door usage and other environmental affecting factors may have helped to reduce the copper content in the copper-silver alloy coated door handles at FamilieLægerne Espergærde, as compared to the ones at the Southwest Regional Wound Care Center. To our knowledge, the cleaning procedure should not have influenced the surface chemistry, since door handles were not subject to extensive disinfection in these environments. However, considering the reduction in copper content in the copper-silver alloy coated door handles after the field test in the Danish clinic, the lifetime (durability) of the coating could be safely estimated to 72 weeks. At that point, the door handles should be recoated to maintain a constant efficacy. During usage, a complementary cleaning procedure to remove dirt and filth (detected as presence of carbon at the surface) and to ensure direct contact between bacteria and the alloy coating would be recommended. Periodical cleaning would both increase efficacy and lifetime of the coating, and in turn the presence of the coating would reduce the amount and need for harsh cleaning chemicals and extensive disinfection interventions.